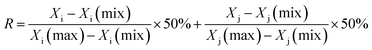Open Access Article
Open Access ArticleAn integrated approach based on phytochemistry, network pharmacology and metabolomics reveals the mechanism of action of Xanthium strumarium L. for allergic rhinitis†
Tie Yao‡
ab,
Jiankun Yan‡bd,
Yang Lib,
Jiaxin Wangb,
Miao Qiaob,
Xintong Hub,
Xuliu Shibd,
Shijie Cao*c and
Feng Qiu *ab
*ab
aSchool of Traditional Chinese Materia Medica, Shenyang Pharmaceutical University, Shenyang 110016, China. E-mail: fengqiu20070118@163.com
bSchool of Chinese Materia Medica, Tianjin State Key Laboratory of Modern Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin 301617, China
cTianjin State Key Laboratory of Modern Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin 301617, China. E-mail: caoshijie@tjutcm.edu.cn
dAnalysis Center of College of Science & Technology, Hebei Agricultural University, Cangzhou 061100, China
First published on 11th November 2020
Abstract
Xanthium strumarium L. (XS) is a traditional Chinese medicine (TCM) that has been widely used in Chinese medicine prescription for allergic rhinitis (AR). However, the action mechanisms of XS on the therapeutic effects on AR remain elusive. Herein, an integrated approach of phytochemistry, network pharmacology and metabolomics was first applied to uncover the action mechanisms of XS for AR. The therapeutic effect of XS extract on AR was evaluated in rat models of ovalbumin (OVA)-induced AR. The cytokine levels in rat serum and histopathological changes of nasal mucosa were assessed after oral treatment with XS. Chemical compositions of XS were elucidated by phytochemical methods, and active ingredients were identified via ADME-TOX screening in silico. Network pharmacology was performed to establish and analyze the compound-target-disease network so as to find the possible mechanism of XS in treating AR. In addition, metabolomics analysis was applied to investigate the changes in the endogenous metabolite levels that result from XS treatments. As result, the XS extract significantly increased the serum concentrations of IL-2 and reduced the levels of serum IL-4, while XS could ameliorate inflammation in the nasal sub-mucosal area, indicating that XS has significant therapeutic effects on AR model rats. Furthermore, a total of 119 compounds were isolated from XS, and 59 of these compounds were identified as active ingredients through ADME-TOX screening in silico. An in-depth analysis of the network pharmacology implied that the active ingredients of XS could regulate the inflammatory response via “multi-component, multi-target” patterns. In combination with the results of metabolomics, we found that the active ingredients of XS have a beneficial effect on AR through regulating the metabolism of arachidonic acid, which was reflected by medicating the Fc epsilon RI signaling pathway, and the neuroactive ligand–receptor interaction pathway, as well as the key proteins in arachidonic acid metabolism, such as PTGS2, PTGS1, PTGES and ALOX5. Additionally, molecular docking showed that multiple compounds have better binding with PTGS2 and ALOX5, which might be two crucial targets. Overall, these results suggest that the treatment of XS for AR is realized by regulating the metabolism of arachidonic acid via a combination form. This study provides the basis for clinical applications of XS.
1. Introduction
Allergic rhinitis (AR) is a chronic upper respiratory inflammatory disease that is characterized by sneezing, rhinorrhea and nasal congestion.1,2 Owing to the increasing incidence of AR worldwide over the last decades, it has become one of the most common inflammatory diseases. Although AR did not endanger the lives of patients, the repeated recrudesce and multiple complications seriously affect the quality of peoples' lives.3,4 AR belongs to the immunoglobulin E (IgE)-mediated type hypersensitivity reaction, when the allergen exposures stimulate infiltration inflammatory cells within the nasal mucosa, such as eosinophils, basophils, mast cells and T cells, which release multiple cytokines including prostaglandins, leukotrienes, and histamines contributing to the development of AR.5 In general, pharmacotherapy is a common treatment for AR, but most drugs (such as antihistamines and steroids) have serious side effects.6Traditional Chinese medicine (TCM) has been widely used in China during thousands of years, which has obvious advantages of fewer side effects compared to modern medicine for treating multi-factorial diseases.7 Herbal medicine generally consists of tens to hundreds of components, and is capable of systematically controlling various diseases via potentially synergistic component interactions.8 However, the complex chemical composition also brings great difficulties to interpret the molecular mechanisms of herbal medicine for the treatment of diseases. Fortunately, Andrew L. Hopkins proposed the network pharmacology approach in 2007, which coincides with the tenet of the overall regulation of TCM.9 In recent years, as the bioinformatics and systems biology have rapidly progressed, the network pharmacology approach became a dominant paradigm to decipher the complex pharmacological mechanism of action of effective substances of Chinese medicines.
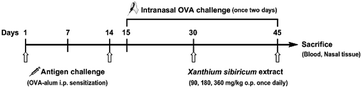
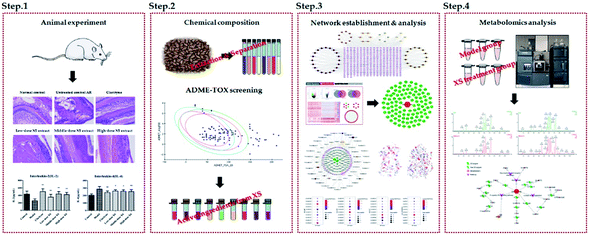
The fruits of Xanthium strumarium L. (XS) have been widely used in Chinese medicine prescriptions for the treatment of AR.10,11 XS is composed of multiple components and has been shown to possess extensive pharmacological effects on anti-inflammatory diseases.12,13 However, the action mechanisms of XS on the therapeutic effects on AR remain elusive. In the present work, we exert an integrated research strategy to decipher the molecular mechanism of XS in treating AR. The flowchart of the whole study design is illustrated in Fig. 1.
 | ||
| Fig. 1 The flowchart of integrated phytochemistry, network pharmacology and metabolomics strategy for deciphering the action mechanisms of XS on AR. | ||
2. Materials and methods
2.1 Chemicals and materials
Clarityne (drug approval number: H10970410) was purchased from Bayer Co., Ltd (Shanghai, China). Ovalbumin (OVA) was purchased from Yuanye Bio-Technology Co., Ltd (Shanghai, China). Aluminum hydroxide gel was purchased form Hat Biotechnology Co., Ltd (Xi'an, China). XS was purchased from Tong Ren Tang Co., Ltd (Beijing, China) and identified by Professor Lijuan Zhang, Tianjin University of Traditional Chinese Medicine. A voucher specimen (herbarium no. 20151015) has been deposited in the lab. All compounds used in the experiments were isolated and identified from XS. All reagents were HPLC or analytical grade, and were purchased from Tianjin Concord Chemical Company.2.2 Animals
Sprague-Dawley (SD) male rats (8 weeks old, weighing 180 ± 20 g) were purchased from the National Quality Inspection Center of Experimental Animals (Beijing, China; Certificate No. SCXK 2016-0006). The animals were housed under a comfortable condition of temperature (23 ± 2 °C), humidity (50 ± 5%) and 12 hours light/dark cycle before the experiments were performed. The protocol was strictly in accordance with the Guide for the Care and Use of Laboratory Animals (US National Research Council, 2011), and approved by the Animal Experiments Ethics Committee of Tianjin University of Traditional Chinese Medicine (authorization number: TJAB-TJU20180016).2.3 Allergic rhinitis model and treatment
After a week for acclimatization, forty-two rats were randomly divided into the following six groups: (1) the negative control group that was challenged with saline, (2) the AR group that was challenged with OVA, (3) the Clarityne oral treatment group that was treated with Clarityne dissolved in CMC-Na (0.5%) via intragastric administration 1 hour before intranasal OVA challenge, and (4) the XS extract treatment groups that were treated with low-, middle- and high-dose extracts dissolved in CMC-Na (0.5%) and given intragastric administration 1 hour before intranasal OVA challenge, respectively. The procedure for allergen sensitization and challenge is summarized in Fig. 2. Briefly, rats were sensitized with OVA (0.3 mg intraperitoneally) as an antigen, which was administered together with aluminum hydroxide (30 mg) in saline (1 mL intraperitoneally) every 2 days for 14 days. All groups except the negative control were sensitized with intranasal instillation with 10 μL of 20 mg mL−1 OVA on days 15 to 30. From day 31, the Clarityne (0.9 mg kg−1), low-dose extract (90 mg kg−1), middle-dose extract (180 mg kg−1) and high-dose extract (360 mg kg−1) dissolved in CMC-Na (0.5%) were treated via intragastric administration 1 hour before intranasal OVA challenge, respectively.2.4 Sample collection
Blood samples were collected from the aorta abdominalis 24 hours after the final OVA challenge, and centrifuged at 3000 rpm for 10 min, and then the supernatant was stored in a freezer at −80 °C. Rats were sacrificed after the blood collection. The heads were then stripped of the eyes, skin, and muscle, and the mandibles were excised to obtain the nasal bone parts, which were immersed in 10% neutral formalin for histological evaluation.2.5 Histological examination
The nasal bone parts were fixed in 10% neutral formalin for 3 days, and then decalcified in 5% trichloroacetic acid for 15 days. The nasal cavity was transversely sectioned at the level of the incisive papilla of the hard palate, and the tissue block was embedded in paraffin. Some of the sections were stained with hematoxylin and eosin (H&E) to examine the tissue histology.2.6 Compound-target-disease network construction
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC| ≥ 1.5. The DEGs were input into a String database to obtain protein–protein interactions (PPI). The hub genes were identified from a large PPI network by MCODE and NetworkAnalyzer. The MCODE can analyze the network based on the given parameter, and it assigns the weight to the vertex in the local neighborhood density from the dense regions by using vertex weighing, cluster prediction and optimal post-processing. The NetworkAnalyzer was used to analyze biological networks and calculate the network topology parameters, including the diameter of a network, the average number of neighbors, and the number of connected pairs of nodes. The R value was used to screen the hub genes from DEGs by the following formula:
FC| ≥ 1.5. The DEGs were input into a String database to obtain protein–protein interactions (PPI). The hub genes were identified from a large PPI network by MCODE and NetworkAnalyzer. The MCODE can analyze the network based on the given parameter, and it assigns the weight to the vertex in the local neighborhood density from the dense regions by using vertex weighing, cluster prediction and optimal post-processing. The NetworkAnalyzer was used to analyze biological networks and calculate the network topology parameters, including the diameter of a network, the average number of neighbors, and the number of connected pairs of nodes. The R value was used to screen the hub genes from DEGs by the following formula:where Xi is the degree, Xj is the betweenness centrality, and R is an indicator to evaluate the hub genes.
2.7 Metabolomics analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, the supernatant layer was transferred to a clean tube and evaporated to dryness. The residue was re-dissolved with 300 μL of 50% acetonitrile, vortexed for 2 min, and then centrifuged at 14
000 rpm for 10 min, the supernatant layer was transferred to a clean tube and evaporated to dryness. The residue was re-dissolved with 300 μL of 50% acetonitrile, vortexed for 2 min, and then centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for another 10 min. Finally, 200 μL supernatant was transferred to the analysis vial, and then analyzed by UPLC-Q-TOF/MS. Furthermore, serum samples were singled out from each group, which were mixed together as quality control (QC) samples. Before analysis, QC samples were continuously injected six times to equilibrate the UPLC system.
000 rpm for another 10 min. Finally, 200 μL supernatant was transferred to the analysis vial, and then analyzed by UPLC-Q-TOF/MS. Furthermore, serum samples were singled out from each group, which were mixed together as quality control (QC) samples. Before analysis, QC samples were continuously injected six times to equilibrate the UPLC system.The mass spectrometry detection was performed on a Xevo G2-S Q-TOF mass spectrometer (Waters, Milford, MA, USA) equipped with an electrospray ionization (ESI) source monitored in positive and negative ion mode. The detailed parameters were as follows: capillary voltage of 3.0 kV for positive mode and 2.5 kV for negative mode; extraction cone voltage of 3.0 V; sample cone voltage of 40 V; desolvation temperature of 400 °C, and source temperature of 100 °C; cone gas flow of 50 L h−1; desolvation gas flow of 600 L h−1. In the MSE mode, the trap collision energy of the low-energy function was set at 0 eV, while the ramp trap collision energy of the high-energy function was set at 15–60 eV. In MS2 mode, the samples were detected at the collision energy of 20 eV and 40 eV. Data in the centroid mode were set from 50 to 1000 Da.
2.8 ELISA analysis
The levels of serum cytokine (IL-2 and IL-4) were measured by commercial ELISA kits (Solarbio, China) according to the protocols provided from the manufacturer.2.9 Statistical analysis
Data were expressed as the mean ± standard deviation. All of the statistical analyses were carried out using SPSS software. Multiple group comparisons were performed using a one-way ANOVA test with the post hoc test. The Student's t-test was used for comparing the differences between the two groups. P < 0.05 was considered to indicate statistical significance.3. Results
3.1 The efficacy of the XS extract on AR in model rats
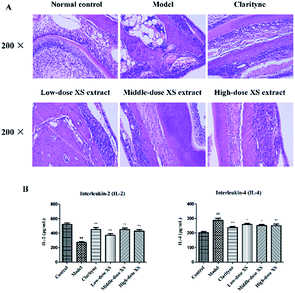
As shown in Fig. 3B, compared with the control group, the IL-4 levels in the model group were significantly increased, while IL-2 was obviously decreased, indicating that the AR model was successfully induced. The level of IL-4 in the middle- and high-dose XS extract treatment groups were significantly reduced compared with the model group, and IL-2 was markedly increased. Simultaneously, the results of the H&E staining showed that infiltration, vascular congestion and proliferation of the inflammatory cells in the model group were significantly higher than those in the control group, but the XS extract could significantly ameliorate these abnormalities (Fig. 3A). These data suggest that 75% XS extract could improve inflammation in the AR rat model.3.2 Chemical constituents of XS
A total of 119 chemical compositions were isolated and identified from 75% XS extract, which were divided into seven classes of compounds in combination with the information available, including 19 anthraquinones, 44 phenylpropanoids, 18 phenolic acids, 10 flavonoids, 9 alkaloids, 6 triterpenes and steroids, as well as 13 others. Integrating the above data, these compounds constituted the chemical components database, their detailed classification and structures in Fig. S1.†3.3 Compound-target network construction
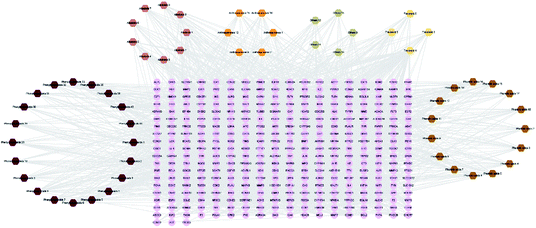
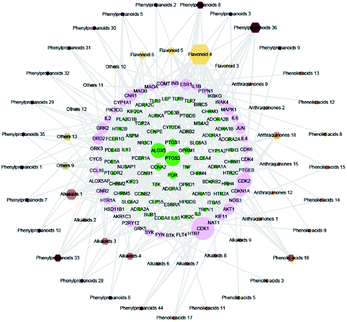
The 59 potential active ingredients were filtered out from the 119 components of XS by the criterion of ADME-TOX on the Discovery Studio 2018 software (Fig. 4). Subsequently, 345 targets were predicted to be associated with the 59 potential active ingredients. In order to visualize the relationship between the potential active compounds and targets, the compound-target network was constructed. The network comprised 404 nodes (59 compounds and 345 potential targets) and 2419 edges (compound-target and protein–protein interactions) (Fig. 5).3.4 Disease network construction
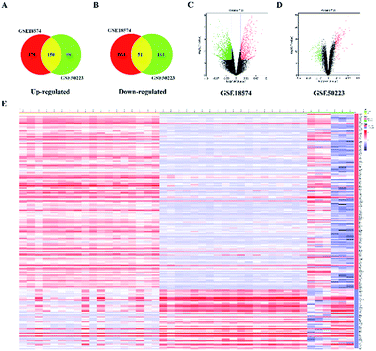
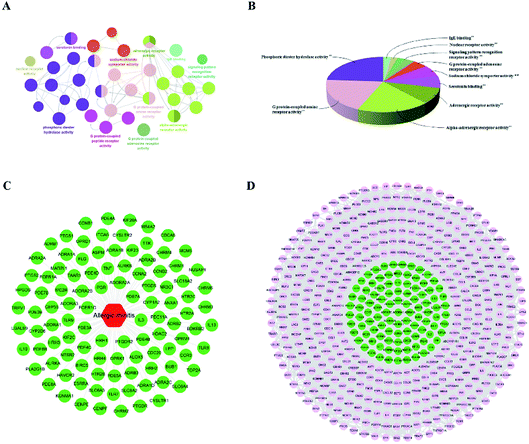
The genes associated with AR were collected from two sources: first, 83 genes related to AR were collected from DrugBank, TTD database and IPA software. Second, the microarray data analysis was applied to explore the genes that were associated with AR. A total of 201 overlapping DEGs were screened out, including 150 up-regulated and 51 down-regulated DEGs, as shown in Fig. 6A and B. The volcano plot and heatmap showed the fold-changes and DEGs between the two groups (Fig. 6C–E). The DEGs were submitted to the String database to obtain the PPI network. Then, MCODE plug-in was used to perform the clustering analysis for the PPI network (Fig. 7), and the top three modules containing 40 nodes were screened out from the PPI network. In the meantime, NetworkAnalyzer was used to calculate the topology parameters of the PPI network of DEGs. The nodes with a high degree and large betweenness centrality value were considered as important genes in the network. The top 25 genes were screened out via the R value, which was used to evaluate the importance of the genes as defined in the R equation, and as shown in Table 1. The 22 overlapping genes between MCODE and NetworkAnalyzer were considered to be the significant genes for AR. Finally, 104 proteins were collected as the targets associated with AR, and were used to construct the disease network, as shown in Fig. 8C. Subsequently, ClueGO was used to further identify the molecular functions of these 104 targets. The molecular functions mainly consisted of ten groups, such as the alpha-adrenergic receptor activity, G protein-coupled amine receptor activity and serotonin binding, which indicated that most targets were related to the stress response, signaling receptor activity and serotonin binding (Fig. 8A and B). To further explore the genes associated with AR, these targets were submitted to the String database to extend with a threshold value greater than 0.9 (highest confidence). Finally, an expanded PPI network associated with AR was constructed, as shown in Fig. 8D, and was composed of 511 nodes (104 direct correlations and 407 indirect correlations) and 9796 edges.| Swiss prot | Genes/proteins | Description | Up/down | Degree | BCa | R |
|---|---|---|---|---|---|---|
| a BC was the abbreviation of betweenness centrality. | ||||||
| Q12834 | CDC20 | Cell division cycle protein 20 homolog | Up | 50 | 0.33696 | 0.6584 |
| P20248 | CCNA2 | Cyclin-A2 | Up | 53 | 0.13701 | 0.5761 |
| P02649 | APOE | Apolipoprotein E | Down | 4 | 0.90000 | 0.5288 |
| P16410 | CTLA4 | Cytotoxic T-lymphocyte protein 4 | Up | 3 | 0.83333 | 0.4822 |
| P14635 | CCNB1 | G2/mitotic-specific cyclin-B1 | Up | 45 | 0.07355 | 0.4639 |
| O95067 | CCNB2 | G2/mitotic-specific cyclin-B2 | Up | 44 | 0.03752 | 0.4343 |
| O43683 | BUB1 | Mitotic checkpoint serine/threonine-protein kinase BUB1 | Up | 38 | 0.02227 | 0.3681 |
| Q96GD4 | AURKB | Aurora kinase B | Up | 37 | 0.02088 | 0.3578 |
| P11388 | TOP2A | DNA topoisomerase 2-alpha | Up | 37 | 0.02031 | 0.3574 |
| Q13257 | MAD2L1 | Mitotic spindle assembly checkpoint protein MAD2A | Up | 36 | 0.01816 | 0.3466 |
| Q53HL2 | CDCA8 | Borealin | Up | 34 | 0.00806 | 0.3218 |
| O95235 | KIF20A | Kinesin-like protein KIF20A | Up | 32 | 0.00486 | 0.3008 |
| Q8IZT6 | ASPM | Abnormal spindle-like microcephaly-associated protein | Up | 31 | 0.02118 | 0.3002 |
| O14965 | AURKA | Aurora kinase A | Up | 31 | 0.00491 | 0.2912 |
| Q99661 | KIF2C | Kinesin-like protein KIF2C | Up | 31 | 0.00397 | 0.2907 |
| P43489 | TNFRSF4 | Tumor necrosis factor receptor superfamily member 4 | Up | 2 | 0.50000 | 0.2874 |
| O15392 | BIRC5 | Baculoviral IAP repeat-containing protein 5 | Up | 30 | 0.01214 | 0.2856 |
| Q9BXS6 | NUSAP1 | Nucleolar and spindle-associated protein 1 | Up | 29 | 0.00303 | 0.2709 |
| Q02224 | CENPE | Centromere-associated protein E | Up | 28 | 0.00210 | 0.2608 |
| P33981 | TTK | Dual specificity protein kinase TTK | Up | 27 | 0.00281 | 0.2516 |
| Q02241 | KIF23 | Kinesin-like protein KIF23 | Up | 27 | 0.00279 | 0.2516 |
| P49454 | CENPF | Centromere protein F | Up | 26 | 0.00118 | 0.2410 |
| Q53EZ4 | CEP55 | Centrosomal protein of 55 kDa | Up | 25 | 0.00429 | 0.2332 |
| P01034 | CST3 | Cystatin-C | Down | 2 | 0.40000 | 0.2318 |
| P33992 | MCM5 | DNA replication licensing factor MCM5 | Up | 23 | 0.03189 | 0.2293 |
3.5 Compound-target-disease network construction and network topological analysis
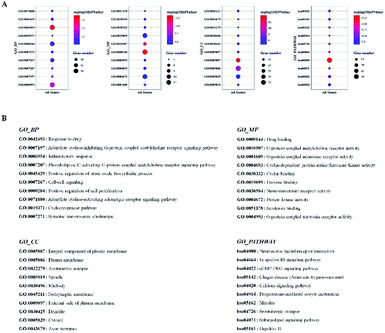
The compound-target network and AR disease network were merged by Cytospace software to construct a compound-target-disease network that was composed of 166 nodes (52 compounds and 114 targets) and 388 edges, as shown in Fig. 9. The composite network illuminated the relationship among the herbs, candidate compounds, and AR. Upon further analysis of the compound-target-disease network, we found that 35 compounds had a degree value ≥3, which indicated that most compounds interacted with the multi-target. For instance, apigenin (flavonoid 4) interacted with 26 targets, stylopine (alkaloids 1) interacted with 13 targets, and atractyligenin (others 13) interacted with 11 targets. Meanwhile, we found some gene targets that were hit by multi-compounds, such as PTGE2 and ALOX5, which were targeted by 11 and 14 compounds, respectively. In addition, CDK1 and CDK2 were not hit by multiple compounds; however, they had a high value degree, which indicated that they play important roles in the link of the PPI network.Before further analysis of the compound-target-disease network, molecular docking was applied to verify the interaction between the important targets and compounds. Molecular docking was performed according to the value of the degree with PTGS2 and ALOX5 as the crucial targets. As shown in Fig. 10, the binding patterns between the compounds and the target were good. Furthermore, previous numerous reports indicated that most compounds in the network regulated the related proteins, which indicated that the compound-target-disease network was reliable.14–19 Subsequently, the 114 targets in the compound-target-disease network were submitted to the DAVID v6.8 database to further identify the biological functions. In Fig. 11, the results were divided into four strata: molecular function, biological process, cellular component and KEGG pathway analysis. In particular, the molecular functions mainly consisted of drug binding, G-protein coupled acetylcholine receptor activity, G-protein coupled adenosine receptor activity and cyclin-dependent protein serine/threonine kinase activity, which indicated that most targets were related to the signaling receptor activity and drug binding. Additionally, the biological process of the targets was mainly related to the inflammatory response, positive regulation of the nitric oxide biosynthetic process and response to the drug. These results suggest that the targets are related to AR at different levels, indicating that XS might produce a therapeutic effect on AR through multiple ways.
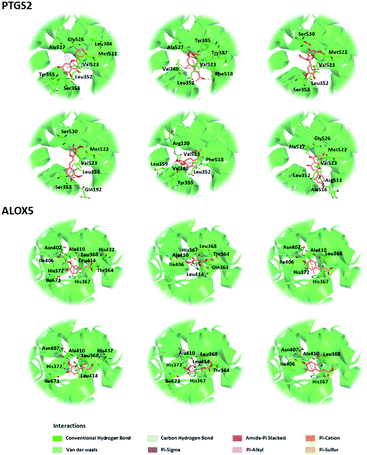 | ||
| Fig. 10 Three-dimensional model of the potential active compounds within the binding domains of PTGS2 (PDB code: 3O8Y) and ALOX5 (PDB code: 5IKR). | ||
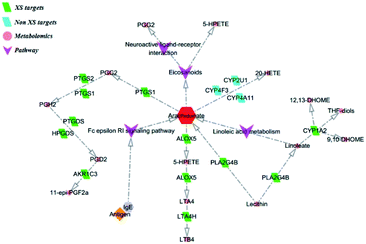
3.6 Metabolite identification and metabolic pathway
Metabolites were identified by the method described in Metabolomics data processing. These MSMS fragment ions of the metabolites were compared to the human metabolome databases (HMDB: http://www.hmdb.ca/). Eight biomarkers were confirmed, and the detailed information is shown in Table 2. The interrelated pathways of the above-mentioned biomarkers were analyzed by Metaboanalyst 4.0 (https://www.metaboanalyst.ca/). Finally, the changes in the metabolism of arachidonic acid, linoleic acid and α-linolenic acid were revealed after XS treatment.| No. | TR (min) | Ion form | m/z | Formula | Metabolite | VIP | P-Value | Trend | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10.39 | [M + H]+ | 758.5693 | C42H80NO8P | PC (14:0/20:2 (11Z, 14Z)) | 5.797 | 0.0161 | ↓ | 25.72 |
| 2 | 1.39 | [M − H]− | 260.0214 | C9H11NO6S | 2-Methoxyacetaminophen sulfate | 2.564 | 0.0002 | ↑ | 2.44 |
| 3 | 2.9 | [M − H]− | 212.0033 | C8H7NO4S | Indoxyl sulfate | 24.025 | 0.0136 | ↓ | 2.44 |
| 4 | 4.04 | [M − H]− | 333.2074 | C20H30O4 | 15-Deoxy-PGD2 | 2.31 | 0.0143 | ↓ | 10.9 |
| 5 | 4.53 | [M − H]− | 335.2224 | C20H32O4 | 5,20-DiHETE | 7.015 | 0.0002 | ↑ | 2.64 |
| 6 | 4.66 | [M − H]− | 337.2401 | C20H34O4 | 12-Keto-tetrahydro-leukotriene B4 | 3.765 | 0.0017 | ↑ | 3.83 |
| 7 | 5.23 | [M − H]− | 333.2056 | C20H30O4 | 5-Oxo-12-HETE | 2.884 | 0.0284 | ↑ | 6.07 |
| 8 | 8.34 | [M − H]− | 329.2480 | C22H34O2 | Docosapentaenoic acid | 5.885 | 0.0001 | ↑ | 2.23 |
4. Discussion
XS has been used as a TCM for treating AR, which has a good curative effect, and fewer side effects compared to hormone drugs. However, XS is similar to other TCMs and has a complex chemical composition, which makes it hard to understand its molecular mechanism, therefore limiting its worldwide use. So far, only a few studies showed that the mechanism of XS against AR might be associated with the inhibition of Ca2+ uptake and histamine release, increase cAMP, and inhibition of pro-inflammatory cytokines.20 In recent years, network pharmacology has been used in many successful cases for revealing the molecular mechanism of TCM.21,22 Although network pharmacology has advantages in elucidating the mechanisms of TCM, there are still some limitations. In most studies, the active ingredients are obtained from the herb database, while ingredients that are not recorded are often neglected, resulting in the primary ingredients and targets predicted by network pharmacology possibly deviating from the truth. Herein, we first apply the phytochemistry combined with network pharmacology and metabolomics to reveal the action mechanisms of XS in treating AR.In the current study, the therapeutic effect of XS on AR was first validated in animal models. The results of the animal experiment showed that the XS extract has a good therapeutic effect on AR, which provided a strong basis for subsequent studies. Subsequently, the chemical compositions of XS were elucidated by phytochemical methods. Among XS, 59 compounds were screened out by in silico ADME-TOX as potentially active ingredients. A combination of network pharmacology and metabolomics was performed to speculate the action mechanisms of XS in treating AR.
The compound-target-disease network showed that most compounds interacted with multi-targets, consistent with the concept of “multi-component, multi-target” in TCM. GO enrichment analysis uncovered the molecular mechanisms of XS in the treatment of AR. The GO BP terms showed that the gene targets were mainly enriched in the inflammatory response, cyclooxygenase pathway, and positive regulation of the nitric oxide biosynthetic process, which indicate that XS could participate in the regulation of the inflammation response in the treatment of AR. Consistent with the GO BP terms, in comparison with the model group, XS significantly increased the level of IL-2, while reducing the level of IL-4 in the animal experiment.
Furthermore, the pathway enrichment analysis is mainly involved with the Fc epsilon RI signaling pathway, Toll-like receptor signaling pathway and T cell receptor signaling pathway. Numerous reports demonstrated that the allergens could activate the Toll-like receptor signaling pathway, T cell receptor signaling pathway and B cell receptor signaling pathway, causing the aggregation of the T cells and B cells, thereby activating the Fc epsilon RI signaling pathway, and then results in inflammation.23,24 The results indicate that the treatment of XS against AR is a synergistic effect. In addition, we found some gene targets with a high value of degree in the network, which play key roles in treating AR. For instance, PTGS2 is responsible for the production of inflammatory prostaglandins, and ALOX5 is a key enzyme catalyzing the first step in leukotriene biosynthesis.25,26 As these targets are involved in the production of inflammatory mediators, we speculate that XS may inhibit the production of inflammatory mediators to treat AR. Subsequently, the results of the metabolomics analysis proved our hypothesis. Eight differential metabolites were identified between the model group and treatment group, which are mainly enriched in the arachidonic acid (AA) metabolism pathway. Arachidonic acid, as a precursor of bioactive substances, is involved in the regulation of various biological functions. The main metabolic pathway of AA is the cyclooxygenase (COXs) pathway, lipoxygenase (LOX) pathway and CYP-450 pathway in the organism, and the metabolites of AA play an important role in the inflammatory process.27–30 Therefore, the regulation of enzyme activity in the AA metabolism pathway can regulate inflammation, and the inflammatory response and process.31,32 Subsequently, we further reconstructed a metabolic network based on the above results, as shown in Fig. 12. This metabolic network indicates that XS can interact with multiple enzymes in the AA metabolic pathway to regulate the metabolism of AA. Taken together, we conclude that the treatment of XS for AR is realized by regulating the arachidonic acid metabolism via a combination form. In addition, we found some targets related to cell proliferation, such as CDK1, CDK2, CDK4, TTK and ADRA1D. Whether these targets are related to the treatment of AR with XS needs to be further investigated.
5. Conclusions
In the current work, a comprehensive integration of phytochemistry, network pharmacology, and metabolomics was taken first to systematically investigate the molecular mechanism of XS for AR. This integrated approach successfully illuminates the molecular mechanism of XS in treating AR. The present work may provide valuable evidence for further clinical applications of XS.Author contributions
Feng Qiu, Shijie Cao and Tie Yao conceived and designed the study. Tie Yao, Jiankun Yan, Yang Li, and Jiaxin Wang performed most of the experiments. Tie Yao, Miao Qiao, and Xintong Hu analyzed and interpreted the data. Xuliu Shi provided the technical support. Tie Yao drafted the article. All authors made the final approval of the version to be submitted.Abbreviations
| AA | Arachidonic acid |
| ADME-TOX | Absorption, distribution, metabolism, elimination and toxicity |
| ALOX5 | Arachidonate 5-lipoxygenase |
| AR | Allergic rhinitis |
| BC | Betweenness centrality |
| BP | Biological process |
| CDK | Cyclin-dependent kinase |
| DEGs | Differential expressed genes |
| ELISA | Enzyme-linked immunosorbent assay |
| GO | Gene ontology |
| H&E | Hematoxylin and Eosin |
| IgE | Immunoglobulin E |
| IL-2 | Interleukin-2 |
| IL-4 | Interleukin-4 |
| MMFF | Merck molecular force field |
| OVA | Ovalbumin |
| PPI | Protein–protein interaction |
| PTGES | Prostaglandin E synthase 3 |
| PTGS1 | Prostaglandin G/H synthase 1 |
| PTGS2 | Prostaglandin G/H synthase 2 |
| TCM | Traditional Chinese medicine |
| TTK | Dual specificity protein kinase TTK |
| XS | Xanthium strumarium |
Conflicts of interest
The authors declare that they have no conflict of interest.Notes and references
- M. S. Blaiss, E. O. Meltzer, M. J. Derebery and J. M. Boyle, Allergy Asthma Proc., 2007, 28(Suppl 1), S4–S10 CrossRef.
- L. Klimek, P. Hogger and O. Pfaar, HNO, 2012, 60, 611–617 CrossRef CAS.
- M. Westman, P. Stjarne, A. Asarnoj, I. Kull, M. van Hage, M. Wickman and E. Toskala, J. Allergy Clin. Immunol., 2012, 129, 403–408 CrossRef.
- P. W. Hellings and J. L. Ceuppens, Allergy, 2004, 59, 914–919 CrossRef CAS.
- J. S. Cho, J. H. Kang, I. H. Han, J. Y. Um, I. H. Park, S. H. Lee and H. M. Lee, Clin. Exp. Otorhinolaryngol., 2015, 8, 243–249 CrossRef CAS.
- N. P. Tran, V. John and M. S. Blaiss, Allergy, Asthma Immunol. Res., 2011, 3, 148–156 CrossRef CAS.
- F. Cheung, Nature, 2011, 480, S82–S83 CrossRef CAS.
- S. Li and B. Zhang, Chin. J. Nat. Med., 2014, 11, 110–120 CrossRef.
- A. L. Hopkins, Nat. Chem. Biol., 2008, 4, 682–690 CrossRef CAS.
- Y. Y. Kung, Y. C. Chen, T. J. Hwang, T. J. Chen and F. P. Chen, Allergy, 2006, 61, 1316–1318 CrossRef CAS.
- S. K. Huang, Y. L. Ho and Y. S. Chang, J. Ethnopharmacol., 2015, 173, 212–216 CrossRef.
- H. Jiang, L. Yang, G. X. Ma, X. D. Xing, M. L. Yan, Y. Y. Zhang, Q. H. Wang, B. Y. Yang, H. X. Kuang and X. D. Xu, Fitoterapia, 2017, 117, 11–15 CrossRef CAS.
- Y. S. Shi, Y. B. Liu, S. G. Ma, Y. Li, J. Qu, L. Li, S. P. Yuan, Q. Hou, Y. H. Li, J. D. Jiang and S. S. Yu, J. Nat. Prod., 2015, 78, 1526–1535 CrossRef CAS.
- J. B. Park, J. Nutr. Biochem., 2009, 20, 800–805 CrossRef CAS.
- R. Rezg, B. Mornagui, S. EI-Fazaa and N. Gharbi, Toxicology, 2008, 250, 27–31 CrossRef CAS.
- A. M. Vetrano, D. E. Heck, T. M. Mariano, V. Mishin, D. L. Laskin and J. D. Laskin, J. Biol. Chem., 2005, 280, 35372–35381 CrossRef CAS.
- S. Yano, D. Umeda, T. Yamashita, Y. Ninomiya, M. Sumida, Y. Fujimura, K. Yamada and H. Tachibana, Eur. J. Nutr., 2007, 46, 257–263 CrossRef CAS.
- M. Nardini, F. Leonardi, C. Scaccini and F. Virgili, Free Radic. Biol. Med., 2001, 30, 722–733 CrossRef CAS.
- S. I. Jang, B. H. Kim, W. Y. Lee, S. J. An, H. G. Choi, B. H. Jeon, H. T. Chung, J. R. Rho, Y. J. Kim and K. Y. Chai, Arch Pharm. Res., 2004, 27, 923–929 CrossRef CAS.
- W. X. Fan, L. X. Fan, C. Y. Peng, Q. Zhang, L. Wang, L. Li, J. L. Wang, D. Y. Zhang, W. Peng and C. J. Wu, Molecules, 2019, 24, 359 CrossRef.
- Y. B. Li, Y. M. Li, W. L. Lu, H. B. Li, Y. M. Wang, H. M. Luo, Y. Y. Wu, W. Y. Dong, G. Bai and Y. J. Zhang, Front. Pharmacol., 2018, 9, 597 CrossRef.
- C. Wang, Q. Ren, X. T. Chen, Z. Q. Song, Z. C. Ning, J. H. Gan, X. L. Ma, D. R. Liang, D. G. Guan, Z. L. Liu and A. P. Lu, Front. Pharmacol., 2018, 9, 841 CrossRef.
- M. Volkova, Y. Zhang, A. C. Shaw and P. J. Lee, J. Gerontol. Biol. Med. Sci., 2012, 67, 247–253 CrossRef.
- J. Bousquet, P. K. Jeffery, W. W. Busse, M. Johnson and A. M. Vignola, Am. J. Respir. Crit. Care Med., 2000, 161, 1720–1745 CrossRef CAS.
- S. F. Kim, D. A. Huri and S. H. Snyder, Science, 2005, 310, 1966–1970 CrossRef CAS.
- N. C. Gilbert, S. G. Bartlett, M. T. Waight, D. B. Neau, W. E. Boeglin, A. R. Brash and M. E. Newcomer, Science, 2011, 311, 217–219 CrossRef.
- T. G. Brock, R. W. Mcnish and M. Peters-Golden, J. Biol. Chem., 1999, 274, 11660–11666 CrossRef CAS.
- M. Abramovitz, E. Wong, M. E. Cox, C. D. Richardson, L. I. Chun and P. J. Vickers, Eur. J. Biochem., 1993, 215, 105–111 CrossRef CAS.
- Z. M. Chu, K. D. Croft, D. A. Kingsbury, J. R. Falck, K. M. Reddy and L. J. Beilin, Clin. Sci., 2000, 98, 277–282 CrossRef CAS.
- S. E. Wenzel, Pharmacotherapy, 1997, 17, 3S–12S CAS.
- J. Hellmann, Y. Tang, M. J. Zhang, T. Hai, A. Bhatnagar, S. Srivastava and M. Spite, Prostag. Other Lipid Mediat., 2015, 116–117, 49–56 CrossRef CAS.
- L. Li, H. Zeng, L. Shan, X. Yuan, Y. Li, R. Liu and W. Zhang, J. Ethnopharmacol., 2012, 143, 732–739 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06763f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |