Open Access Article
Open Access ArticleElaborating piperazinyl-furopyrimidine based scaffolds as phosphoinositol-3-kinase enzyme alpha (PI3Kα) inhibitors to combat pancreatic cancer†
Mai A. Mansour *a,
Deena S. Lasheen
*a,
Deena S. Lasheen b,
Hatem M. Gaberc and
Khaled A. M. Abouzid
b,
Hatem M. Gaberc and
Khaled A. M. Abouzid *bd
*bd
aPharmaceutical Chemistry Department, Faculty of Pharmacy, Badr University in Cairo, Egypt. E-mail: maiali92@yahoo.com
bPharmaceutical Chemistry Department, Faculty of Pharmacy, Ain Shams University, Abbassia, Cairo 11566, Egypt. E-mail: khaled.abouzid@pharma.asu.edu.eg
cNational Organization for Drug Control and Research, Egypt
dDepartment of Organic and Medicinal Chemistry, Faculty of Pharmacy, University of Sadat City, Sadat City, Menoufia, Egypt
First published on 28th August 2020
Abstract
Phosphoinositol-3-kinase enzyme (PI3K) plays a crucial role in driving oncogenic growth in various mammalian cells, particularly pancreatic cells. In the current study a series of novel furo[2,3-d]pyrimidine based-compounds were designed and synthesized as potential PI3K-α inhibitors. In accordance to the structure–activity relationship (SAR) studies of known PI3K-α inhibitors, different linkers including amide, urea and ether were attached to a piperazinyl furo[2,3-d]pyrimidine core. The synthesized compounds that revealed moderate PI3K-α inhibitory activity were tested for their anti-proliferative activities against pancreatic carcinoma on the PANC-1 cell line. Compounds 7b and 8a showed the highest anti-proliferative activity with IC50 values of 4.5 μM and 6 μM, respectively and relatively, the best in vitro PI3K inhibition ability within the newly synthesized compounds. Additionally, all the newly synthesized final compounds were tested on 60 human cancer cell lines. A docking study was carried out on the PI3K-α active site showing a comparable binding mode to that of FDA approved PI3K-α inhibitors. These newly discovered lipid kinase inhibitors could be considered as potential candidates for the development of new targeted anticancer agents.
Introduction
PI3K belongs to the lipid kinase family, responsible for phosphorylating the 3′-position (–OH) moiety of the inositol ring, yielding phosphatidylinositol-3,4,5-trisphosphate (PIP3), a messenger that in turn activates another serine–threonine protein kinase B (AKT) and the cellular messenger, mTOR.1 This family is divided into three main classes (class I, II and III) based on their different structures and substrate specificities. Class I, our subject of concern, contains four different isoforms, that possess diverse catalytic p110 and regulatory subunits.2 Stimulated by Tyr kinase, G protein-coupled receptors and Ras, class I PI3Ks use phosphatidylinositol-4,5-bisphosphate as their substrate and form a complex with a regulatory subunit, either a p85 isoform (for p110α, p110β and p110δ) or p101 or p87 (for p110γ).3Dysregulation in the PI3K pathway is associated with many types of cancer due to its role in various cellular processes, including cell survival, proliferation and differentiation.4–6 In pancreatic malignancies, separate studies showed that mutations in the K-ras gene occurred in the majority of pancreatic ductal adenocarcinomas (PDAC).7 This oncogenic process causes the over-activation of the PI3K pathway,4 particularly the catalytic subunit p110α of the PI3K-α isoform, which contributes to the super-activation of K-ras in the newly formed pancreatic ductal preneoplastic lesions.8 Inactivating this single isoform ceased the irreversible transition of exocrine acinar cells into pancreatic preneoplastic ductal lesions.9 Not to mention that, the downstream effector, AKT2, an oncogene encoding the protein kinase B, has also been found to be amplified in many pancreatic carcinomas,10 making AKT activate in approximately (59%) of the pancreatic cancers.11
In the meantime, pancreatic cancer represents the seventh leading cause of cancer associated death, world widely,12 and expected to be the second cause in the developed world by 2030.13 This can be attributable to its low 5 year relative survival rate (9%) and poor prognosis.14 However, PDAC, the most common malignancy of all pancreatic cancers (90%)15 has surgical resection as the only recent chance for cure and prolonged survival.16 Gemcitabine17 and other aggressive chemotherapeutic combinations such as, FOLFIRINOX (folinic acid, 5-fluorouracil [5-FU], irinotecan, oxaliplatin), albumin-bound paclitaxel (nab-paclitaxel) plus gemcitabine18 delivered another intolerable protocol of treatment,19 which revealed chemoresistance within weeks of treatment initiation.20
To improve treatment responses, targeting key regulators involved in the aberrant PI3K signaling pathway in PDAC has been implemented in clinical trials. Potent pan-class I PI3K inhibitor such as Copanlisib (I), Buparlisib (II), and ZSTK474 (III) as well as the FDA approved selective PI3K-α inhibitor, Alpelisib,21 have been incorporated in a recent ongoing clinical study22 (Fig. 1).
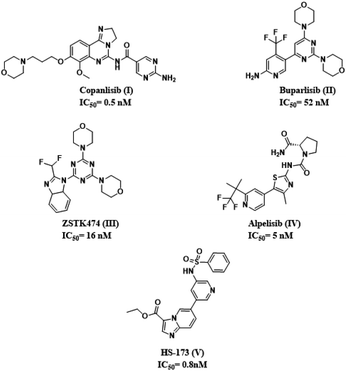 | ||
| Fig. 1 Chemical structures of PI3K-α inhibitors used as clinical candidates in pancreatic cancer treatment with their corresponding IC50 against PI3K-α. | ||
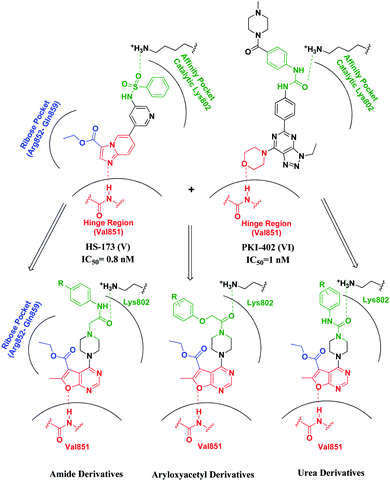
Upon exploring literature surveys and previously reported SAR studies as well as binding interactions of known potent PI3Kinase-α inhibitors, such as binding modes of Alpelisib (PDB code 4JPS)23 and the ester-containing imidazopyridine HS-173 (IC50 = 0.8 nM)24 (V), which as well as established potent anti-tumor efficacy in vivo xenograft and orthotopic pancreatic tumor models,25 revealed three essential pharmacophoric features in a triangle arrangement.26 (1) A hinge binder (HB) that interacts through the formation of the essential hydrogen bond with the backbone valine residue at the hinge region (Val851);23 (2) an affinity pocket binder (APB) that fits into the cavity with possible hydrogen bonding to the catalytic lysine residue (Lys802, Tyr836, Ile932, Ile848, Ile800, Asp933, Pro778 and Met772);23,27 (3) ribose pocket binder (RPB), a lipophilic group, that extends into the ribose binding pocket of ATP, also called region 1, made of 8 amino acids (Arg852–Gln859) maintaining isoform specificity.28,29 In this study, we designed and synthesized a series of novel potential furopyrimidines targeting pancreatic cancer via the PI3K-α inhibition based on mimicking the purine portion of adenosine-5′-triphosphate (ATP)30 providing binding to the hinge region of the kinase by hydrogen bonds with the backbone amino acids.
In order to design novel analogues and clarify the structure–activity relationships, we suggested the following modifications on the potent inhibitor HS-173 (V). The imidazopyridine heterocyclic ring forming the key hinge region hydrogen bond to the backbone-NH of Val851 through the nitrogen of imidazole the moiety was replaced with the fused furo[2,3-d]pyrimidine ring, aiming to form such interaction through hydrogen bonding via the oxygen atom of the furan ring. Whereas, the ester group attached to C3 was preserved to occupy the ribose pocket. A flexible aliphatic piperazine moiety was attached at position 4, due to its expected anti-tumor31 and kinase inhibitory activity.32,33 Besides, for better enzymatic and cellular potency, various amide and urea moieties were introduced to our target compounds aiming to provide the key interaction with the Lys802 amino acid in the catalytic region as observed in PKI402 (VI) and its triazolopyrimidines series, where the amides proceeded as a solubilizing group (Fig. 2).34
Results and discussion
Chemistry
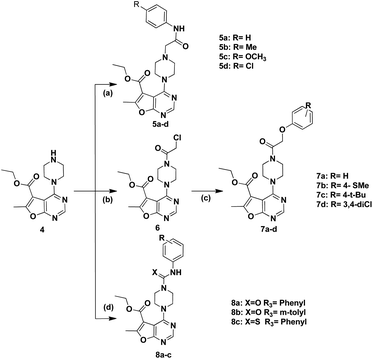
The synthetic pathways for preparation of the target compounds were outlined in Schemes 1 and 2. In Scheme 1, the synthesis of furo[2,3-d]pyrimidine target compounds was started via the preparation of ethyl 5-amino-4-cyano-2-methylfuran-3-carboxylate (1) through reacting ethyl 2-chloroacetoacetate with malononitrile in sodium ethoxide following to the reported method.35 Cyclization of compound (1) was afforded via its reflux with formic acid and acetic anhydride for 48 hours36 to yield the cyclized furo[2,3-d]pyrimidinone derivative (2). Afterwards, chlorination of the cyclized derivative (2) was achieved by reflux in phosphorus oxychloride, followed by coupling with anhydrous piperazine in isopropanol to attain the key intermediate (4)37 Scheme 1.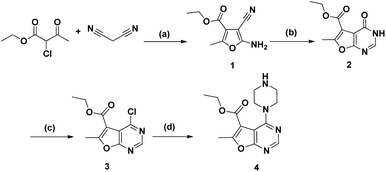 | ||
| Scheme 1 Reagents and conditions: (a) NaOEt, rt, 6 h, 77%; (b) HCOOH, acetic anhydride, reflux, 48 h, 80%; (c) POCL3, reflux, 3 h, 79%; (d) anhydrous piperazine, i-PrOH, reflux, 7 h, 60%. | ||
The synthesis of the final compounds (5a–d) involved nucleophilic attack of the intermediate 4 on respective 2-chlorophenyl acetamide derivatives.38 Whereas, according to Scheme 2, the synthesis of aryloxyacetyl derivatives, initially involved reacting chloroacetyl chloride with 4, to afford the intermediate 6, followed by Williamson's reaction with the appropriate phenols to achieve the compounds entitled (7a–d).39 Finally, urea derivatives were afforded by nucleophilic addition of intermediate 4 to isocyanate and isothiocyanate derivatives, performed in chloroform at room temperature (Scheme 2).40
Biological evaluation
At this concentration, the piperazinyl furo[2,3-d]pyrimidine-5-carboxylate derivatives (7a, 7b) incorporating the aryloxyacetyl moiety and (8a, 8b, 8c) incorporating the substituted arylurea/thiourea moiety attached to the parent scaffold, revealed the moderate inhibition between (51–69%) against the PI3K-α kinase activity.
Upon investigating the structure–activity relationship among the newly synthesized furo[2,3-d]pyrimidines, the phenyl acetamide derivatives (5a–d) showed weak inhibitory activity (11–28%). The unsubstituted derivative (5a) relatively exhibited the weakest inhibitory activity; while final compounds (5b–d) having lipophilic substituents presented relative higher inhibition.
A significant increase in enzymatic inhibition was revealed in the 6-methylfuro[2,3-d]pyrimidines series (7a–7d) bearing the aryloxyacetyl moiety which is most probably attributed to the formation of the essential H-bond with the catalytic Lys802, which was proved to possess a crucial role in the phosphor transfer reaction and inhibitory activity.41 Furthermore, it seemed that such linker permits more free movement of the terminal aromatic ring that extends to occupy the affinity pocket. The unsubstituted and thiomethyl derivative (7a, 7b) showed best inhibition percentage within the series. However, incorporating a bulky tert butyl substituent at the terminal phenyl ring generally reduced the inhibitory activity. It can be postulated that the addition of bulky substituents hindered the aryloxyacetyl derivatives from fitting within the affinity pocket.
Upon the incorporation of the arylurea or thioarylurea linkers with the key intermediate, in final derivatives (8a–c), moderate enzyme inhibition was conserved as the free movement of the terminal aromatic ring is quite maintained, with a noteworthy increase in the inhibitory activity by the unsubstituted urea derivative (8a) that reached 69%.
| PANC-1 cancer cell line | |
|---|---|
| Cpd ID | IC50 μM |
| 7a | 10.70 ± 0.55 |
| 7b | 4.50 ± 0.72 |
| 7d | 10.20 ± 0.90 |
| 8a | 6.00 ± 0.41 |
| 8b | 9.98 ± 0.66 |
| 8c | 17.00 ± 0.85 |
| Paclitaxel | 0.008 ± 0.23 |
Yet, compound 7c was able to show the best mean inhibition within the furopyrimidine final compounds, with a value of 21.54%; however it displayed low PI3K-alpha inhibition 25%, indicating its possible activity against other kinases. A significant growth inhibition was achieved against the renal UO-31 cancer cell line with a value of 51.97%.
Molecular modelling
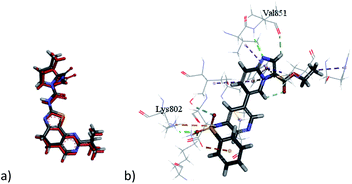
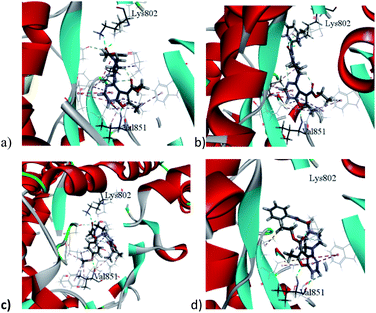
Docking 6-methylfuro[2,3-d]pyrimidines of the target compounds revealed that derivatives (7a, d) and (8b–c) fulfilled the basic key interactions of PI3K-α inhibitors, where the oxygen atom of furan ring forms a H-bond with Val851 at the hinge region and the and another hydrogen bond is formed with the catalytic Lys802 through the oxygen atom of either the aryloxyacetyl or urea series. The ester moiety saturated the ribose pocket by hydrophobic interaction with different residues whether with Ser854, or Gln859. Compounds with amide linkage (5a–d) missed the key interaction with Lys802 residue, resulting in lower docking scores. These results may also deduce their PI3K inhibitory inactivity in the kinase assay.
Interestingly, compound (7b) showed a flipped orientation, preserving the required key interaction in a different approach. The thiomethyl moiety formed a hydrogen bond with the Val851 residue instead of the furopyrimidine's oxygen atom and the NCO moiety formed a hydrogen bond with the catalytic Lys802 amino acid while the furopyrimidine ring bound to the ribose pocket. Although compound (7c), the aryloxyacetyl derivative, was expected to show similar inhibitory activity, yet the docking study shown that the tert-butyl group led to repositioning of the furopyrimidine core, which prevented it from binding to the hinge region, and the NCO group formed the hydrogen bond with Val851 instead, moreover, it missed the key hydrogen bonding with Lys802, explaining the loss of inhibitory activity of this compound (Fig. 4).
The urea derivative (8a) (PI3K-α inhibition; 69%) established highest docking scores within all the target compounds. Yet, it performed a slightly different orientation, where the terminal aromatic ring formed a pi–pi T shaped bond with Trp780 in the specificity pocket.
Conclusion
Novel furo[2,3-d]pyrimidine derivatives were designed, synthesized and evaluated for their in vitro PI3K-α inhibitory activity and their anti-proliferative activity against the PANC-1 cell line as well as NCI 60 cell line panel. Compounds bearing a urea linker or aryloxyacetyl moieties showed moderate PI3K-α inhibition (45–69%) at 10 μM concentration except for the aryloxyacetyl derivative compound (7c) due to its bulky p-tert-butyl substituent. The amide-based derivatives (5a–5d) showed poor PI3K-α inhibition (0–28%) at 10 μM concentration. In addition, compounds (7b and 8a) displayed good in vitro ability to inhibit PANC-1 cell line proliferation with IC50 values of 4.5 and 6.0 μM respectively. These outcomes were further clarified using molecular docking studies which revealed the ability of the urea-based derivatives to form a network of key interactions, known to be essential for PI3K-α inhibition. However, their amide-based analogues missed a key interaction with Lys802 residue. Moreover, the introduction of the thiomethyl moiety to the ether based compounds led to repositioning the furo[2,3]pyrimidine ring, maintaining the key interactions characterizing PI3K-α inhibitors.In silico predictive ADME study for targeted compounds
In order to further examine physicochemical properties of the synthesized compounds, computer-aided ADMET study was implemented by using Accelrys Discovery Studio 2.5 software.These studies are based on a protocol that calculates a long list of traditional molecular descriptors, semiempirical quantum mechanics (QM) descriptors, density functional QM descriptors, as well as user-built QSAR models. As a result, a prediction various molecular properties can be obtained based on the given molecular chemical structure. Parameters including atom-based log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P98 (A
P98 (A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P98), aqueous solubility level (AQ SOl LEV), 2D polar surface area (ADMET 2D PSA), and blood–brain barrier level (BBB LEV), were calculated to expect compounds pharmacokinetics. Anticipation of the aqueous solubility level was made for the target compounds, showing fluctuating values (2–3) between good and low solubility (Table 3), which are ideal for oral bioavailability.
P98), aqueous solubility level (AQ SOl LEV), 2D polar surface area (ADMET 2D PSA), and blood–brain barrier level (BBB LEV), were calculated to expect compounds pharmacokinetics. Anticipation of the aqueous solubility level was made for the target compounds, showing fluctuating values (2–3) between good and low solubility (Table 3), which are ideal for oral bioavailability.
| Cpd ID | aA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P98 P98 |
bAqueous solubility level | cPSA_2D | dIntestinal absorption level | eBBB level |
|---|---|---|---|---|---|
a Lipophilicity descriptor.b Aqueous solubility level; 4 = optimal, 3 = good, 2 = low solubility, 1 = very low but soluble, 0 = extremely low.c Polar surface area; compounds must have log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p value not greater than 5.0 to attain a reasonable probability of being well absorbed.d Absorption level; 3 = very low absorption, 2 = low absorption, 1 = moderate, 0 = good absorption.e Blood–brain barrier level; 4 = undefined, 3 = low penetration 2 = medium penetration, 1 = high penetration. p value not greater than 5.0 to attain a reasonable probability of being well absorbed.d Absorption level; 3 = very low absorption, 2 = low absorption, 1 = moderate, 0 = good absorption.e Blood–brain barrier level; 4 = undefined, 3 = low penetration 2 = medium penetration, 1 = high penetration. |
|||||
| HS-173 | 2.934 | 2 | 88.702 | 0 | 3 |
| 5a | 2.059 | 3 | 98.123 | 0 | 3 |
| 5b | 2.545 | 2 | 98.123 | 0 | 3 |
| 5c | 2.042 | 3 | 107.053 | 0 | 3 |
| 5d | 2.723 | 2 | 98.123 | 0 | 3 |
| 7a | 2.181 | 3 | 94.243 | 0 | 3 |
| 7b | 2.723 | 3 | 94.243 | 0 | 3 |
| 7c | 3.582 | 2 | 94.243 | 0 | 3 |
| 7d | 3.51 | 2 | 94.243 | 0 | 3 |
| 8a | 2.254 | 3 | 98.123 | 0 | 3 |
| 8b | 2.74 | 2 | 98.123 | 0 | 3 |
| 8c | 2.117 | 2 | 68.012 | 0 | 3 |
PSA is another drug bioavailability aspect, in which actively absorbed molecules with PSA > 140 are considered of poor in oral bioavailability. Here in, the synthesized compounds PSA fell within the range 68–107.1, predicting the presence of good passive oral absorption. Furthermore, results are offered as a 2D ADMET plot, drawn using calculated PSA_2D against A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P98 properties. Blood–brain barrier (BBB) and human intestinal absorption (HIA) plots were presented as well. In BBB plot, almost all the synthesized compounds were located between the 99% and 95% ellipses with a low level of BBB penetration, indicating low CNS adverse effects, unlike the HS-173, which is predicted to highly penetrate the BBB. While, in HIA plot, all the target compounds fell within the 99% ellipse, indicating good intestinal absorption with an absorption level 0 (ESI†).
P98 properties. Blood–brain barrier (BBB) and human intestinal absorption (HIA) plots were presented as well. In BBB plot, almost all the synthesized compounds were located between the 99% and 95% ellipses with a low level of BBB penetration, indicating low CNS adverse effects, unlike the HS-173, which is predicted to highly penetrate the BBB. While, in HIA plot, all the target compounds fell within the 99% ellipse, indicating good intestinal absorption with an absorption level 0 (ESI†).
Experimental
Chemistry and analysis
Starting materials and reagents were purchased from Sigma-Aldrich (USA), and Loba Chemie Organics and were used without further purification. Solvents were purchased from Fisher scientific or Sigma-Aldrich and used without further purification. Anhydrous acetone was dried by distillation. Reactions were monitored by analytical thin layer chromatography (TLC), on silica gel 60 F254 that is packed on aluminum sheets, purchased from Merck, and were visualized under UV light (254 nm). Intermediates monitoring was carried out using melting points, or 1H NMR which matched the reference data. Melting points were measured on an electrothermal apparatus in closed capillary tubes using Gallenkamp melting point apparatus and were uncorrected. 1H NMR spectra were recorded on a Bruker 400 MHz spectrometer in δ scale (ppm), using CDCl3 as solvent and TMS as internal standard signal at the Faculty of Pharmacy, Ain-Shams University. Whereas 13C NMR spectra were recorded a 100 MHz. EI-MS spectra were recorded on direct probe controller inlet part to single quadrupole mass analyzer in (Thermo Scientific GCMS) model (ISQ LT) using Thermo X-Calibur software at the regional center for mycology and biotechnology (RCMB) Al-Azhar University. Elemental analyses were performed at the regional center of mycology and biotechnology at Al-Azhar University. Compounds 1,35 2, 3 (ref. 39) and the chlorophenylacetamide derivatives,44,45 were prepared according to the reported procedures.1H-NMR (CDCl3-d1, 400 MHz): δ (ppm) 8.28 (s, 1H, pyrimidine H), 4.28 (q, J = 7.1 Hz, 2H, CH2CH3), 3.5 (t, J = 5 Hz, 4H, piperazine H), 2.86 (t, J = 5 Hz, 4H, piperazine H), 2.57 (s, 3H, CH3), 2.35 (s, 1H, NH D2O exchangeable), 1.31 (t, J = 7.1 Hz, 3H, CH2CH3); FT-IR (ύ max, cm−1) 3419 (NH), 2956 (C–H aliphatic), 2921 (C–H aromatic), 1719 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1589 (C
O), 1589 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C).
C).
General procedure. To a solution of ethyl 6-methyl-4-(piperazin-1-yl)furo[2,3-d]pyrimidine-5-carboxylate (4) (1 equiv., 0.283 g, 0.97 mmol), potassium carbonate (4.5 equiv., 0.604 g, 4.37 mmol) and potassium iodide (4.5 equiv., 0.72 g, 4.37 mmol) were added and appropriate 2-chloro-N-phenylacetamide derivatives (1 equiv., 0.97 mmol) in acetone (30 mL). The reaction was refluxed for 30–36 h and monitored using TLC (DCM
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AcOEt = 1
AcOEt = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).The mixture was vacuum filtered and poured on water (50 mL), filtered then recrystallized from absolute ethanol.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1591 (C
O), 1591 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 423): m/z, 423 [M+, (23.6%)] 323.4 (100%); anal. calcd for C22H25N5O4: C, 62.40; H, 5.95; N, 16.54; found: C, 62.31; H, 5.74; N, 16.71.
C); MS: (Mw: 423): m/z, 423 [M+, (23.6%)] 323.4 (100%); anal. calcd for C22H25N5O4: C, 62.40; H, 5.95; N, 16.54; found: C, 62.31; H, 5.74; N, 16.71.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1593 (C
O), 1593 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 437): m/z, 457 [M+, (20.87%)] 329.09 (100%); anal. calcd for C23H27N5O4: C, 63.14; H, 6.22; N, 16.01; found: C, 63.14; H, 6.20; N, 16.01.
C); MS: (Mw: 437): m/z, 457 [M+, (20.87%)] 329.09 (100%); anal. calcd for C23H27N5O4: C, 63.14; H, 6.22; N, 16.01; found: C, 63.14; H, 6.20; N, 16.01.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1593 (C
O), 1593 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 451): m/z, 451 [M+, (22.4%)] 329 (100%); anal. calcd for C23H27N5O5: C, 60.92; H, 6.00; N, 15.44; found: C, 60.92; H, 5.93; N, 15.41.
C); MS: (Mw: 451): m/z, 451 [M+, (22.4%)] 329 (100%); anal. calcd for C23H27N5O5: C, 60.92; H, 6.00; N, 15.44; found: C, 60.92; H, 5.93; N, 15.41.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1589 (C
O), 1589 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). MS: (Mw: 457): m/z, 457 [M+, (21.27%)] 285.57 (100%); anal. calcd for C22H24ClN5O4: C, 57.71; H, 5.28; N, 15.29; found: C, 57.89; H, 5.48; N, 15.57.
C). MS: (Mw: 457): m/z, 457 [M+, (21.27%)] 285.57 (100%); anal. calcd for C22H24ClN5O4: C, 57.71; H, 5.28; N, 15.29; found: C, 57.89; H, 5.48; N, 15.57.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1593 (C
O), 1593 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 366): m/z, 366 [M+, (44.09%)] 330 (100%); anal. calcd for C16H19ClN4O4: C, 52.39; H, 5.22; N, 15.27; found: C, 52.47; H, 5.38; N, 15.13.
C); MS: (Mw: 366): m/z, 366 [M+, (44.09%)] 330 (100%); anal. calcd for C16H19ClN4O4: C, 52.39; H, 5.22; N, 15.27; found: C, 52.47; H, 5.38; N, 15.13.General procedure. To a solution of ethyl 4-(4-(2-chloroacetyl)piperazin-1-yl)-6-methylfuro[2,3-d]pyrimidine-5-carboxylate (6) in (30 mL) acetone, potassium carbonate (4.5 equiv., 0.604 g, 4.37 mmol) and potassium iodide (4.5 equiv., 0.72 g, 4.37 mmol) were added (1 equiv., 0.283 g, 0.97 mmol) along with appropriate phenolic derivatives (3,4-dichlorophenol, 4-(methylthio)phenol, 4-tert-butylphenol, and phenol) (1 equiv., 0.97 mmol) in acetone 30 mL. The reaction was refluxed for 20–30 h and monitored using TLC (DCM
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AcOEt = 1
AcOEt = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The mixture was vacuum evaporated and poured on water, filtered then recrystallized from absolute ethanol.
1). The mixture was vacuum evaporated and poured on water, filtered then recrystallized from absolute ethanol.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O),1587 (C
O),1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 424): m/z, 424 [M+, (29.74%)] 285.79 (100%); anal. calcd for C22H24N4O5: C, 62.25; H, 5.70; N, 13.20; found: C, 62.57; H, 5.82; N, 13.47.
C); MS: (Mw: 424): m/z, 424 [M+, (29.74%)] 285.79 (100%); anal. calcd for C22H24N4O5: C, 62.25; H, 5.70; N, 13.20; found: C, 62.57; H, 5.82; N, 13.47.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1591 (C
O), 1591 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 470): m/z, 470 [M+, (14%)] 316.87 (100%); anal. calcd for C23H26N4O5 S: C, 58.71; H, 5.57; N, 11.91; found: C, 58.45; H, 5.79; N, 11.82.
C); MS: (Mw: 470): m/z, 470 [M+, (14%)] 316.87 (100%); anal. calcd for C23H26N4O5 S: C, 58.71; H, 5.57; N, 11.91; found: C, 58.45; H, 5.79; N, 11.82.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O),1585 (C
O),1585 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 480): m/z, 480 [M+, (11.9%)] 375.2 (100%); anal. calcd for C26H32N4O5: C, 64.98; H, 6.71; N, 11.66; found: C, 64.89; H, 6.58; N, 11.59.
C); MS: (Mw: 480): m/z, 480 [M+, (11.9%)] 375.2 (100%); anal. calcd for C26H32N4O5: C, 64.98; H, 6.71; N, 11.66; found: C, 64.89; H, 6.58; N, 11.59.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1590 (C
O), 1590 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 492): m/z, 492 [M+, (23.37%)] 321.18 (100%); anal. calcd for C22H22Cl2N4O5: C, 53.56; H, 4.49; N, 11.36; found: C, 53.56; H, 4.79; N, 11.36.
C); MS: (Mw: 492): m/z, 492 [M+, (23.37%)] 321.18 (100%); anal. calcd for C22H22Cl2N4O5: C, 53.56; H, 4.49; N, 11.36; found: C, 53.56; H, 4.79; N, 11.36.General procedure. To a solution of ethyl 6-methyl-4-(piperazin-1-yl)furo[2,3-d]pyrimidine-5-carboxylate (4) (1 equiv., 0.2 g, 0.69 mmol) in chloroform (40 mL), the respective aryl isocyanate or aryl isothiocyanate (1 equiv., 0.69 mmol) were added and stirred for 6 h at room temperature. The chloroform was vacuum evaporated and the precipitate was recrystallized from ethanol.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1591 (C
O), 1591 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 409): m/z, 409 [M+, (18.95%)] 222.16 (100%); anal. calcd for C21H23N5O64: C, 61.60; H 5.66; N, 17.10; found: C, 61.78; H, 5.89; N, 16.89.
C); MS: (Mw: 409): m/z, 409 [M+, (18.95%)] 222.16 (100%); anal. calcd for C21H23N5O64: C, 61.60; H 5.66; N, 17.10; found: C, 61.78; H, 5.89; N, 16.89.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1589 (C
O), 1589 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); MS: (Mw: 423): m/z, 423 [M+, (23.7%)] 419.87 (100%); anal. calcd for C22H25N5O4: C, 62.40; H, 5.95; N, 16.54; found: C, 62.49; H, 6.14; N, 16.73.
C); MS: (Mw: 423): m/z, 423 [M+, (23.7%)] 419.87 (100%); anal. calcd for C22H25N5O4: C, 62.40; H, 5.95; N, 16.54; found: C, 62.49; H, 6.14; N, 16.73.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1587 (C
O), 1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1231 (C
C), 1231 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S); MS: (Mw: 425): m/z, 425 [M+, (48.18%)] 392.3 (100%); anal. calcd for C21H23N5O3S: C, 59.28; H, 5.45; N, 16.46; found: C, 59.52; H, 5.63; N, 16.72.
S); MS: (Mw: 425): m/z, 425 [M+, (48.18%)] 392.3 (100%); anal. calcd for C21H23N5O3S: C, 59.28; H, 5.45; N, 16.46; found: C, 59.52; H, 5.63; N, 16.72.Biological evaluation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PS served as the standardized substrate and Adapta Universal Kinase Assay Kit was used (ESI†).
PS served as the standardized substrate and Adapta Universal Kinase Assay Kit was used (ESI†).Molecular docking
Molecular modeling was carried out using C-Docker 2.5 software in the interface of Accelry's Discovery Studio 2.5 (Accelrys Inc., San Diego, CA, USA) at Faculty of Pharmacy, Ain Shams University, drug design laboratory. The X-ray crystal structure of PI3K-α co-crystallized with the imidazopyridine-based compound (I) was obtained from the Protein Data Bank at the Research Collaboration for Structural Bioinformatics (RCSB) website [https://www.rcsb.org] (PDB code: 4ZOP) and loaded in Accelry's Discovery (ESI†).Conflicts of interest
There are no conflicts to declare.Notes and references
- F. M. Elmenier, D. S. Lasheen and K. A. M. Abouzid, Phosphatidylinositol 3 kinase (PI3K) inhibitors as new weapon to combat cancer, Eur. J. Med. Chem., 2019, 183, 111718 CrossRef CAS PubMed.
- S. Jean and A. A. Kiger, Classes of phosphoinositide 3-kinases at a glance, J. Cell Sci., 2014, 127, 923–928 CrossRef CAS PubMed.
- B. Vanhaesebroeck, J. Guillermet-Guibert, M. Graupera and B. Bilanges, The emerging mechanisms of isoform-specific PI3K signalling, Nat. Rev. Mol. Cell Biol., 2010, 11, 329–341 CrossRef CAS PubMed.
- C. Sheridan and J. Downward, Inhibiting the RAS-PI3K Pathway in Cancer Therapy, in Enzymes, Elsevier Inc., 1st edn, 2013, vol. 34, pp. 107–136 Search PubMed.
- P. Liu, H. Cheng, T. M. Roberts and J. J. Zhao, Targeting the phosphoinositide 3-kinase pathway in cancer, Nat. Rev. Drug Discovery, 2009, 8, 627–644 CrossRef CAS PubMed.
- X. Shi, J. Wang, Y. Lei, C. Cong, D. Tan and X. Zhou, Research progress on the PI3K/AKT signaling pathway in gynecological cancer (review), Mol. Med. Rep., 2019, 19, 4529–4535 CAS.
- M. Löhr, G. Klöppel, P. Maisonneuve, A. B. Lowenfels and J. Lüttges, Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis, Neoplasia, 2005, 7, 17–23 CrossRef PubMed.
- R. Baer, C. Cintas, N. Therville and J. Guillermet-Guibert, Implication of PI3K/Akt pathway in pancreatic cancer: when PI3K isoforms matter?, Adv. Biol. Regul., 2015, 59, 19–35 CrossRef CAS PubMed.
- R. Baer, C. Cintas, M. Dufresne, S. Cassant-Sourdy, N. Schönhuber, L. Planque, H. Lulka, B. Couderc, C. Bousquet, B. Garmy-Susini, B. Vanhaesebroeck, S. Pyronnet, D. Saur and J. Guillermet-Guibert, Pancreatic cell plasticity and cancer initiation induced by oncogenic Kras is completely dependent on wild-type PI 3-kinase p110α, Genes Dev., 2014, 28, 2621–2635 CrossRef PubMed.
- B. A. Ruggeri, L. Huang, M. Wood, J. Q. Cheng and J. R. Testa, Amplification and Overexpression of the AKT2 Oncogene in a Subset of Human Pancreatic Ductal, Adenocarcinomas, 1998, 86, 81–86 Search PubMed.
- M. G. Schlieman, B. N. Fahy, R. Ramsamooj, L. Beckett and R. J. Bold, Incidence, mechanism and prognostic value of activated AKT in pancreas cancer, Br. J. Cancer, 2003, 89, 2110–2115 CrossRef CAS PubMed.
- F. Bray, J. Ferlay and I. Soerjomataram, Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, Ca-Cancer J. Clin., 2018, 394–424 CrossRef PubMed.
- L. Rahib, B. D. Smith, R. Aizenberg, A. B. Rosenzweig, J. M. Fleshman and L. M. Matrisian, Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States, Cancer Res., 2014 DOI:10.1158/0008-5472.CAN-14-0155.
- R. L. Siegel, K. D. Miller and A. Jemal, Cancer statistics, 2020, Ca-Cancer J. Clin., 2020, 70, 7–30 CrossRef PubMed.
- M. Orth, P. Metzger, S. Gerum, J. Mayerle, G. Schneider, C. Belka, M. Schnurr and K. Lauber, Pancreatic ductal adenocarcinoma: biological hallmarks, current status, and future perspectives of combined modality treatment approaches, Radiat. Oncol., 2019, 14, 141 CrossRef PubMed.
- S. Gillen, T. Schuster, C. Meyer zum Büschenfelde, H. Friess and J. Kleeff, Preoperative/Neoadjuvant Therapy in Pancreatic Cancer: A Systematic Review and Meta-analysis of Response and Resection Percentages, PLoS Med., 2010, 7, e1000267 CrossRef PubMed.
- H. A. Burris, M. J. Moore, J. Andersen, M. R. Green, M. L. Rothenberg, M. R. Modiano, M. C. Cripps, R. K. Portenoy, A. M. Storniolo, P. Tarassoff, R. Nelson, F. A. Dorr, C. D. Stephens and D. D. Von Hoff, Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial, J. Clin. Oncol., 1997, 15, 2403–2413 CrossRef CAS PubMed.
- I. Garrido-Laguna and M. Hidalgo, Pancreatic cancer: from state-of-the-art treatments to promising novel therapies, Nat. Rev. Clin. Oncol., 2015, 12, 319–334 CrossRef CAS PubMed.
- K. E. Behrns, FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer, Surgery, 2011, 364, 1817–1825 Search PubMed.
- M. Amrutkar and I. P. Gladhaug, Pancreatic Cancer Chemoresistance to Gemcitabine, Cancers, 2017, 9, 157 CrossRef PubMed.
- A. Markham, Alpelisib: First Global Approval, Drugs, 2019, 79, 1249–1253 CrossRef CAS PubMed.
- J. R. W. Conway, D. Herrmann, T. R. J. Evans, J. P. Morton and P. Timpson, Combating pancreatic cancer with PI3K pathway inhibitors in the era of personalised medicine, Gut, 2019, 68, 742–758 CrossRef CAS PubMed.
- P. Furet, V. Guagnano, R. A. Fairhurst, P. Imbach-weese, I. Bruce, M. Knapp, C. Fritsch, F. Blasco, J. Blanz, R. Aichholz, J. Hamon, D. Fabbro and G. Caravatti, Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation, Bioorg. Med. Chem. Lett., 2013, 23, 3741–3748 CrossRef CAS PubMed.
- O. Kim, Y. Jeong, H. Lee, S.-S. Hong and S. Hong, Design and Synthesis of Imidazopyridine Analogues as Inhibitors of Phosphoinositide 3-Kinase Signaling and Angiogenesis, J. Med. Chem., 2011, 54, 2455–2466 CrossRef CAS PubMed.
- M. Rumman, K. H. Jung, Z. Fang, H. H. Yan, M. K. Son, S. J. Kim, J. Kim, J. H. Park, J. H. Lim, S. Hong and S.-S. Hong, HS-173, a novel PI3K inhibitor suppresses EMT and metastasis in pancreatic cancer, Oncotarget, 2016, 7, 78029–78047 CrossRef PubMed.
- P. Wu and Y. Hu, Small molecules targeting phosphoinositide 3-kinases, Medchemcomm, 2012, 3, 1337–1355 RSC.
- S. Maheshwari, M. S. Miller, R. O'Meally, R. N. Cole, L. M. Amzel and S. B. Gabelli, Kinetic and structural analyses reveal residues in phosphoinositide 3-kinase α that are critical for catalysis and substrate recognition, J. Biol. Chem., 2017, 292, 13541–13550 CrossRef CAS PubMed.
- Y. Liu, W.-Z. Wan, Y. Li, G.-L. Zhou and X.-G. Liu, Recent development of ATP-competitive small molecule phosphatidylinostitol-3-kinase inhibitors as anticancer agents, Oncotarget, 2017, 8, 7181–7200 CrossRef PubMed.
- M. S. Miller, P. E. Thompson and S. B. Gabelli, Structural determinants of isoform selectivity in pi3k inhibitors, Biomolecules, 2019, 9, 1–34 CrossRef PubMed.
- M. A. Aziz, R. A. T. Serya, D. S. Lasheen and K. A. M. Abouzid, Furo[2,3-d]pyrimidine based derivatives as kinase inhibitors and anticancer agents, Future J. Pharm. Sci., 2016, 2, 1–8 CrossRef.
- R. S. Viswas, S. Pundir and H. Lee, Design and synthesis of 4-piperazinyl quinoline derived urea/thioureas for anti-breast cancer activity by a hybrid pharmacophore approach, J. Enzyme Inhib. Med. Chem., 2019, 34, 620–630 CrossRef CAS PubMed.
- H. M. Shallal and W. A. Russu, Discovery, synthesis, and investigation of the antitumor activity of novel piperazinylpyrimidine derivatives, Eur. J. Med. Chem., 2011, 46, 2043–2057 CrossRef CAS PubMed.
- S. Ponnurangam, D. Standing, P. Rangarajan and D. Subramaniam, Tandutinib inhibits the Akt/mTOR signaling pathway to inhibit colon cancer growth, Mol. Cancer Ther., 2013, 12, 598–609 CrossRef CAS PubMed.
- C. M. Dehnhardt, A. M. Venkatesan, E. Delos Santos, Z. Chen, O. Santos, S. Ayral-Kaloustian, N. Brooijmans, R. Mallon, I. Hollander, L. Feldberg, J. Lucas, I. Chaudhary, K. Yu, J. Gibbons, R. Abraham and T. S. Mansour, Lead optimization of N-3-substituted 7-morpholinotriazolopyrimidines as dual phosphoinositide 3-kinase/mammalian target of rapamycin inhibitors: discovery of PKI-402, J. Med. Chem., 2010, 53, 798–810 CrossRef CAS PubMed.
- M. Bakavoli, B. Feizyzadeh and M. Rahimizadeh, Investigation of hydrazine addition to functionalized furans: synthesis of new functionalized 4,4′-bipyrazole derivatives, Tetrahedron Lett., 2006, 47, 8965–8968 CrossRef CAS.
- M. A. Aziz, R. A. T. Serya, D. S. Lasheen, A. K. Abdel-Aziz, A. Esmat, A. M. Mansour, A. N. B. Singab and K. A. M. Abouzid, Discovery of Potent VEGFR-2 Inhibitors based on Furopyrimidine and Thienopyrimidne Scaffolds as Cancer Targeting Agents, Sci. Rep., 2016, 6, 24460 CrossRef CAS PubMed.
- K. Matsuno, Y. Nomoto, M. Ichimura, S. Ide and S. Oda, Nitrogenous heterocyclic compounds, US Pat., US6423716, 2000.
- I. Bharatam and Chakraborti Group, Mono-Alkylation of a Phenol with 1,2-Dibromoethane via Williamson Ether Synthesis; Substituted Phenolic Ether, National Institute of Pharmaceutical Education and Research, http://cssp.chemspider.com/Article.aspx?id=253 Search PubMed.
- B. Li, W. Gao-wei, Z. Xiao-juan and G. Wen-xian, Synthesis of 2-phenyl-(4-pyrrolidinylethoxyphenyl)ethan-1-one, Chem. Res. Appl., 2012, 24, 936–939 CrossRef.
- H. H. Fahmy, A. El-masry and S. H. Ali Abdelwahed, Synthesis and Preliminary Antimicrobial Screening of New Benzimidazole Heterocycles, Arch. Pharmacal Res., 2001, 24, 27–34 CrossRef CAS PubMed.
- P. Wipf and R. J. Halter, Chemistry and biology of wortmannin, Org. Biomol. Chem., 2005, 3, 2053–2061 RSC.
- R. Gradiz, H. C. Silva, L. Carvalho, M. F. Botelho and A. Mota-Pinto, MIA PaCa-2 and PANC-1 - pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors, Sci. Rep., 2016, 6, 1–14 CrossRef PubMed.
- R. H. Shoemaker, The NCI60 human tumour cell line anticancer drug screen, Nat. Rev. Cancer, 2006, 6, 813–823 CrossRef CAS PubMed.
- D. C. P. Singh, S. R. Hashim and R. G. Singhal, Synthesis and antimicrobial activity of some new thioether derivatives of quinoxaline, E-J. Chem., 2011, 8, 635–642 CrossRef CAS.
- S. A. Katke, S. V Amrutkar, R. J. Bhor and M. V Khairnar, Synthesis of Biologically Active 2-Chloro-N-Alkyl/Aryl Acetamide Derivatives, Int. J. Pharma Sci. Res., 2011, 2, 148–156 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06428a |
| This journal is © The Royal Society of Chemistry 2020 |