Open Access Article
Open Access ArticleEffects of ozone treatment on the antioxidant capacity of postharvest strawberry†
Huijie Zhanga,
Kunlun Lib,
Xiaojun Zhangc,
Chenghu Dongd,
Haipeng Jid,
Runhui Kee,
Zhaojun Banf,
Yunfeng Hua,
Shaohua Ling and
Cunkun Chen *d
*d
aCollege of Food Science and Engineering, Tianjin University of Science and Technology, Tianjin, China
bInstitute of Plant Protection, Tianjin Academy of Agricultural Sciences, Tianjin, China
cCollege of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
dNational Engineering Technology Research Center for Preservation of Agricultural Products, Key Laboratory of Postharvest Physiology and Storage of Agricultural Products, Ministry of Agriculture of China, Tianjin, China. E-mail: chencunkun@126.com
eChina National Research Institute of Food & Fermentation Industries Co., Ltd, Beijing, China
fZhejiang University of Science and Technology, Hangzhou, China
gDepartment of Food and Biological Engineering, Beijing Vocational College of Agriculture, Beijing, China
First published on 15th October 2020
Abstract
Strawberries are highly popular around the world because of their juicy flesh and unique taste. However, they are delicate and extremely susceptible to peroxidation of their membrane lipids during storage, which induces water loss and rotting of the fruit. This study investigated the effects of ozone treatment on the physiological traits, active oxygen metabolism, and the antioxidant properties of postharvest strawberry. The results revealed that the weight loss (WL) and respiration rate (RR) of strawberry were inhibited by ozone treatment (OT), while the decline of firmness (FIR) and total soluble solids (TSS) were delayed. Ozone also reduced the generation rate of superoxide radical anions 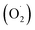 , and the content of hydrogen peroxide (H2O2) enhanced the activity of superoxidase (SOD), catalase (CAT), ascorbate peroxidase (APX), and monodehydroascorbate reductase (MDHAR), as well as promoted the accumulation of ascorbic acid (ASA), glutathione (GSH), and ferric reducing/antioxidant power (FRAP). In addition, a total of 29 antioxidant-related proteins were changed between the OT group and control (CK) group as detected by label-free proteomics during the storage time, and the abundance associated with ASA–GSH cycle was higher in the OT group at the later stage of storage, and the qRT-PCR results were consistent with those of proteomics. The improvement of the antioxidant capacity of postharvest strawberry treated with ozone may be achieved by enhancing the activity of the antioxidant enzymes and increasing the expression of the antioxidant proteins related to the ascorbic acid–glutathione (ASA–GSH) cycle.
, and the content of hydrogen peroxide (H2O2) enhanced the activity of superoxidase (SOD), catalase (CAT), ascorbate peroxidase (APX), and monodehydroascorbate reductase (MDHAR), as well as promoted the accumulation of ascorbic acid (ASA), glutathione (GSH), and ferric reducing/antioxidant power (FRAP). In addition, a total of 29 antioxidant-related proteins were changed between the OT group and control (CK) group as detected by label-free proteomics during the storage time, and the abundance associated with ASA–GSH cycle was higher in the OT group at the later stage of storage, and the qRT-PCR results were consistent with those of proteomics. The improvement of the antioxidant capacity of postharvest strawberry treated with ozone may be achieved by enhancing the activity of the antioxidant enzymes and increasing the expression of the antioxidant proteins related to the ascorbic acid–glutathione (ASA–GSH) cycle.
1. Introduction
Strawberries are rich in antioxidant active substances as well as flavonoids, anthocyanins, ascorbic acid, and phenols, which play an important role in their storage quality and good aspects for a healthy diet.1–3 Strawberries have antioxidant capacity, possessing a positive effect on preventing tumors and improving chronic diseases.4–7 Simultaneously, strawberry extracts, by activating redox-sensitive cellular signal molecules, induce multiple pathways that allow them to participate in mitochondrial biosynthesis and antioxidant defense, and regulate ROS levels, DNA damage, protein and lipid metabolism, and improve physical conditions.8–11 Improving the nutritional value of fruit has become a new goal of the biotechnology strategy. Therefore, the content of compounds related to human health in strawberries can be adjusted through the cultivation of new varieties, dietary collocation, and investigating the optimal eating time.12–14 There are many factors that affect the quality of strawberries. In addition to the actual strawberry variety, other factors are related to the post-harvest processing methods, and environmental and agronomic factors.15–18 Also, as a commercial crop, strawberries are widely cultivated in many parts of the world, including Europe and Asia.19 However, the skin of the strawberry is susceptible to mechanical damage, water loss, and pathogen infection, while the nutritional quality can be easily and rapidly reduced during storage, causing serious economic losses.2,20 Also, the traditional fresh-keeping bacteriostats are not suitable for the preservation of postharvest strawberries due to residual problems.Ozone is a strong oxidant with bactericidal properties and no residue and has been widely used in the storage of agricultural products, as it can help maintain a higher storage quality of fruit in terms of firmness,21 water loss,22 total soluble solids and titratable acids,23 respiration, and ethylene content.24 This is because ozone not only effectively inhibits the growth of microorganisms and reduces pesticide residues,25,26 but it also activates the antioxidant defense system of the fruit to remove active oxygen produced during physiological metabolism.27,28 The ozone-activated fruit antioxidant defense is mainly manifested in both enzymatic and non-enzymatic reactions. On the one hand, this induces the accumulation of non-enzyme antioxidant active substances, such as ascorbic acid (ASA), glutathione (GSH), and flavonoids.29–31 On the other hand, it enhances the activities of antioxidant enzymes, such as superoxidase (SOD), catalase (CAT), and ascorbate peroxidase (APX).32–34 Zhang et al. demonstrated that ozone treatment suppressed decreases in the ASA content and CAT enzyme activity to maintain the quality of postharvest strawberries.35 Chen et al. also found that the content of total phenols, flavonoids, and anthocyanins was increased in ozone treatment compared with the CK group to extend the storage time of postharvest strawberries.36 In addition, the content of ASA and the capacity of total antioxidants were increased in the OT group.37 However, studies on the antioxidant effects of ozone on postharvest strawberries are still relatively rare, especially on the regulation of the active oxygen metabolism and improvement of the antioxidant capacity.
The development of omics technology has provided a great aid to study the mechanism of postharvest agricultural products,38–40 especially the recent application of proteomics in fruit preservation, revealing the molecular level regulation mechanism of aging.41,42 Pan et al. discovered 5 protein candidate genes of sweet orange involved in carotenoid biosynthesis and regulation based on proteome and transcriptome analyses.43 Li used proteomics to study the aging mechanism of lychees and found that changed proteins were linked to active oxygen scavenging, glycolysis, tricarboxylic acid cycle, and ATP synthesis.44 Also, Jiao revealed the molecular mechanism of postharvest soybean sprouts induced by sodium nitroprusside was related to proteins from antioxidant progress, lipid peroxidation, and flavonoid synthesis through iTRAQ proteomics.45 Proteomics has an essential role to play in understanding the physiological and metabolic mechanisms of plants.
Our previous research also found that 10.72 mg m−3 was a suitable ozone concentration to prolong the storage time of postharvest strawberry, and the phenylalanine metabolic mechanism, which was explained using label-free quantitative proteomics.36 However, few studies have systematically elucidated the effects of ozone on the ASA–GSH cycle and reactive oxygen metabolism in postharvest strawberries. Therefore, the ‘Jingtaoxiang’ strawberry was used as an experimental material and was treated with ozone at 10.72 mg m−3 concentration to investigate the antioxidant mechanism. The effects of ozone treatment on the weight loss (WL), respiration rate (RR), firmness (FIR), total soluble solids (TSS), superoxide anion 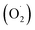 , hydrogen peroxide (H2O2), SOD, CAT, ferric reducing/antioxidant power (FRAP), and ASA–GSH cycles of strawberry fruit during storage were studied, and the active oxygen metabolism was investigated by unlabeled proteomics and verified by qRT-PCR technology.
, hydrogen peroxide (H2O2), SOD, CAT, ferric reducing/antioxidant power (FRAP), and ASA–GSH cycles of strawberry fruit during storage were studied, and the active oxygen metabolism was investigated by unlabeled proteomics and verified by qRT-PCR technology.
2. Materials and methods
2.1. Plant material
Fresh strawberries (Jingtaoxiang) were picked at the Modern Agricultural Science and Technology Innovation Base (Tianjin, China) and delivered to the National Agricultural Products Preservation Engineering Research Center (Tianjin) laboratory within 1 h after harvesting. After harvesting, the strawberries were of the same size, with no disease, no mechanical damage, and all were seven-point cooked.2.2. Treatments and storage conditions
A total of 36 boxes of picked strawberries, about 300 g per box, were divided into two groups: a control (CK) group and a 10.72 mg m−3 ozone treatment (OT) group. The strawberries were placed in an ozone precision-controlled fumigation device (2 m × 1.5 m × 0.8 m, with a storage capacity of 1200 L) and treated for 10 h under different concentrations of ozone (0, 10.72 mg m−3) every 7 d at 4 ± 1 °C with 75–80% relative humidity (RH), which was independently developed by the National Agricultural Products Preservation Engineering Technology Research Center.46 The storage period of the strawberries was 28 d, and samples were collected for physical and chemical detection after 0, 7, 14, 21, and 28 d, for protein analysis after 0, 7, and 21 d.2.3. Measurement of weight loss, firmness, respiration rate, and total soluble solids
The weight loss (WL) was determined using the method of Souza et al.47 First, 3 boxes of strawberries were randomly selected in each group at the initial time, and measurements were made at both the initial and storage time nodes. The percentage of WL was calculated according to the equation described by Souza et al.The methods for assessing strawberry firmness (FIR) and respiration rate (RR) were performed as per our previous study.24 The results of FIR are expressed in newton (N) and the RR as mg kg −1 h−1 CO2.
The content of total soluble solids (TSS) was detected according to the method of Fuggate,48 and the TSS content is expressed as %.
2.4. Measurement of the superoxide anion generation rate
The superoxide anion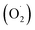 generation rate was determined according to Wang's method with minor modifications.49 A 1 g sample was homogenized with 5 ml of a buffer consisting of phosphoric acid (pH 7.8) 1.0 mM EDTA, 0.3% Triton X-100, and 2% polyvinylpyrrolidone (PVP). After centrifugation at 12
generation rate was determined according to Wang's method with minor modifications.49 A 1 g sample was homogenized with 5 ml of a buffer consisting of phosphoric acid (pH 7.8) 1.0 mM EDTA, 0.3% Triton X-100, and 2% polyvinylpyrrolidone (PVP). After centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min, the supernatant (1.0 ml) was mixed with 1.0 mM hydroxylamine hydrochloride and 50 mM potassium phosphate buffer (pH 7.8), shaken at 25 °C for 1 h, and then 17 mM sulfamic acid and 7 mM a-naphthylamine were added. The reaction was allowed to take place for 20 min, and the absorbance was measured with an ultraviolet spectrophotometer at a wavelength of 530 nm. The rate of superoxide anion formation on a fresh weight basis is expressed as U g−1.
000 × g for 20 min, the supernatant (1.0 ml) was mixed with 1.0 mM hydroxylamine hydrochloride and 50 mM potassium phosphate buffer (pH 7.8), shaken at 25 °C for 1 h, and then 17 mM sulfamic acid and 7 mM a-naphthylamine were added. The reaction was allowed to take place for 20 min, and the absorbance was measured with an ultraviolet spectrophotometer at a wavelength of 530 nm. The rate of superoxide anion formation on a fresh weight basis is expressed as U g−1.
2.5. Measurement of hydrogen peroxide content
The hydrogen peroxide (H2O2) content was quantified according to the method of Huang et al.50 Fresh sample (1.0 g) was homogenized with 10 ml of cold acetone; then after centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min, 0.1 ml of 5% titanium sulfate and 0.2 ml of ammonia were added to 1 ml of the extract, followed by centrifugation for another 10 min. The precipitate was dissolved in 3 ml of 1 mM sulfuric acid solution and then centrifuged at 12
000 × g for 30 min, 0.1 ml of 5% titanium sulfate and 0.2 ml of ammonia were added to 1 ml of the extract, followed by centrifugation for another 10 min. The precipitate was dissolved in 3 ml of 1 mM sulfuric acid solution and then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min. The supernatant was measured for absorbance at 420 nm. The H2O2 content of the sample was calculated using the standard curve of H2O2. The H2O2 content is expressed as mmol kg−1.
000 × g for 10 min. The supernatant was measured for absorbance at 420 nm. The H2O2 content of the sample was calculated using the standard curve of H2O2. The H2O2 content is expressed as mmol kg−1.
2.6. Measurement of ferric reducing/antioxidant power
The ferric reducing/antioxidant power (FRAP) was determined according to the total antioxidant capacity assay kit (FRAP method) produced by NanJing JianCheng Bioengineering Institute. Fresh samples were taken according to the procedures of the instructions, and the absorbance of the fresh samples at 593 nm was determined by using a microplate reader.2.7. Measurement of the key antioxidant enzyme
The activity of superoxide dismutase (SOD, EC 1.15.1.1) was determined according to the method of Abdel et al.,51 and the absorbance of the reactant was measured at a wavelength of 560 nm to determine the SOD activity. The amount of enzyme that SOD inhibited the reduction of nitro blue tetrazolium (NBT) by 50% was defined as 1 unit of SOD.The activity of catalase (CAT, EC 1.11.1.6) was determined according to the method of Xu et al.27 The CAT activity measured the absorbance of H2O2 that reacted with the enzyme extract at 240 nm, and the enzyme activity of 1 unit of CAT was defined as the amount of enzyme that reduced the absorbance by 0.01 at 240 nm min−1.
2.8. Determination of the key substances and enzyme activities in the ascorbic acid–glutathione (ASA–GSH) cycle
The activity of the ascorbate peroxidase (APX, EC 1.11.1.11) was determined according to the method of Xu et al.27 The APX reaction solution included 2.6 ml of 50 mM PBS (pH 7.5), 0.1 ml of enzyme extract, and 0.3 ml of 2 mM H2O2. The change in absorbance was measured at 290 nm, and APX activity was defined as the amount of enzyme that reacted with 1 mol L−1 ASA in 1 min.The ASA and GSH contents were determined according to the methods of Yin et al.52 and Israr et al.,53 respectively. The change in absorbance was measured at 525 nm to determine the ASA concentration, and the absorbance of GSH was determined at 412 nm. The contents of ASA and GSH were calculated according to the standard curve, and the unit is expressed as mg kg−1.
The enzyme activity of monodehydroascorbate reductase (MDHAR) was determined according to the MDHAR test kit produced by Shanghai Enzyme Biotechnology Co., Ltd. Fresh samples were taken according to the manufacturer's instructions, and the MDHAR activity was calculated by measuring the absorbance at 340 nm. One enzyme unit was defined as the amount of enzyme that oxidized 1 nmol of NADH per milligram of protein per minute at 25 °C.
2.9. Extraction of the protein
Detailed procedures for the extraction and identification of strawberry proteins, analysis, and validation of the data, and bioinformatics analysis are described in previously published articles.46Strawberry peptides were analyzed on a Q-Exactive HF mass spectrometer (Thermo Scientific, Waltham, MA, USA) and Ultimate 3000 system (Thermo Scientific). Tandem mass spectra were searched against Mascot 2.0 (Matrix Science, London, UK) using a UniProt sequence database. Raw spectral data were processed using SEQUEST software to extract the peaks. The obtained peak lists were analyzed using the Proteome Discoverer (Thermo Scientific, version 1.4) against strawberry RNA-seq sequences combined with the sequences of common impurities. The false discovery rates (FDR) were set to 0.01. A database for strawberry and all plant protein sequences can be searched using the UniProt sequence database, including Blast and UniProt annotation information in the protein database.
2.10. RNA extraction and quantitative reverse transcription PCR (qRT-PCR) analysis
Based on the differential expression of proteins in functional categories, 29 key proteins were selected for expression verification using qRT-PCR. The specific primers were designed using Primer 3.0 (ESI Table 1†), and the method of detection was described in our previous study.46 Total RNA was extracted from three independent samples using the MiniBEST Plant RNA Extraction Kit (TAKARA, Japan). Relative quantification of the candidate genes by qRT-PCR was carried out on an ABI StepOnePlus Real-Time PCR System and analyzed using the comparative Ct method with 18S rRNA (GenBank no. LOC101312917) as the internal control. The reaction was carried out using a green two-step qRT-PCR SuperMix (Thermo) following the manufacturer's instructions, and three repetitions were performed. Equation 2−ΔΔCt was used to calculate the relative transcription levels.2.11. Data analysis
All the data statistics were analyzed as the mean ± SD. One-way analysis of variance (ANOVA) was performed using IBM SPSS Statistics ver. 22 (SPSS Inc, Armonk, New York, USA) software. The differences between the mean values were analyzed by the least significant difference (LSD) method at 5% significance level (*p < 0.05), 1% significance level (**p < 0.01), and 0.1% significance level (***p < 0.001), and *p < 0.05 was considered to show statistical significance. Error bars represent the standard errors of the experimental data for three replicates.3. Results and discussion
3.1. Effect of ozone on the sensory physiological index
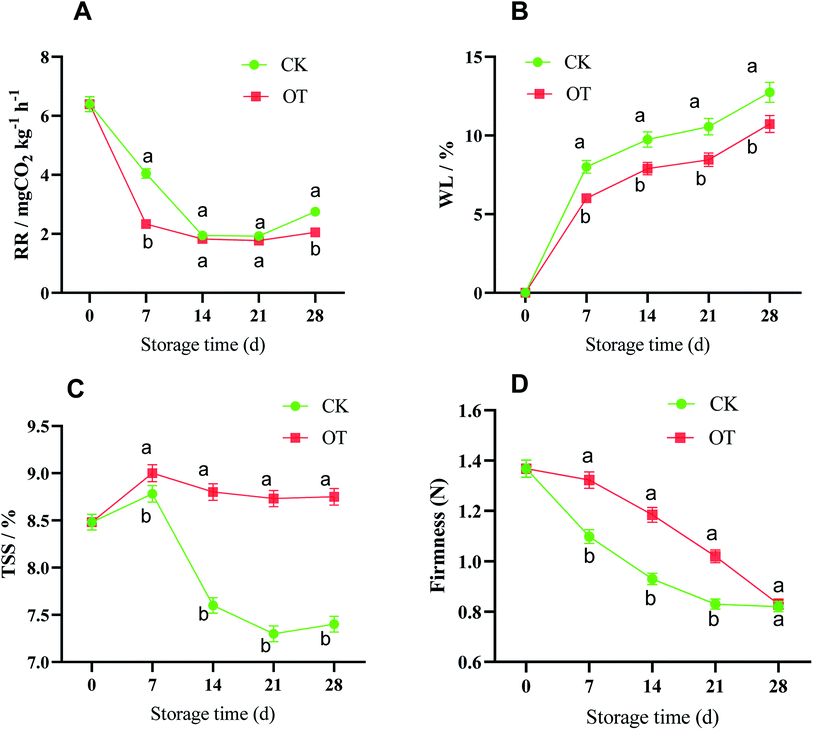
It can be seen from Fig. 1 that the strawberries stored for 0 d were less mature. On the 7th day, although the strawberries in the CK group had matured faster than in the OT, the strawberries in the two groups were not much different and were still a little green. By the 14th day of storage, all the strawberries of the CK group in the low-temperature storage had reached full ripeness, and the color had also turned dark red. However, the lower part of most strawberry pulp in the OT group turned red, and the skin of the leaf pedicle was slightly lighter. On the 21st day of storage, the strawberries in the control group had tended to rot and soften, while the strawberries in the treatment group were still fresh and bright red in color. At the end of storage, the CK group lost water and shrank, softened, and changed color. This may have been due to the evaporation of water in the tissues caused by the very thin skin of the strawberry and the strong respiration of the fruit, leading to them rotting. Although the strawberries in the treatment group tended to lose water, there was still a certain degree of hardness.The respiration rate (RR), as an important indicator, reflecting the consumption of nutrients and the aging of the fruit, as shown in Fig. 2A. The change in the RR in strawberry fruit in the CK group and the OT group was consistent, but was suppressed in the OT group compared with the CK group, especially on day 7, where the RR of the OT group (2.32 mg kg−1 h−1 CO2) was 42.57% lower than that of the CK group (4.04 mg kg−1 h−1 CO2). A lower RR was also detected in ozone-treated papaya fruit (Carica papaya L.), cantaloupes, and coriander compared with the controls.24,27,34
Weight loss (WL) is a key sensory feature for evaluating vegetable and fruit quality,22 and is closely related to the transpiration and respiration of postharvest vegetable and fruit.54,55 In this study, the WL of both the CK and OT groups increased with the storage time, especially in the first 7 d, which may be related to the accelerated ventilation during refrigeration in the cold storage. Also, the OT group could effectively suppress the dehydration of the strawberry compared with the CK group during the whole storage, which may be why the respiration of fruit treated with ozone was inhibited and the energy embolism was reduced. Chen et al. also found a significant positive correlation between the reduction of respiration in their ozone-treated group and the WL of postharvest Toona sinensis, and the RR and WL were lower in the OT group compared with the CK group.22
Firmness (FIR) directly reflects the changes in the ripening and softening degree and storage quality of strawberry fruit after harvest because they are very susceptible to water loss and wilting.56 The trend of FIR was a decrease during the whole storage time in both the CK and OT groups; however, the value in the OT group was always significantly higher than that in the CK group, except on day 28, indicating that the strawberry pulp after ozone treatment was firmer. Ali believed that ozone had the advantages of delaying maturity and maintaining hardness in papaya fruit.57 The same phenomenon was also found in postharvest ‘Iedzenu’ apples.58 Ioannis concluded that ozone treatment significantly reduced kiwi fruit pulp softening and cell wall disintegration.59
Total soluble solids (TSS) have a positive effect on the formation of flavor and nutrients of fresh fruit,60,61 and the main component of these is sugars, followed by a small number of acids, vitamins, minerals, pectin, and some volatile compounds. Also, the decomposition of the TSS leads to a rapid decline in postharvest strawberry quality in middle and late storage.62 As shown in Fig. 2C, the TSS of strawberry increased first and then decreased in both the CK and OT groups throughout the storage period, with a peak on day 7. However, the TSS of the CK group decreased rapidly after the peak time, while the content was maintained at a high level in the OT group. Tests on ozone-treated papaya and mulberry also reached the same conclusion.23,57
A lower RR contributes to the accumulation of TSS, maintenance of FIR, and reduction of WL in postharvest fruit, all of which contribute to the appearance of postharvest fruit.55,63–65 A good appearance and retention of nutrients are not only beneficial to the market economic effects of postharvest strawberry, but also to the health of consumers. In this research, it was indeed found that ozone could inhibit the respiration of postharvest strawberry and maintain high quality based on the analysis of physical traits, but the mechanism of this was not clear. Gas exchange in the plant was limited under OT and this may be one of the reasons,66 but the mechanism of ROS metabolism, one of the most important factors that affect the aging and quality of postharvest fruit as confirmed in many studies,67,68 may be needed to unveil this phenomenon. Next, we explored the effect of OT on the key reactive oxygen species and their rate-limiting proteins.
3.2. Effect of ozone on active oxygen and on the activity of antioxidant enzymes
ROS is the fundamental process of energy production by photosynthesis and respiration,69 and the accumulation of these is a response to various adverse environmental conditions.70 The spoilage of harvested fruit and vegetables is closely related to the content of superoxide anions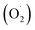 , hydroxyl radicals (OH˙), hydrogen peroxide (H2O2), and other ROS substances.68,71 Also, high concentrations of hydrogen peroxide can damage the host cell membrane, lead to lipid peroxidation, and cause oxidative stress in plants.72 Reducing the ROS content and improving the antioxidant properties have been reported to be the most effective method to extend the storage period and maintain the nutritional quality of harvested fruit and vegetables.73,74
, hydroxyl radicals (OH˙), hydrogen peroxide (H2O2), and other ROS substances.68,71 Also, high concentrations of hydrogen peroxide can damage the host cell membrane, lead to lipid peroxidation, and cause oxidative stress in plants.72 Reducing the ROS content and improving the antioxidant properties have been reported to be the most effective method to extend the storage period and maintain the nutritional quality of harvested fruit and vegetables.73,74
SOD, a dismutase that catalyzes 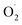 to H2O2, is the first line of defense to remove reactive oxygen species in plant cells,75 and to maintain the balance of reactive oxygen species and protect the structure of the cell membrane.76 CAT is a key enzyme for scavenging reactive oxygen species in plants, and can cooperate with POD to catalyze the decomposition of H2O2 into water and oxygen.77
to H2O2, is the first line of defense to remove reactive oxygen species in plant cells,75 and to maintain the balance of reactive oxygen species and protect the structure of the cell membrane.76 CAT is a key enzyme for scavenging reactive oxygen species in plants, and can cooperate with POD to catalyze the decomposition of H2O2 into water and oxygen.77
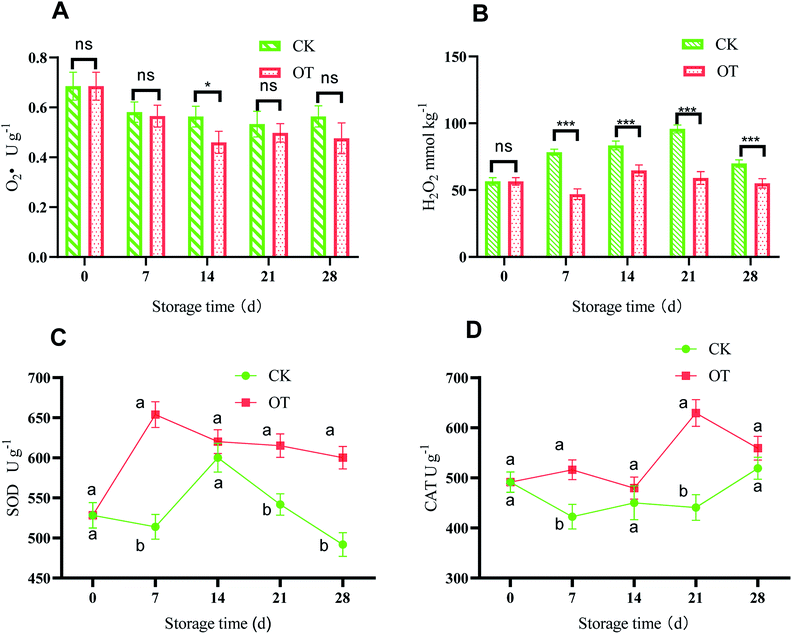
The changes in 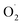 and H2O2 in different treatments are shown in Fig. 3A and B. The trend for the production of
and H2O2 in different treatments are shown in Fig. 3A and B. The trend for the production of 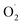 was decreased in both the CK and OT groups at the beginning of storage, and the generation rate of
was decreased in both the CK and OT groups at the beginning of storage, and the generation rate of 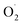 in the OT group was lower than that of the CK group during the entire storage period. At the same time, the content of H2O2 in the CK group continued to increase in the first 21 d and was significantly higher than that in the OT group during the entire storage period. This showed that the production of
in the OT group was lower than that of the CK group during the entire storage period. At the same time, the content of H2O2 in the CK group continued to increase in the first 21 d and was significantly higher than that in the OT group during the entire storage period. This showed that the production of 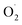 and H2O2 in postharvest strawberry was inhibited by ozone treatment, which may be due to the induction of antioxidant enzymes by ozone to increase the metabolism of reactive oxygen species.
and H2O2 in postharvest strawberry was inhibited by ozone treatment, which may be due to the induction of antioxidant enzymes by ozone to increase the metabolism of reactive oxygen species.
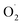 and H2O2 are the main active oxygen components in plants. The decrease in the
and H2O2 are the main active oxygen components in plants. The decrease in the 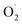 production rate in the treatment group and the CK group in the early storage period may be related to the low temperature. Kan et al. also found that low temperature can control the accumulation of active oxygen in peach to maintain quality.78 The generation rate of
production rate in the treatment group and the CK group in the early storage period may be related to the low temperature. Kan et al. also found that low temperature can control the accumulation of active oxygen in peach to maintain quality.78 The generation rate of 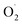 in the OT group was lower than that in the CK group, which may be due to ozone-induced higher SOD activity in the harvested fruit, which led to the conversion of
in the OT group was lower than that in the CK group, which may be due to ozone-induced higher SOD activity in the harvested fruit, which led to the conversion of 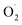 to H2O2 under the catalysis of SOD. Tomasz et al. found that an appropriate concentration of ozone treatment could maintain higher SOD activity in postharvest raspberry, and the
to H2O2 under the catalysis of SOD. Tomasz et al. found that an appropriate concentration of ozone treatment could maintain higher SOD activity in postharvest raspberry, and the 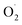 and H2O2 produced by the fruit were significantly lower than those of the control.28
and H2O2 produced by the fruit were significantly lower than those of the control.28
CAT and SOD are the criteria for evaluating antioxidant properties, and their high enzyme activity can help resist fruit aging. As shown in Fig. 3C, with the extension of storage time, the SOD activity of strawberry in the treatment group increased rapidly in the first 7 d and reached a maximum of 653.92 U g−1 on day 7, which was significantly higher than for the control (p < 0.05). After 7 d, the SOD activity of the treatment group decreased, but it was still significantly higher than that of the CK group. It can be seen that the SOD activity of strawberry was improved significantly in the OT group compared with the CK group and also its high peak appeared earlier. This may be because SOD is subjected to a certain degree of oxidative stress,79 and the appropriate ozone concentration stimulates the enzyme activity. From a molecular point of view, the increase of SOD activity may be due to the induction of the SOD gene and its isozyme gene expression by O3.80
Fig. 3D shows that the activity of CAT in the OT group was higher than that in the CK group during the entire storage period, especially on day 21, where the CAT of the OT group was 1.45 times that of the CK group. This indicated that ozone treatment stimulated the catalase activity of postharvest strawberry during the storage period. Zhang et al. found that ozonation inhibited the decrease in CAT and POD activities during storage, and improved the storage quality and extended the storage life of strawberry.35 Boonkorn et al. also found that the CAT activity of oranges treated with ozone was significantly higher than that of the CK group stored at room temperature for 3 d, which was consistent with our results, indicating that the CAT activity of postharvest strawberry could be enhanced by ozone.81
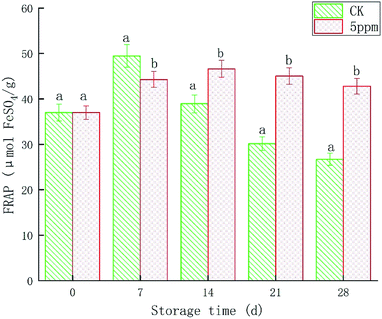
It can be seen from Fig. 4 that the total antioxidant capacity showed a trend of first increasing and then decreasing with the extension of storage time. The total antioxidant capacity of the treatment group was at a high level during days 7–28, and peaked on day 14 at 46.61 μmol FeSO4 g−1. Compared with the initial storage period, the OT group increased by 25.93%, and was significantly higher than that of the CK group of 38.94 μmol FeSO4 g−1 (p < 0.05). Except for the initial value, the total antioxidant capacity of the treatment group remained at between 42.81–46.61 μmol FeSO4 g−1. The FRAP of the CK group appeared on day 7, and was 49.48 μmol FeSO4 g−1, which was significantly higher than for the OT group (p < 0.05). As the FRAP of the CK group continued to decline, it was reduced to 26.70 μmol FeSO4 g−1 at the end of storage, which was significantly lower than that of the OT group (p < 0.05). OT can not only increase the FRAP of strawberry fruit, but also delay the time when the peak appears, especially at the end of storage, thus showing certain advantages. Alothman et al. also reached a similar conclusion.82 Fresh-cut pineapple and bananas were treated with ozone for more than 20 min, which significantly increased the total phenol and total flavonoids content of the fruit, which enables the fruit to obtain higher DPPH free radical scavenging capacity and FRAP. This conclusion was also supported by Sudheer et al.83
3.3. Effect of ozone treatment on the ASA–GSH cycle of postharvest strawberry
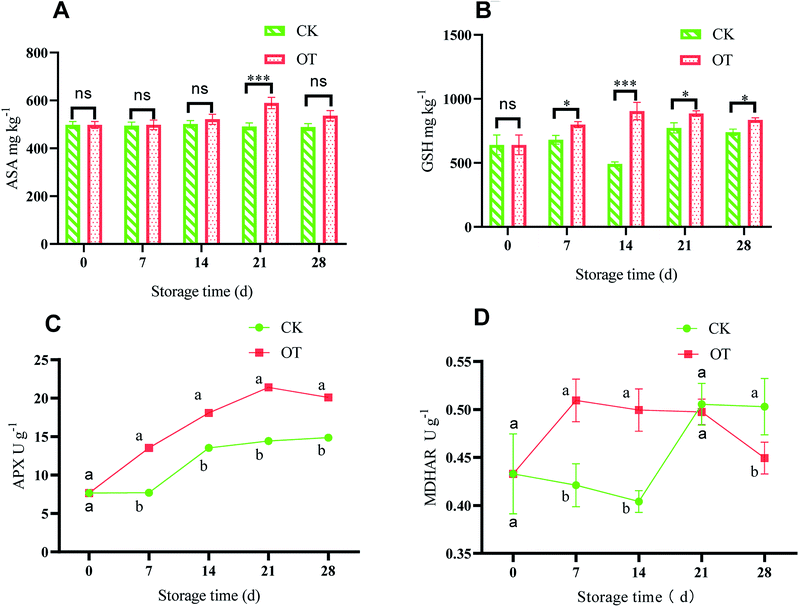
The ASA–GSH cycle is considered an important antioxidant system to scavenge ROS in plants, especially when CAT is insufficient, while H2O2 produced in cells is mainly eliminated by the ASA–GSH cycle.84,85 ASA can directly remove H2O2 to water under the catalysis of APX,86,87 while at the same time, ASA is oxidized to monodehydroascorbic acid (MDHA) and dehydroascorbic (DHA).88 Monodehydroascorbate reductase (MDHAR) catalyzes MDHA to ASA under the condition of electrons provided by the reduced form of nicotinamide-adenine dinucleotide (NADH) or nicotinamide-adenine dinucleotide phosphate (NADPH), which can promote ASA regeneration. Also, DHAR catalyzes the oxidized DHA to ASA under the action of GSH.89 Further, the oxidized glutathione (GSSG) is converted to GSH with an electron provided by NAD(P)H under the action of GR.90 Therefore, the balance between ROS production and elimination in the plant is maintained by enzymatic and non-enzymatic systems.91ASA with the function of eliminating various reactive oxygen plays a key role in improving antioxidant capacity.92 As shown in Fig. 5A, with the extension of storage time, the ASA content of strawberry in the CK group did not change much, while it first increased and then decreased in the OT group. Besides, the ASA content in the OT group was significantly higher than that in the CK group on days 21 and 28 during the storage (p < 0.05). Therefore, ozone treatment can significantly promote the production of ASA in strawberries during the storage, especially on days 21 and 28 (p < 0.05), which is beneficial to the cycle of ASA. The results of Ali were consistent with ours, and the peak value of ASA content in papaya treated with O3 was also delayed.57 Pérez et al. also found that the content of ASA in postharvest strawberry treated with ozone was increased compared with in their CK group.93
As a rich antioxidant in plant tissues, GSH participates in the detoxification of active oxygen, and H2O2 can be eliminated through the ASA–GSH cycle.94 As shown in Fig. 5B, with the extension of storage time, the GSH content in the OT group first increased and then remained stable, and it was higher than that in the CK group, indicating that production of GSH in postharvest strawberry was induced by ozone. Dumont also found that the total ASA and GSH contents of the three Euramerican poplar genotypes increased under the action of ozone.95 Also, the contents of ASA and GSH in the passion fruit liana were also stimulated by fumigation with ozone.96
APX is a key enzyme to eliminate large amounts of H2O2 in plants, and catalyzes ASA to MDHA under the oxidation of H2O2.97 Pang showed that the ASA–GSH cycle is an important part of a plant's active oxygen scavenging system, and APX is the key enzyme of the ASA–GSH cycle.90 As shown in Fig. 5C, the APX activity of the OT group continued to increase at first and reached a maximum of 21.41 U g−1 at day 21, which was significantly higher than that in the CK group (p < 0.05). Although the APX activity showed an increasing trend with storage time in the CK group, this was significantly lower than that of the OT group during the entire storage period. This indicated that O3 treatment could improve the APX activity and induce the cycle of ASA–GSH of postharvest strawberry. Goffi found that gradual cooling and ozone treatment increased APX activity levels in kiwifruit, and the activity of APX was positively correlated with the content of ASA.33 The APX activity of tangerine was also increased after ozone fumigation of tangerine.81
MDHAR is an important member of ASA metabolism. ASA is oxidized to MDHA, and MDHA is reduced to ASA by MDHAR.98 As shown in Fig. 5D, the activity of MDHAR in the early storage period was significantly higher in the OT group than in the CK group, but there was no significant difference at the later stage of storage. This indicated that MDHAR activity in the postharvest strawberry was induced by ozone at first, which was beneficial to the ASA–GSH cycle in the early storage time. The increased MDHAR activity was beneficial to the maintenance of ASA content in tomato fruit.99
On the whole, the productions of 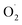 and H2O2 in the OT group were lower than in the CK group, and the ROS-removal enzyme activity and the content of antioxidants in the OT group were higher than those in the CK group. Ozone treatment improved the ability of eliminating ROS by promoting postharvest strawberry antioxidant systems. This may be due to the increase in free radicals produced by the appropriate concentration of ozone that could induce the strawberry antioxidant system. Huan also found that the antioxidant enzyme system was induced by the production of ROS.74,100 At the same time, the activity of antioxidant enzymes, such as APX and MDHAR, produced more non-enzymatic antioxidants, such as ASA and GSH.101 However, it is unknown whether the expression levels of these key enzymes are also induced by ozone.
and H2O2 in the OT group were lower than in the CK group, and the ROS-removal enzyme activity and the content of antioxidants in the OT group were higher than those in the CK group. Ozone treatment improved the ability of eliminating ROS by promoting postharvest strawberry antioxidant systems. This may be due to the increase in free radicals produced by the appropriate concentration of ozone that could induce the strawberry antioxidant system. Huan also found that the antioxidant enzyme system was induced by the production of ROS.74,100 At the same time, the activity of antioxidant enzymes, such as APX and MDHAR, produced more non-enzymatic antioxidants, such as ASA and GSH.101 However, it is unknown whether the expression levels of these key enzymes are also induced by ozone.
3.4. Effect of ozone treatment on antioxidant-related proteins of postharvest strawberry
The extraction quality data for the strawberry samples are shown in Table 1.| Sample | Weight (g) | Optical density | Protein concentration (g L−1) | Total volume (μL) | Total protein (μg) | Volume of electrophoresis (μL) |
|---|---|---|---|---|---|---|
| a Values are mean ± standard deviation of three replicates; where CK 0 is the initial time, and CK 7 and CK 21 are the CK group at day 7 and day 21; OT 7 and OT 21 are the OT group at day 7 and day 21. | ||||||
| CK 0 | 0.8 ± 0.01 | 1.026 ± 0.014 | 1.74 ± 0.04 | 150 | 261 ± 6 | 14.43 ± 0.33 |
| CK 7 | 0.8 ± 0.02 | 1.054 ± 0.031 | 1.88 ± 0.06 | 150 | 287 ± 9 | 12.55 ± 0.39 |
| OT 7 | 1.0 ± 0.05 | 1.213 ± 0.062 | 2.86 ± 0.13 | 150 | 423 ± 19.5 | 9.01 ± 0.42 |
| CK 21 | 0.8 ± 0.01 | 1.059 ± 0.019 | 1.98 ± 0.08 | 150 | 297 ± 12 | 12.57 ± 0.51 |
| OT 21 | 1.0 ± 0.03 | 1.177 ± 0.052 | 2.64 ± 0.12 | 150 | 414 ± 18 | 9.43 ± 0.41 |
The antioxidant capacity of plants is not only related to the activity of antioxidant proteins as many studies have proved that the increased expression of antioxidant genes and proteins could also improve the antioxidant capacity of plants. Wang et al. found that the specific PbrSODs mRNA expression levels of PbrCDS5, PbrCDS6, PbrFSD1, and PbrMSD2 after pear ripening are considered to be positively correlated with SOD activity. Through 1-MCP fumigation, the expression levels of PbrCDS5 and PbrFSD1 were upregulated, while ethephon treatment inhibited their expression, which was consistent with the decrease in SOD activity and the increase in ROS levels.102 It was also found that the SOD activity and the expression of the mSOD1 gene were high in the fruit of cucumber transferred with cassava mSOD1 compared with the CK group, which shows it played a defensive role against oxidative stress.103 The expression level of CAT and enzyme activity in transgenic plants were increased under the regulation of transcription factor (PtrbHLH), leading to a decrease in ROS accumulation under low-temperature stress.104 The overexpression of AO (ascorbate oxidase) in transgenic tobacco plants led to a decrease in H2O2 content and induced increased levels of ASA cycle-related genes, such as APX, DHAR, and GR, to delay dark-induced senescence.105 The activity and transcriptional abundance of APX and GR in cantaloupe were increased within 1 day post-spore-inoculation to activate the process of H2O2 removal in the early stage of fruit treatment.106 The relative expressions of MDHAR and DHAR were positively correlated with the accumulation of ASA in the peel and pulp of citrus fruit, respectively, which helped to increase the circulatory capacity of ASA–GSH.107 MDHAR allele was also considered as a candidate gene for increasing the ASA content in tomato.99 Eltelib's group found that the expression of MDHAR could be detected in overripe acerola and the transcription level of DHAR was highest in the middle stage of the fruit maturity.108 GSH can reduce H2O2 to water under the catalysis of GPX.109 Liang found that there were significantly more GST upregulated genes (28) than downregulated genes (3) in kiwifruit under the treatment of exogenous melatonin, which may be due to the increase in the GST transcription level to reduce the generation of free radicals and to improve the plant's oxidation resistance.110 The antioxidant capacity of tobacco was enhanced by the overexpression of GST and GR genes simultaneously.111 The transcription levels of PpAPXs, PpMDHARs, PpDHARs, and PpGRs were upregulated and the accumulation of ASA was stimulated in postharvest peach fruit treated with melatonin to remove H2O2 and to reduce oxidative damage.88 In addition, Liang et al. showed that the pattern of ASA metabolism in kiwifruit was consistent with the changing pattern of mRNA expression levels of AO2, APX3, GR1, and DHAR1.112 Different treatments improved the antioxidant capacity of fruits and vegetables, and there was a certain correlation between antioxidant enzymes and transcription abundance. The activity and transcription levels of SOD, CAT, APX, and GR, as well as the expression levels of AO, APX, DHAR, GR, and GST, increased, leading to a decrease in ROS accumulation. The regulation of AO, APX, GR, MDHAR, and DHAR mRNA expression levels was consistent with the change law for the ASA content. The upregulation of their transcription levels promoted the accumulation of ASA, eliminated H2O2, reduced the generation of free radicals, and enhanced the antioxidant capacity.
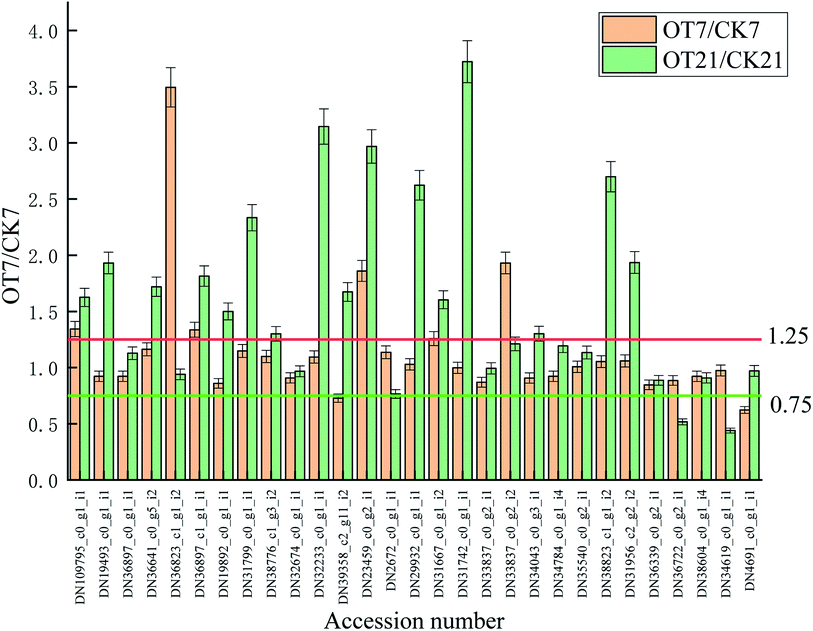
As shown in Table 2 and Fig. 6, a total of 29 antioxidant-related proteins were changed, of which 3 proteins were identified as L-ascorbate oxidase (L-AO); 4 proteins were identified as ascorbate peroxidase (APX); 2 proteins were dehydroascorbate reductase (DHAR); 1 protein was identified as monodehydroascorbate reductase (MDHAR); 2 proteins were glutathione peroxidase (GPX); 10 proteins were identified as glutathione S transferase (GST); 1 protein was identified as glutathione reductase (GR); 3 proteins were identified as superoxide dismutase (SOD); 1 protein was identified as catalase (CAT); and 2 proteins were identified as peroxidase (POD).
| Accession | Blast-hit | Blast-hit-description | FC(CK 7/CK 0) | FC(CK 21/CK 0) | FC(OT 7/CK 0) | FC(OT 21/CK 0) | FC(OT 7/CK 7) | FC(OT 21/CK 21) |
|---|---|---|---|---|---|---|---|---|
| a Where the accession is the number of proteins, and the Blast-hit protein is the most homologous protein obtained by comparison with the nonredundant database of the National Center for Biotechnology Information (NCBI); CK 0 is the initial time, and CK 7, OT 7, CK 21, and OT 21 are for the CK group and OT group on day 7 and 21, respectively. | ||||||||
| DN109795_c0_g1_i1 | A0A0B2QFU7 | L-Ascorbate oxidase like | 0.958 | 0.711 | 0.802 | 0.935 | 0.837 | 1.316 |
| DN19493_c0_g1_i1 | A0A0V0IGF6 | Putative L-ascorbate oxidase-like | 1.108 | 0.882 | 0.857 | 1.192 | 0.774 | 1.352 |
| DN36897_c0_g1_i1 | A0A1S4AXF2 | L-Ascorbate oxidase homolog | 1.450 | 1.225 | 1.500 | 1.388 | 1.034 | 1.133 |
| DN36641_c0_g5_i2 | A0A067XJ96 | Cytosolic ascorbate peroxidase | 0.850 | 0.783 | 0.647 | 1.183 | 0.761 | 1.511 |
| DN36823_c1_g1_i2 | M9QZ26 | Chloroplast ascorbate peroxidase (fragment) | 0.651 | 0.761 | 0.936 | 0.642 | 1.437 | 0.843 |
| DN36897_c1_g1_i1 | O48919 | Cytosolic ascorbate peroxidase | 0.650 | 0.515 | 0.610 | 0.767 | 0.938 | 1.489 |
| DN19892_c0_g1_i1 | M9QZ14 | Dehydroascorbate reductase (fragment) | 0.650 | 0.453 | 0.654 | 1.119 | 1.006 | 2.473 |
| DN31799_c0_g1_i1 | S4VM76 | Dehydroascorbate reductase | 0.721 | 0.574 | 0.644 | 0.618 | 0.894 | 1.077 |
| DN38776_c1_g3_i2 | A0A1S4CLG2 | Monodehydroascorbate reductase 5, mitochondrial-like isoform X2 | 1.102 | 1.086 | 1.345 | 1.686 | 1.221 | 1.552 |
| DN32674_c0_g1_i1 | M9R5M8 | Mitochondrial ascorbate peroxidase (fragment) | 1.000 | 0.541 | 0.553 | 0.794 | 0.553 | 1.467 |
| DN32233_c0_g1_i1 | M5WVT4 | Glutathione peroxidase | 1.059 | 0.510 | 0.696 | 1.324 | 0.657 | 2.596 |
| DN39358_c2_g11_i2 | K9LWY5 | Glutathione reductase | 0.968 | 1.018 | 1.144 | 2.113 | 1.182 | 2.076 |
| DN23459_c0_g2_i1 | A0A061G4E6 | Glutathione peroxidase | 1.065 | 1.261 | 1.196 | 1.717 | 1.122 | 1.362 |
| DN2672_c0_g1_i1 | W9S5P8 | Putative glutathione S-transferase | 0.322 | 0.238 | 0.253 | 0.244 | 0.785 | 1.025 |
| DN29932_c0_g1_i1 | A0A193KWW8 | Glutathione S-transferase U22 | 1.589 | 1.267 | 1.456 | 1.678 | 0.916 | 1.325 |
| DN31667_c0_g1_i2 | A0A061FY71 | Glutathione S-transferase tau 9 | 1 | 1 | 1 | 1 | 1.485 | 2.884 |
| DN31742_c0_g1_i1 | M5WTI5 | Glutathione S-transferase Riant1 | 0.932 | 0.770 | 1.114 | 1.136 | 1.195 | 1.475 |
| DN33837_c0_g2_i1 | A0A1C7A0M2 | Phi class glutathione S-transferase | 1.000 | 1.064 | 1.000 | 1.191 | 1.000 | 1.120 |
| DN33837_c0_g2_i2 | A0A1C7A0M2 | Phi class glutathione S-transferase | 0.754 | 0.895 | 1.200 | 1.486 | 1.591 | 1.660 |
| DN34043_c0_g3_i1 | A0A061FH05 | Glutathione S-transferase TAU 19 | ND | 0.195 | 0.592 | 0.535 | ND | 2.738 |
| DN34784_c0_g1_i4 | W9QWL1 | Glutathione S-transferase L3 | 0.992 | 1.088 | 0.976 | 1.248 | 0.984 | 1.147 |
| DN35540_c0_g2_i1 | A0A1J6IGD0 | Putative glutathione S-transferase | 0.692 | 0.670 | 0.850 | 0.546 | 1.228 | 0.815 |
| DN38823_c1_g1_i2 | A0A0X9LEN0 | Glutathione S-transferase 1 | 2.241 | 2.184 | 1.770 | 4.759 | 0.790 | 2.179 |
| DN31956_c2_g2_i2 | B2CP37 | Superoxide dismutase [Cu–Zn] | 2.400 | 2.160 | 2.060 | 4.100 | 0.858 | 1.898 |
| DN36339_c0_g2_i1 | M9R044 | Superoxide dismutase (fragment) | 0.695 | 0.644 | 0.678 | 0.622 | 0.976 | 0.966 |
| DN36722_c0_g2_i1 | H9TEU1 | Superoxide dismutase [Cu–Zn] | 1.701 | 1.896 | 1.753 | 1.922 | 1.031 | 1.014 |
| DN38604_c0_g1_i4 | M9R036 | Catalase (fragment) | 0.718 | 1.154 | 0.611 | 0.309 | 0.850 | 0.267 |
| DN34619_c0_g1_i1 | M5W6X8 | Peroxidase | 0.479 | 7.041 | 0.562 | 3.858 | 1.172 | 0.548 |
| DN4691_c0_g1_i1 | M5WIW4 | Peroxidase | 1.268 | 1.049 | 0.894 | 1.244 | 0.705 | 1.186 |
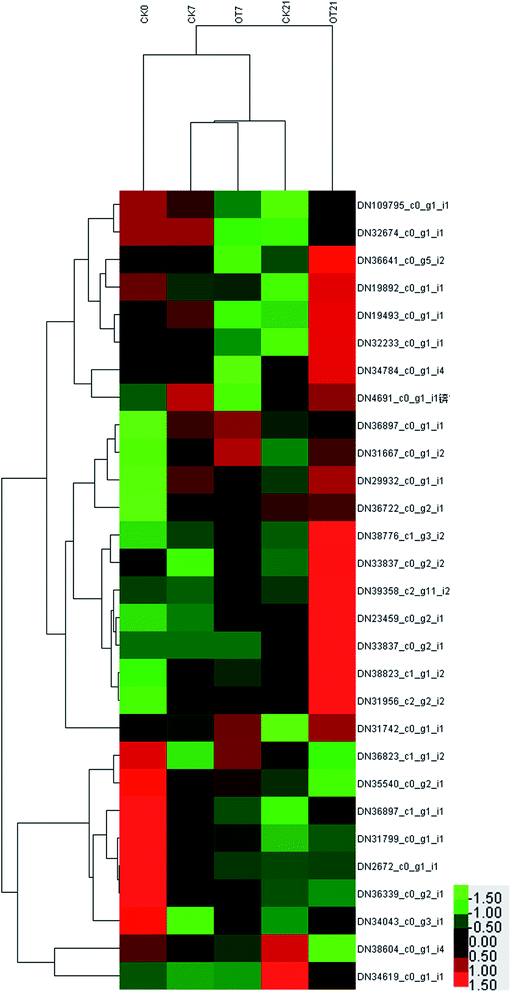
Compared with the initial time, the expressions of 9 proteins in the CK group were downregulated on the 7th day of storage, which comprised 2 APX proteins, 2 DHAR proteins, 2 GST proteins, and 1 SOD protein, 1 CAT protein, and 1 POD protein. Also, the expressions of 11 proteins were downregulated in the OT group on day 7 compared with the initial time, respectively: comprising 3 APX proteins, 2 DHAR proteins, 1 GPX protein, 2 GST proteins, 1 SOD protein, 1 CAT protein, and 1 POD protein. The abundances of DN34619_c0_g1_i1 (POD), DN38604_c0_g1_i4 (CAT), DN36339_c0_g2_i1 (SOD), DN2672_c0_g1_i1 (GST), DN31799_c0_g1_i1 (DHAR) and DN36897_c1_g1_i1 (APX) were lessened in both the CK and OT group on day 7. At the same time, the number of upregulated proteins in both the CK and OT groups that were the same was five, comprising: DN36722_c0_g2_i1 (SOD), DN31956_c2_g2_i2 (SOD), DN38823_c1_g1_i2 (GST), DN29932_c0_g1_i1 (GST), DN36897_c0_g1_i1 (AO). These results suggested that the expression of proteins related to antioxidant behavior significantly occurred during the early storage. Also, as shown in the trend clustering graph, the changed proteins between the OT group and the CK group were not large during the early storage period, which can infer that the strawberry protein changes in the early storage period may be mainly caused by low temperature.
On the 21st day, there were 10 proteins and 7 proteins in the CK, and OT groups that were downregulated on day 21 compared with initial time, 5 of which were the same proteins. Also, 6 proteins identified as POD, SOD, GST, and GPX were upregulated in both the CK and OT groups, and the expressions of another 5 proteins related to ASA–GSH were found to be more abundant in the OT group compared with the initial time. The results indicated that the ozone mainly induced the ASA–GSH cycle-related proteins of postharvest strawberry at the end of the storage. In addition, as shown in Fig. 7C and Table 2, the expressions of 17 proteins were more abundant in the OT group compared with the CK group, while only 2 proteins were downregulated. Interestingly, the downregulated proteins in the OT group were CAT (DN38604_c0_g1_i4) and POD (DN34619_c0_g1_i1), while 16 of the upregulated proteins were related to the ASA–GSH cycle and another 1 protein was SOD; these results revealed that the expressions of CAT and POD of strawberry after harvest were inhibited, and ozone mainly activated the ASA–GSH pathway to reduce the accumulation of H2O2. This study thus demonstrated that the appropriate concentration of ozone treatment can stimulate the fruit ASA–GSH cycle to achieve the purpose of removing active oxygen.96 H2O2 itself is the reaction substrate of CAT and the content of H2O2 is positively correlated with the expression of CAT,113,114 which may be the reason for both the reduction of the H2O2 content and the expression of CAT in the later storage period. Besides, POD not only had the ability to catalyze the removal of hydrogen peroxide but was also related to the decline of the fruit quality and plant senescence.115,116 Studies have shown that POD is one of the key enzymes for the browning of postharvest fruit,117 and Ting et al. also found that both POD activity and POD genes were closely related to the browning of fresh-cut lotus root.118 The changes of CAT and SOD protein and enzyme activity were not synchronized; indicating that the storage quality is regulated by both enzyme activity and protein expression.
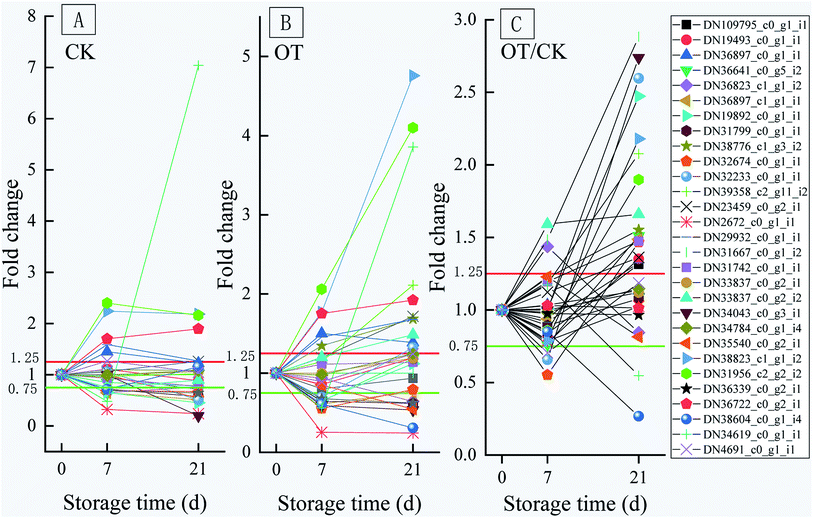 | ||
| Fig. 7 Variation trend of the oxidation-related proteins in the OT group and CK group throughout the entire storage period; (A) is the CK group; (B) is the OT group; (C) is the OT group/CK group. | ||
On the whole, the expression of antioxidant proteins was changed with the storage time. On the 7th day of storage, there was no significant difference in the expression of antioxidant proteins between the CK group and the OT group. However, the abundance of most proteins related to the ASA–GSH cycle in the OT group was significantly high than that in the CK group on day 2, which was consistent with the trend of the expression of proteins related to phenylalanine metabolism in our previous study. This indicated that the ozone treatment has a positive effect on the expression of the ASA–GSH cycle proteins of postharvest strawberry in the late storage period, and that the changes in the expression of antioxidant proteins in the early storage may be related to low temperature.
3.5. Analysis of the q-RTPCR results of postharvest strawberry protein
As shown in Fig. 8, 34 antioxidant-related proteins were detected in the q-RTPCR verification experiment. DN109795_c0_g1_i1, DN36897_c1_g1_i1 and DN23459_c0_g2_i1 were upregulated in the q-RTPCR results and not significantly changed in the proteomics data. DN38604_c0_g1_i4 showed no change in mRNA expression on day 7 and day 21, while the abundance was off, which was downregulated in the proteome results. DN32233_c0_g1_i1 and DN34864_c0_g1_i1 were downregulated in the proteomic results and upregulated in the mRNA results. The abundance of DN33837_c0_g2_i2 in the proteome data was increased on day 21, but the fold change in mRNA level was 1.22. In general, most results detected at the mRNA level were consistent with those of the proteomics.4. Conclusion
Ozone effectively inhibited the increase in strawberry weight loss and respiration rate, maintained firmness, and delayed the decrease in the total soluble solids content to achieve postharvest strawberries with high sensory and economic value, which may be related to ozone regulating the ROS metabolism level of the postharvest strawberries. The production of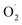 and H2O2 in the postharvest strawberries subjected to ozone were reduced compared with in the CK group, which may be because ozone activated the antioxidant defense system of strawberry, including the activities of SOD, CAT, APX, MDHAR, and the contents of ASA and GSH. In addition, 17 proteins were more abundant in the OT group than in the CK group at the end of storage, 16 proteins of which were related to the ASA–GSH cycle, but the expression of CAT and POD in the antioxidant enzyme system was downregulated during the whole storage time, which indicated that ozone kept the ROS concentration of postharvest strawberry low mainly by activating the ASA–GSH cycle, especially in late storage.
and H2O2 in the postharvest strawberries subjected to ozone were reduced compared with in the CK group, which may be because ozone activated the antioxidant defense system of strawberry, including the activities of SOD, CAT, APX, MDHAR, and the contents of ASA and GSH. In addition, 17 proteins were more abundant in the OT group than in the CK group at the end of storage, 16 proteins of which were related to the ASA–GSH cycle, but the expression of CAT and POD in the antioxidant enzyme system was downregulated during the whole storage time, which indicated that ozone kept the ROS concentration of postharvest strawberry low mainly by activating the ASA–GSH cycle, especially in late storage.
Conflicts of interest
The authors declare there is no competing financial interest.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2019YFC1606504); National Natural Science Foundation of China Youth Science Fund Project (31501547); Innovation Team of the Tianjin Forestry & Pomology Research System (ITTFPRS2018009, ITTFPRS2018010); Tianjin Innovative Experimental Project for Young Scientists (2020009); Key Laboratory of Storage of Agricultural Products, Ministry of Agriculture and Rural Affairs (kf2019006; kf2019008); Beijing Financial Support Characteristic High Level Vocational College Construction Special Project-Food Nutrition and Safety Application Technology Collaborative Innovation Center Project (PXM2020-157102-000025).References
- F. Giampieri, T. Y. F. Hernandez, M. Gasparrini, J. M. A. Suarez, S. Afrin, S. Bompadre, J. L. Quiles, B. Mezzetti and M. Battino, Food Funct., 2015, 6, 1386–1398 RSC.
- S. Petrasch, S. J. Knapp, J. A. L. van Kan and B. B. Ulate, Mol. Plant Pathol., 2019, 20, 877–892 CrossRef.
- J. Severo, I. R. de Oliveira, R. Bott, C. Le Bourvellec, C. M. G. C. Renard, D. Page, F. C. Chaves and C. V. Rombaldi, LWT, 2017, 85, 390–393 CrossRef CAS.
- J. M. A. Suarez, D. Dekanski, S. Ristić, N. V. Radonjić, N. D. Petronijević, F. Giampieri, P. Astolfi, A. M. G. Paramás, C. S. Buelga, S. Tulipani, J. L. Quiles, B. Mezzetti and M. Battino, PLoS One, 2011, 6, e25878 CrossRef.
- C. Forni, R. Braglia, N. Mulinacci, A. Urbani, M. Ronci, A. Gismondi, C. Tabolacci, B. Provenzano, A. Lentini and S. Beninati, Mol. BioSyst., 2014, 10, 1255–1263 RSC.
- Y. Zhang, N. P. Seeram, R. Lee, L. Feng and D. Heber, J. Agric. Food Chem., 2008, 670–675 CrossRef CAS.
- T. Y. F. Hernandez, M. Gasparrini, S. Afrin, S. Bompadre, B. Mezzetti, J. L. Quiles, F. Giampieri and M. Battino, Crit. Rev. Food Sci. Nutr., 2016, 56, s46–s50 CrossRef.
- A. Z. Tulio, C. Chang, I. Edirisinghe, K. D. White, J. E. Jablonski, K. Banaszewski, A. Kangath, R. K. Tadapaneni, B. B. Freeman and L. S. Jackson, J. Agric. Food Chem., 2012, 60, 5803–5812 CrossRef CAS.
- T. Forbes, F. Giampieri, M. Gasparrini, S. Afrin, L. Mazzoni, M. Cordero, B. Mezzetti, J. Quiles and M. Battino, Nutrients, 2017, 9, 621 CrossRef.
- F. Giampieri, J. M. A. Suarez, M. D. Cordero, M. Gasparrini, T. Y. F. Hernandez, S. Afrin, C. S. Buelga, A. M. G. Paramás, P. Astolfi, C. Rubini, A. Zizzi, S. Tulipani, J. L. Quiles, B. Mezzetti and M. Battino, Food Chem., 2017, 234, 464–471 CrossRef CAS.
- F. Giampieri, M. Gasparrini, T. Y. F. Hernandez, L. Mazzoni, F. Capocasa, S. Sabbadini, J. M. A. Suarez, S. Afrin, C. Rosati, T. Pandolfini, B. Molesini, J. F. S. Sevilla, I. Amaya, B. Mezzetti and M. Battino, J. Agric. Food Chem., 2018, 66, 581–592 CrossRef CAS.
- L. Mazzoni, P. P. Lopez, F. Giampieri, J. M. A. Suarez, M. Gasparrini, T. Y. F. Hernandez, J. L. Quiles, B. Mezzetti and M. Battino, J. Sci. Food Agric., 2016, 92, 365–371 CrossRef.
- Y. Huang, E. Park, I. Edirisinghe and B. M. B. Freeman, Food Funct., 2016, 7, 4745–4752 RSC.
- D. Xiao, A. Sandhu, Y. Huang, E. Park, I. Edirisinghe and B. M. B. Freeman, Food Funct., 2017, 8, 3970–3979 RSC.
- M. M. El-Mogy, R. A. Ludlow, C. Roberts, C. T. Müller and H. J. Rogers, J. Berry Res., 2019, 9, 297–307 CAS.
- Y. He, S. K. Bose, M. Wang, T. Liu, W. Wang, H. Lu and H. Yin, J. Berry Res., 2019, 9, 1–16 Search PubMed.
- M. M. El-Mogy, M. R. Ali, O. S. Darwish and H. J. Rogers, J. Berry Res., 2019, 9, 333–348 CAS.
- L. Di Vittori, L. Mazzoni, M. Battino and B. Mezzetti, Sci. Hortic., 2018, 233, 310–322 CrossRef CAS.
- G. R. Aristya and R. Alyza, Biogenesis : J. Ilm. Biol., 2019, 7, 1 CrossRef.
- Y. Wei, Y. Wei, F. Xu and X. Shao, Postharvest Biol. Technol., 2018, 136, 139–144 CrossRef CAS.
- N. Tzortzakis, T. Taybi, R. Roberts, I. Singleton, A. Borland and J. Barnes, Postharvest Biol. Technol., 2011, 61, 152–159 CrossRef CAS.
- S. Lin, C. Chen, H. Luo, W. Xu, H. Zhang, J. J. Tian, R. Ju and L. Wang, Postharvest Biol. Technol., 2019, 154, 1–10 CrossRef CAS.
- Q. Han, H. Gao, H. Chen, X. Fang and W. Wu, Food Chem., 2017, 221, 1947–1953 CrossRef CAS.
- C. Chen, H. Zhang, X. Zhang, C. Dong, W. Xue and W. Xu, Postharvest Biol. Technol., 2020, 163, 111124 CrossRef CAS.
- C. Liu, T. Ma, W. Hu, M. Tian and L. Sun, Int. J. Food Sci. Technol., 2016, 51, 1099–1109 CrossRef CAS.
- H. L. Xue, Y. Bi, R. Hussain, H. J. Wang, L. M. Pu, M. N. Nan, X. Y. Cheng, Y. Wang and Y. C. Li, Food Chem., 2018, 254, 193–200 CrossRef CAS.
- D. Xu, M. Shi, B. Jia, Z. Yan, L. Gao, W. Guan and Q. Wang, J. Food Process. Preserv., 2019, 43, e14020 Search PubMed.
- T. Piechowiak and M. Balawejder, Food Chem., 2019, 298, 125093 CrossRef CAS.
- Q. Wu, X. Zhu, H. Gao, Z. Zhang, H. Zhu, X. Duan, H. Qu, Z. Yun and Y. Jiang, Postharvest Biol. Technol., 2019, 153, 1–12 CrossRef CAS.
- Z. Zhao, G. Xu, Z. Han, Q. Li, Y. Chen and D. Li, J. Food Qual., 2013, 36, 190–197 CrossRef CAS.
- M. S. Król, M. Materska, B. Chilczuk, M. Karaś, A. Jakubczyk, I. Perucka and I. Jackowska, Food Chem., 2016, 211, 59–63 CrossRef.
- M. V. Rao, G. Paliyath and D. P. Ormrod, Plant Physiol., 1996, 110, 125–136 CrossRef CAS.
- V. Goffi, A. Magri, R. Botondi and M. Petriccione, J. Sci. Food Agric., 2020, 100, 961–968 CrossRef CAS.
- M. K. Ong, A. Ali, P. G. Alderson and C. F. Forney, Sci. Hortic., 2014, 179, 163–169 CrossRef CAS.
- X. Zhang, Z. Zhang, L. Wang, Z. Zhang, J. Li and C. Zhao, Front. Agric. China, 2011, 5, 356–360 CrossRef.
- C. Chen, H. Zhang, C. Dong, H. Ji, X. Zhang, L. Li, Z. Ban, N. Zhang and W. Xue, RSC Adv., 2019, 9, 25429–25438 RSC.
- A. Onopiuk, A. Półtorak, J. Wyrwisz, M. Moczkowska, A. Stelmasiak, A. Lipińska, A. Szpicer, M. Zalewska, R. Zaremba, M. Kuboń and A. Wierzbicka, CyTA--J. Food, 2016, 15, 563–573 CrossRef.
- I. S. Minas, G. Tanou, A. Krokida, E. Karagiannis, M. Belghazi, M. Vasilakakis, K. K. Papadopoulou and A. Molassiotis, BMC Plant Biol., 2018, 18, 358 CrossRef CAS.
- N. Waldeck, K. Burkey, T. Carter, D. Dickey, Q. Song and E. Taliercio, BMC Genomics, 2017, 18, 498 CrossRef.
- E. Xu, L. Vaahtera, H. Hõrak, D. K. Hincha, A. G. Heyer and M. Brosché, Plant, Cell Environ., 2015, 38, 1418–1433 CrossRef CAS.
- P. M. K. Mathabe, Z. A. Belay, T. Ndlovu and O. J. Caleb, Sci. Hortic., 2020, 261, 108996 CrossRef CAS.
- P. G. Righetti, C. Esteve, A. D'Amato, E. Fasoli, M. L. Marina and M. C. García, Proteomics, 2015, 15, 1639–1645 CrossRef CAS.
- Z. Pan, Y. Zeng, J. An, J. Ye, Q. Xu and X. Deng, J. Proteomics, 2012, 75, 2670–2684 CrossRef CAS.
- T. Li, H. Zhu, Q. Wu, C. Yang, X. Duan, H. Qu, Z. Yun and Y. Jiang, Food Res. Int., 2015, 78, 274–285 CrossRef CAS.
- C. Jiao and Z. Gu, Food Chem., 2019, 292, 372–376 CrossRef CAS.
- C. Chen, X. Zhang, H. Zhang, Z. Ban, L. Li, C. Dong, H. Ji and W. Xue, RSC Adv., 2019, 9, 676–689 RSC.
- L. P. de Souza, L. R. D. A. Faroni, F. F. Heleno, P. R. Cecon, T. D. C. Gonçalves, G. J. da Silva and L. H. F. Prates, LWT–Food Sci. Technol., 2018, 90, 53–60 CrossRef.
- P. Fuggate, C. Wongs-Aree, S. Noichinda and S. Kanlayanarat, Sci. Hortic., 2010, 126, 120–129 CrossRef CAS.
- J. Wang, L. Mao, X. Li, Z. Lv, C. Liu, Y. Huang and D. Li, Sci. Hortic., 2018, 241, 201–208 CrossRef CAS.
- H. Huang, G. Jing, L. Guo, D. Zhang, B. Yang, X. Duan, M. Ashraf and Y. Jiang, Postharvest Biol. Technol., 2013, 84, 22–27 CrossRef CAS.
- A. A. H. A. Latef and C. X. He, Sci. Hortic., 2011, 127, 228–233 CrossRef.
- H. Yin, Q. Chen and M. Yi, Plant Growth Regul., 2008, 54, 45–54 CrossRef CAS.
- M. Israr, S. Sahi, R. Datta and D. Sarkar, Chemosphere, 2006, 65, 591–598 CrossRef CAS.
- R. Lufu, A. Ambaw and U. L. Opara, Postharvest Biol. Technol., 2019, 157, 110982 CrossRef.
- Y. Lin, N. Li, H. Lin, M. Lin, Y. Chen, H. Wang, M. A. Ritenour and Y. Lin, Food Chem., 2020, 306, 125627 CrossRef CAS.
- M. S. Aday and C. Caner, LWT–Food Sci. Technol., 2013, 52, 102–109 CrossRef CAS.
- A. Ali, M. K. Ong and C. F. Forney, Food Chem., 2014, 142, 19–26 CrossRef CAS.
- K. J. Radenkova, V. Radenkovs, K. Kundzins and D. Seglina, Food Sci. Technol. Int., 2019, 25, 252–267 CrossRef.
- I. S. Minas, A. R. Vicente, A. P. Dhanapal, G. A. Manganaris, V. Goulas, M. Vasilakakis, C. H. Crisosto and A. Molassiotis, Plant Sci., 2014, 229, 76–85 CrossRef CAS.
- D. R. Mota, O. F. Mora, J. A. M. Espinoza, L. L. R. Verástegui, F. D. d. L. Sánchez and F. R. Cabrera, Sci. Hortic., 2019, 257, 108716 CrossRef.
- O. U. Linus and L. S. Magwaza, Sci. Hortic., 2015, 184, 179–192 CrossRef.
- A. M. Nasirkandi, A. Alirezalu and M. A. Hachesu, Biocatal. Agric. Biotechnol., 2020, 25, 101613 CrossRef.
- H. A. Bashir and A. B. A. A. Goukh, Food Chem., 2003, 80, 557–563 CrossRef CAS.
- R. Thakur, P. Pristijono, M. Bowyer, S. P. Singh, C. J. Scarlett, C. E. Stathopoulos and Q. V. Vuong, LWT, 2019, 100, 341–347 CrossRef CAS.
- S. Khalid, A. U. Malik, A. S. Khan, M. N. Khan, M. I. Ullah, T. Abbas and M. S. Khalid, Sci. Hortic., 2017, 220, 183–192 CrossRef CAS.
- D. A. Grantz, X. Zhang and T. Carlson, Plant, Cell Environ., 1999, 22, 1201–1210 CrossRef CAS.
- P. Muñoz and S. M. Munné, Plant Physiol., 2018, 176, 1004 CrossRef.
- S. Tian, G. Qin and B. Li, Plant Mol. Biol., 2013, 82, 593–602 CrossRef CAS.
- V. B. Frank and J. F. Dat, Plant Physiol., 2006, 141, 384–390 CrossRef.
- D. Camejo, Á. G. Cedeño and A. Moreno, Plant Physiol. Biochem., 2016, 103, 10–23 CrossRef CAS.
- G. Qin, Q. Wang, J. Liu, B. Li and S. Tian, Proteomics, 2010, 9, 4241–4257 CrossRef.
- P. Sharma, A. B. Jha, R. S. Dubey and M. Pessarakli, J. Bot., 2012, 2012, 1–26 CrossRef.
- K. Mondal, S. P. Malhotra, V. Jain and R. Singh, Physiol. Mol. Biol. Plants, 2009, 15, 327–334 CrossRef CAS.
- C. Huan, L. Jiang, X. An, M. Yu, Y. Xu, R. Ma and Z. Yu, Plant Physiol. Biochem., 2016, 104, 294–303 CrossRef CAS.
- A. Caverzan, A. Casassola and S. P. Brammer, in Abiotic and Biotic Stress in Plants - Recent Advances and Future Perspectives, 2016 Search PubMed.
- P. Mishra and P. Sharma, in Reactive Oxygen, Nitrogen and Sulfur Species in Plants, 2019, pp. 53–88 Search PubMed.
- S. A. Anjum, X. Y. Xie, L. C. Wang, M. F. Saleem, C. Man and W. Lei, Afr. J. Agric. Res., 2011, 6, 2026–2032 Search PubMed.
- J. Kan, H. M. Wang and C. H. Jin, Agric. Sci. China, 2011, 10, 149–158 CrossRef CAS.
- M. A. Green and S. C. Fry, Nature, 2005, 433, 83 CrossRef CAS.
- A. Sarkar, A. A. Singh, S. B. Agrawal, A. Ahmad and S. P. Rai, Ecotoxicol. Environ. Saf., 2015, 15, 101–111 CrossRef.
- P. Boonkorn, H. Gemma, S. Sugaya, S. Setha, J. Uthaibutra and K. Whangchai, Postharvest Biol. Technol., 2012, 67, 25–28 CrossRef CAS.
- M. Alothman, B. Kaur, A. Fazilah, R. Bhat and A. A. Karim, Innovative Food Sci. Emerging Technol., 2010, 11, 666–671 CrossRef CAS.
- S. Sudheer, W. K. Yeoh, S. Manickam and A. Ali, Postharvest Biol. Technol., 2016, 117, 81–88 CrossRef CAS.
- J. Wang and D. Q. Li, Chin. J. Eco-Agric., 2002, 10, 94–96 Search PubMed.
- J. L. Wang, Y. Wang, T. H. Zhao, Y. Cao, Y. L. Liu and M. Duan, Acta Ecol. Sin., 2011, 31, 2068–2075 CAS.
- Y. L. Li, Y. F. Liu and J. G. Zhang, Front. Agric. China, 2010, 4, 84–90 CrossRef.
- O. Blokhina, E. Virolainen and K. V. Fagerstedt, Ann. Bot., 2003, 91, 179–194 CrossRef CAS.
- S. Cao, J. Shao, L. Shi, L. Xu, Z. Shen, W. Chen and Z. Yang, Sci. Rep., 2018, 8, 806 CrossRef.
- J. Wang, Q. Zeng, J. Zhu, G. Liu and H. Tang, Agric., Ecosyst. Environ., 2013, 165, 39–49 CrossRef CAS.
- C. H. Pang and B. S. Wang, Role of Ascorbate Peroxidase and Glutathione Reductase in Ascorbate–Glutathione Cycle and Stress Tolerance in Plants, Springer Netherlands, Dordrecht, 2010 Search PubMed.
- R. Mittler, Trends Plant Sci., 2002, 7, 405–410 CrossRef CAS.
- S. Ali, A. Nawaz, S. Hussain, S. M. Khan, S. Ejaz and S. Ahmad, in Priming and Pretreatment of Seeds and Seedlings, ed. M. Hasanuzzaman and V. Fotopoulos, Springer Singapore, Singapore, 2019, pp. 459–493 Search PubMed.
- A. G. Pérez, C. Sanz, J. J. Ríos, R. Olías and J. M. Olías, J. Agric. Food Chem., 1999, 47, 1652–2656 CrossRef.
- L. A. H. Shereefa and M. Kumaraswamy, J. Saudi Soc. Agric. Sci., 2016, 15, 48–56 Search PubMed.
- J. Dumont, S. K. Saari, M. Keinänen, D. Cohen, N. Ningre, S. K. Soppela, P. Baldet, Y. Gibon, P. Dizengremel, M. N. Vaultier, Y. Jolivet, E. Oksanen and D. Le Thiec, Tree Physiol., 2014, 253–266 CrossRef CAS.
- F. F. Fernandes, M. P. Esposito, M. R. G. da Silva Engela, P. C. Gustavson, C. M. Furlan, Y. Hoshika, E. Carrari, G. Magni, M. Domingos and E. Paoletti, Sci. Total Environ., 2019, 656, 1091–1101 CrossRef CAS.
- A. Sarkar, R. Rakwal, S. B. Agrawal, J. Shibato, Y. Ogawa, Y. Yoshida, G. Kumar Agrawal and M. Agrawal, J. Proteome Res., 2010, 9, 4565–4584 CrossRef CAS.
- M. Suekawa, Y. Fujikawa and M. Esaka, Physiological Role of Ascorbic Acid Recycling Enzymes in Plants BT – Ascorbic Acid in Plant Growth, Development and Stress Tolerance, Springer International Publishing, Cham, 2017 Search PubMed.
- R. Stevens, D. Page, B. Gouble, C. Garchery, D. Zamir and M. Causse, Plant, Cell Environ., 2008, 31, 1086–1096 CrossRef CAS.
- C. R. Reczek and N. S. Chandel, Curr. Opin. Cell Biol., 2015, 33, 8–13 CrossRef CAS.
- N. Tzortzakis, A. Borland, I. Singleton and J. Barnes, Postharvest Biol. Technol., 2007, 45, 317–325 CrossRef CAS.
- L. Wang, L. Wang, Z. Zhang, M. Ma, R. Wang, M. Qian and S. Zhang, Postharvest Biol. Technol., 2018, 143, 68–77 CrossRef CAS.
- H. S. Lee, E. J. Kwon, S. Y. Kwon, Y. J. Jeong, E. M. Lee, M. H. Jo, H. S. Kim, I. S. Woo, A. Shinmyo, K. Yoshida and S. S. Kwak, Mol. Breed., 2003, 11, 213–220 CrossRef CAS.
- J. Geng, T. Wei, Y. Wang, X. Huang and J. H. Liu, Tree Physiol., 2019, 39, 2045–2054 CAS.
- V. Fotopoulos and A. K. Kanellis, Plant Physiol. Biochem., 2013, 73, 154–160 CrossRef CAS.
- H. Xue, Y. Sun, L. Li, Y. Bi, R. Hussain, R. Zhang, H. Long, M. Nan and L. Pu, Sci. Hortic., 2020, 265, 109264 CrossRef CAS.
- E. Alós, M. J. Rodrigo and L. Zacarías, Planta, 2014, 239, 1113–1128 CrossRef.
- H. A. Eltelib, A. A. Badejo, Y. Fujikawa and M. Esaka, J. Plant Physiol., 2011, 168, 619–627 CrossRef CAS.
- B. Poljšak and I. Milisav, Aging, Oxidative Stress and Chronic Degenerative Diseases. A Role for Antioxidants, 2013, vol. 14, pp. 331–356 Search PubMed.
- D. Liang, F. Gao, Z. Ni, L. Lin, Q. Deng, Y. Tang, X. Wang, X. Luo and H. Xia, Molecules, 2018, 23, 584 CrossRef.
- B. Le Martret, M. Poage, K. Shiel, G. D. Nugent and P. J. Dix, Plant Biotechnol. J., 2011, 9, 661–673 CrossRef CAS.
- D. Liang, T. Zhu, Z. Ni, L. Lin, Y. Tang, Z. Wang, X. Wang, J. Wang, X. Lv and H. Xia, PLoS One, 2017, 12, e0172818 CrossRef.
- C. M. Luna, G. M. Pastori, S. Driscoll, K. Groten, S. Bernard and C. H. Foyer, J. Exp. Bot., 2005, 417–423 CAS.
- Y. Y. Du, P. C. Wang, J. Chen and C. P. Song, J. Integr. Plant Biol., 2008, 50, 1318–1326 CrossRef CAS.
- B. Gong, S. Huang, N. Ye, X. Yuan and H. Ma, Hortic., Environ. Biotechnol., 2018, 59, 835–845 CrossRef.
- X. Y. Wang, J. C. Tan and J. Wang, Int. J. Agric. Biol. Eng., 2014, 7, 108–114 Search PubMed.
- J. Massolo, A. Concellón, A. Chaves and A. Vicente, Postharvest Biol. Technol., 2011, 59, 10–15 CrossRef CAS.
- T. Min, J. Xie, M. Zheng, Y. Yi, W. Hou, L. Wang, Y. Ai and H. Wang, Postharvest Biol. Technol., 2017, 123, 69–76 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06448c |
| This journal is © The Royal Society of Chemistry 2020 |