Open Access Article
Open Access ArticleA sandwich-type bacteriophage-based amperometric biosensor for the detection of Shiga toxin-producing Escherichia coli serogroups in complex matrices†
Irwin A. Quintela and
Vivian C. H. Wu *
*
Produce Safety and Microbiology Research Unit, US Department of Agriculture, Agricultural Research Services, Western Regional Research Center, Albany, California, USA. E-mail: vivian.wu@usda.gov
First published on 30th September 2020
Abstract
Immuno-based biosensors are a popular tool designed for pathogen screening and detection. The current antibody-based biosensors employ direct, indirect, or sandwich detection approaches; however, instability, cross-reactivity, and high-cost render them unreliable and impractical. To circumvent these drawbacks, here we report a portable sandwich-type bacteriophage-based amperometric biosensor, which is highly-specific to various Shiga toxin-producing Escherichia coli (STEC) serogroups. Environmentally isolated and biotinylated bacteriophages were directly immobilized onto a streptavidin-coated screen-printed carbon electrode (SPCE), which recognized and captured viable target cells. Samples (50 μL) were transferred to these bacteriophage-functionalized SPCEs (12 min, room temp) before sequentially adding a bacteriophage–gold nanoparticle solution (20 μL), H2O2 (40 mM), and 1,1′-ferrocenedicarboxylic acid for amperometric tests (100 mV s−1) and analysis (ANOVA and LSD, P < 0.05). The optimum biotin concentration (10 mM) retained 94.47% bacteriophage viability. Non-target bacteria (Listeria monocytogenes and Salmonella Typhimurium) had delta currents below the threshold of a positive detection. With less than 1 h turn-around time, the amperometric biosensor had a detection limit of 10–102 CFU mL−1 for STEC O157, O26, and O179 strains and R2 values of 0.97, 0.99, and 0.87, respectively, and a similar detection limit was observed in complex matrices, 10–102 CFU g−1 or mL−1 with R2 values of 0.98, 0.95, and 0.76, respectively. The newly developed portable amperometric biosensor was able to rapidly detect viable target cells at low inoculum levels, thus providing an inexpensive and improved alternative to the current immuno- and laboratory-based STEC screening methods.
1. Introduction
Shiga toxin-producing Escherichia coli (STEC) has been a significant cause of periodic and epidemic foodborne diseases such as gastroenteritis, hemorrhagic colitis (HC), and hemolytic-uremic syndrome (HUS).1–3 An estimate of 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases is reported each year, ranging from mild diarrhea to HUS (10%), and a recent multi-state prospective study showed 259 children had HUS as a complication of STEC O157:H7 infection.4,5 HUS is one of the primary causes of acute kidney injury (AKI), especially in pediatric patients.1,6 Rapid and accurate screening of STEC using highly selective and easy-to-operate tools is one of the most efficient approaches to reduce the incidence of illnesses and hospitalizations through contaminated food products. Early detection and the ability to precisely screen pathogenic microorganisms prevent potential massive product recalls and incurring a severe economic loss.
000 cases is reported each year, ranging from mild diarrhea to HUS (10%), and a recent multi-state prospective study showed 259 children had HUS as a complication of STEC O157:H7 infection.4,5 HUS is one of the primary causes of acute kidney injury (AKI), especially in pediatric patients.1,6 Rapid and accurate screening of STEC using highly selective and easy-to-operate tools is one of the most efficient approaches to reduce the incidence of illnesses and hospitalizations through contaminated food products. Early detection and the ability to precisely screen pathogenic microorganisms prevent potential massive product recalls and incurring a severe economic loss.
Research focus in the field of detection of pathogenic bacteria has been geared towards the development of on-site sensor devices.7 Researchers are developing affordable and on-site systems that are aimed to move away sample processing and testing from a centralized laboratory.8 In the field of immunoassays, the common antibody–antigen interactions in enzyme-linked immunosorbent assay (ELISA) and agglutination kits for the detection of foodborne pathogens are relatively easier to perform but often generate false-positive results and are not capable of differentiating viable from non-viable cells.9,10 Antibodies are commonly integrated as bioreceptors and capture elements with biosensors due to their high affinities to specific targets.11 Binding fragments are also relatively easy to modify using protein engineering and are widely utilized in nanotechnology applications.12 Foodborne pathogens such as STEC O157:H7, Salmonella spp., Vibrio spp., and viruses have been detected by antibodies-based biosensors coupled with various transducers and techniques such as chemiluminescence,13 electrochemiluminescence and fluorescence,14 QCM immunosensor,15 SAW,16 differential pulse voltammetry,17 evanescent wave fiber-optic assay,18 lateral-flow assay,19 and SPR.20
However, though polymerase chain reaction (PCR)-based method can improve the sensitivities of immunoassays (approximately 100-fold), it requires thermocycling platforms, trained staff, and reliable infrastructures which can be challenging in areas with scarce resources.21 Another drawback of molecular or nucleic acid amplification techniques (i.e., PCR) is its destructive nature or the need to break up the cells. In occasions when rare cells are encountered, it would require more than a single test to be carried out; then, it becomes a limiting factor.22
Due to the inherently long turn-around time of conventional pathogen detection methods, biosensors are specifically designed to significantly reduce the processing time between sample uptake and test results at a fraction of the cost of conventional methods. It has been found in the published scientific literature that biosensor ranks as the fourth most popular method and the fastest growing technology in the area of pathogen detection.23 It is comprised of target or analytes-specific biorecognition or bioreceptor elements and a physio-chemical transducer that converts and relays signals to an amplifier and computer.24,25 However, it is important to note that the continuous emergence of rapid pathogen detection necessitates a thorough understanding of the major differences among devices according to the molecular interactions between the target analyte and biorecognition agents for efficiency.26
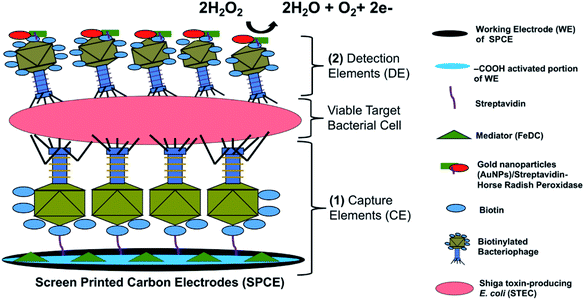
Bacteriophages possess excellent host selectivity and have been used as biorecognition elements for pathogen detection.27 The receptor-binding proteins of bacteriophages recognize bacterial host cells and subsequently inject nucleic acids in the host-cell cytoplasm. Bacteriophages are present in the natural environment and are inexpensive to propagate, and thus are excellent alternatives to antibodies as biological recognition receptors. More importantly, bacteriophages are highly host-specific and very stable, allowing easy handling and storage. Previous studies utilized bacteriophages as biosensor recognition elements, or capture elements for the detection of pathogens such as Salmonella spp.,28,29 Listeria spp.,30 Staphylococcus aureus,31 and E. coli.32 However, the majority of the published study on bacteriophage-based biosensors did not achieve an excellent detection limit (103 CFU mL−1 was reported) even in pure culture setup. This could be attributed to the design of the detection system and the platforms that were utilized. Many of these detection systems employed single-binding event between the biorecognition or capture element of the biosensors and target analyte(s). Though bacteriophages have shown high-specificity toward its bacterial, incorporating bacteriophages onto the detection system as the biorecognition elements may need a secondary binding event to enhance sensitivity, specificity, and reliability. A secondary binding event is often employed in dual-site binding assay or known as a sandwich assay. With these two layers of recognition, the performance of bacteriophage-based can be greatly improved even when used in a more complicated setup and testing various complex matrices. None of the reported bacteriophage-based detection technology employed bacteriophages (two of the same kind) for sandwich capture and detection of viable target bacterial cells. Therefore, here we report a novel approach by utilizing bacteriophages as both capture and detection elements to construct an electrochemical biosensor for STEC detection and achieve an improved detection limit. The primary aim of this study was to develop a sandwich-type bacteriophage-based amperometric biosensor for STEC serogroups and apply the biosensor directly on complex matrices.
2. Experimental methods
2.1. Chemical reagents and apparatus
Phosphate-Buffered Saline (PBS, 10×), Pierce™ 20× TBS Tween™ 20 buffer (TBS-T20), biotin, sulfo-N-hydroxysulfosuccinimide (NHS), streptavidin, potassium ferricyanide K3[FeCN6)], 30% hydrogen peroxide (H2O2), dimethyl sulfoxide (DMSO) solvent, sulfuric acid, ethyl alcohol, Pierce™ Protein-Free Blocking Buffer, polyethylene glycol (PEG), Blocker-Casein blocking buffer, Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Wilmington, DE, USA), sulfosuccinimidobiotin (EZ-Link™ Sulfo-NHS-Biotin), Zeba™ Spin Desalting Columns, 4′-hydroxyazobenzene-2-carboxylic acid (HABA), and streptavidin-coated nanocrystals Qdots (QDs) were purchased from Thermo Fisher Scientific (Waltham, MA). N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), carboxymethyl dextran (CMD) sodium salt, horseradish peroxidase (HRP)-conjugated streptavidin (S-HRP), and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO). 1,1′-Ferrocenedicarboxylic acid (FeDC) was purchased from Strem Chemicals (Newburyport, MA). A solution of gold nanoparticles (AuNPs) with an average diameter of 13 nm was prepared as previously reported.3 Unmodified disposable SPCEs (cat. no. DRP-110, DRP-C110) with three electrodes, a circular carbon working electrode (4 mm), a carbon counter electrode, and a silver reference electrode on a ceramic substrate (34 × 10 × 0.5 mm) were purchased from DropSens (Asturias, Spain). Electrochemical measurements were conducted using PalmSens3 electrochemical portable potentiostat/galvanostat/impedance analyzer (PalmSens, Houten, The Netherlands) that was wirelessly connected via Bluetooth™ and controlled by PSTrace5 software (PalmSens) installed on an Android™ device.2.2. Bacterial strains and bacteriophages
The bacteriophages used in this study were originally isolated from environmental samples and stored at the USDA-Agricultural Research Services Center-Produce Safety and Microbiology Unit, (Albany, CA) belonging to Siphoviridae and Myoviridae families. Representative strains of STEC serogroups, O26:H11 HH8, O157:H7 ATCC 35150, and O179, were utilized as target bacteria. Salmonella Typhimurium ATCC 14028 and Listeria monocytogenes ATCC 19115 were used as non-target groups. All bacterial strains were obtained from the collections of the University of Maine-Pathogenic Microbiology Laboratory (Orono, ME) and the USDA Agricultural Research Services (ARS)-Produce Safety and Microbiology Unit in Albany, CA and USDA ARS in Wyndmoor, PA. Bacterial cells frozen in cryogenic beads (CryoSavers; Hardy Diagnostics, Santa Maria, CA, USA) were activated and revived in Brain Heart Infusion (BHI) broth (Neogen, Lansing, MI, USA) at 37 °C. Overnight cultures were washed in 10 mL of 1× PBS by centrifugation at 5000 × g for 10 min. Pellets were resuspended in 1× PBS and serially diluted up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107. MacConkey Agar with sorbitol, xylose lysine deoxycholate agar, and PALCAM agar (all from Neogen) were used for STEC strains, S. Typhimurium, and L. monocytogenes, respectively.
107. MacConkey Agar with sorbitol, xylose lysine deoxycholate agar, and PALCAM agar (all from Neogen) were used for STEC strains, S. Typhimurium, and L. monocytogenes, respectively.
Titer levels (PFU mL−1) of STEC-specific bacteriophages were determined by a plaque assay. In brief, 100 μL of serially diluted bacteriophage suspensions were mixed with overnight host bacterial cultures (200 μL) and molten tryptic soy agar (5 mL, Neogen) before pouring the mixture into plates and incubated at 37 °C overnight.
2.3. Chemical modification of bacteriophages
Bacteriophage stock solutions (μg mL−1) at various concentrations were biotinylated with sulfosuccinimidobiotin (1–20 mM) and incubated at 4 °C overnight before dialysis against 1× PBS using Zeba™ Spin Desalting Columns to remove excess and unbound biotin. To determine the concentration of bacteriophages, a standard curve based on absorbance (562 nm) was generated by following the manufacturer's instructions (Pierce™ BCA Protein Assay Kit). Incorporated biotin was measured by the spectrophotometric HABA assay at 500 nm absorbance. To monitor the effects of biotin, biotinylated bacteriophages were coupled with streptavidin-coated QDs and viewed under a transmission electron microscope (TEM; FEI Tecnai G2 F20).33 In brief, 2 μL of biotinylated bacteriophage–QDs solution was dropped onto carbon-coated formvar films on copper grids. After negative staining using 1.5% uranyl acetate (pH 4–4.5), the samples were air-dried and viewed under the TEM at 200 kV. To determine the optimum concentration of biotin, the maximum viability retention of bacteriophages was investigated by plaque assays pre and post biotinylation.2.4. Electrochemical behavior and characterization of screen-printed carbon electrodes (SPCE)
Unmodified SPCEs were characterized by recording cyclic voltammograms of 0.5 mM K3[Fe(CN6)] in two separate supporting electrolytes, 0.1 M H2SO4, and 1× PBS at increasing scan rates (50, 100, 200, and 500 mV s−1) under −500 mV to +500 mV vs. counter/reference electrode potential step. Oxidation and reduction peak potentials from the scans were used to identify peak separation ΔEp (ΔEp = Ecp − Eap, where Ecp is the cathodic peak and Eap is anodic peak) of the redox system. To characterize the behaviors of SPCE post modifications, all reagents were individually tested with the cyclic voltammetry (CV) probe, K3[Fe(CN6)]. In brief, reagents (20 μL) including CMD, EDC-NHS, streptavidin, biotinylated bacteriophages, FeDC, AuNP, BSA, casein, protein-free blocking reagents, and PEG were dropped individually onto SPCEs, air-dried, and tested with 0.5 mM K3[Fe(CN6)] at 100 mV s−1.2.5. Bacteriophage-based capture and detection elements
To introduce carboxyl (–COOH) onto the clean working electrode surface, CMD was added (50 mg mL−1) and kept in an orbital shaker at room temperature (25 °C) for 3 h. An equal volume of EDC (0.4 M) and NHS (0.1 M) was added to activate –COOH. Amine-carrying streptavidin (50 μg mL−1) was added and incubated for 40 min to allow carboxyl-to-amine crosslinking. After removing the excess liquid, modified SPCEs were stored in humidified containers before adding biotinylated bacteriophages (>109 PFU mL−1) and incubation at 4 °C overnight. Biofunctionalized SPCEs were blocked with 30% casein (20 μL) at 4 °C overnight, washed twice with TBS-T20, and once with 0.5× PBS prior to storage at 4 °C. Each set of biofunctionalized SPCEs was specific to each of the three STEC serogroups. The capture element refers to biofunctionalized SPCEs and is used hereafter. The bioactivity of the capture element was evaluated after 48 h using an agar diffusion method. Cyclic voltammograms of capture elements and unmodified SPCEs were compared at 100 mV s−1 with 0.5 mM K3[Fe(CN6)].In constructing the detection element solution, biotinylated bacteriophages were mixed with streptavidin-HRP (100 μg mL−1) and AuNP solution (20 μM) and incubated overnight at 4 °C. The viability of bacteriophages was evaluated by conducting the plaque assay and viewing under TEM. All reagents in the biosensor assembly were investigated for noise that contributed to the background signal of the system. In brief, 20 μL of each reagent was added onto clean SPCEs, which were then incubated for 30 s before subjecting to by amperometric tests.
2.6. Electrochemical tests of pure STEC strain cultures and detection limit
Serially diluted STEC samples (50 μL, 10–104 and 108 CFU mL−1) or control (50 μL, 1× PBS) were added onto the functionalized SPCEs, which were then incubated at room temperature (25 °C, 12 min) and washed with 0.5× PBS. Detection element solution (20 μL) was incubated for 10 min, washed twice with TBS-T20 and 0.5× PBS. The mediator (5 μL, 250 mM FeDC in DMSO) was added for 30 s, followed by the addition of H2O2 (40 mM) for another 30 s. Then amperometry was performed at room temperature (25 °C), with a fixed potential of 0.5 V throughout the trial, 0.5 s interval, and 100 s run time. All samples were analyzed in triplicates. The response currents (RC, μA) within a set of potentials over time were measured. Delta (Δ) current, [Δ current = RC of sample (target or non-target) − RC of control] was used to determine the specificity and sensitivity of the assay, with the signal threshold for positive detection defined as a signal-to-noise (S/N) ratio of > 2, wherein the target generated a signal a least two times greater than that from non-target samples. The linear calibration curve (y = mx + b) assumed the response (y) is linearly related to the concentration (x). The detection limit was determined by the statistical significance of the difference of signals (Δ current) between non-target bacteria and the lowest inoculum of target bacteria that had Δ current above the signal threshold for positive detection.2.7. Detection of STEC strains in artificially inoculated and natural complex matrices
Fresh ground beef and pasteurized apple juice were purchased from a local retail store. In brief, weighed fresh ground beef samples (25 g) were transferred into individual stomacher bags (Thermo Fisher Scientific). Overnight bacterial cultures were washed, diluted, and individually spiked on fresh ground beef samples (25 g) to reach the desired final inoculum levels of 10–104 and 108 CFU g−1 or mL−1, followed by 10 s of homogenization using a Pulsifier (Microbiology International, MD, USA) at a low setting. For pasteurized apple juice, 1 mL of inoculum was added into 9 mL of sample and then diluted to obtain the same inoculum levels as the fresh ground beef samples. For both food matrices, 1× PBS was used as a blank, and S. Typhimurium ATCC 14028 was used as a non-target sample.Natural environmental water samples were collected from ponds and water trough used to irrigate fields and farms with observed animal activities (i.e., coyote and deer tracks) (Table S1†). Fifty milliliters of liquid samples were used for testing, while conventional PCR targeting stx genes and the standard plate count method (Table S2†) were conducted for parallel comparison and verification.
2.8. Statistical analysis
Each experiment was conducted on three separate occasions in triplicates. Data were reported as the mean ± S.D. and analyzed by one-way analysis of variance followed by Fisher's least significance difference post-hoc tests using JMP software (Cary, NC, USA). P < 0.05 was considered significant.3. Results and discussion
The detection principle of the developed bacteriophage-based amperometric biosensor is shown in Fig. 1 and the complete system architecture is presented in Fig. S1.†The key novel feature of the sensor presented here is the bacteriophage-based capture and sandwich-type detection of viable target cells. The majority of functional receptors of bacteriophages are located on its tail extremities. Sulfosuccinimido-biotin was used to modify the bacteriophage heads as it reacted with the primary amino groups of the coat proteins concentrated on the bacteriophage heads.28 This modification resulted in an oriented immobilization of the bacteriophages, wherein the tails are directed upward and allows efficient target recognition and capture.34 Once STEC cells were captured by the immobilized bacteriophages, the detection element solution was added, which was comprised of the same bacteriophages but labeled with HRP to accelerate the catalytic reaction and conjugated with AuNPs to amplify signals. The addition of the detection element solution allowed sandwich capture of STEC cells and specific bacteriophage-host binding events. After bacteriophage/STEC/bacteriophage-S-HRP complex formation, a mixture of 40 mM H2O2 and FeDC was added onto the SPCEs. H2O2 served as a substrate for HRP, whereas FeDC acted as a mediator, shuttling electrons between the redox reaction center and the working electrode.35 Horse-Radish Peroxidase (HRP) has been previously used to label long tail fiber proteins of bacteriophages S16 to construct a highly specific and sensitive probe for Salmonella spp. HRP-catalyzed detection assay.36 Moreover, three STEC bacteriophages that can infect O26, O103, O111, O145, and O157 serogroups were chemically labeled with HRP for luminescence-based assay.37 The high binding efficiency of the HRP-labeled bacteriophage to the host cells resulted in high biosensor sensitivity. To our knowledge, this is the first study to use two sets of same bacteriophages for sandwich capture and detection of viable target bacterial cells. An excellent biorecognition element is highly durable, easy to immobilize, cost-efficient, and highly selective and sensitive.31 These key attributes are fulfilled by bacteriophages, both as biorecognition for capturing and reporting viable STEC cells.
3.1. Viability of chemically-modified bacteriophages
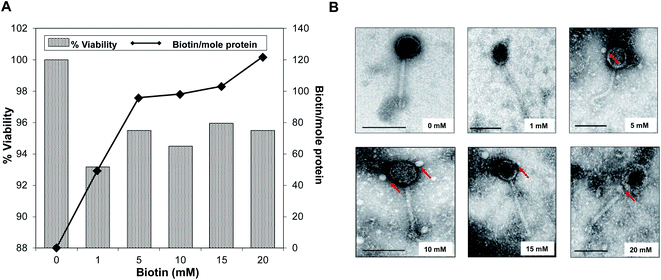
The effects of biotinylation on the bacteriophage viability at 1010 PFU mL−1 titer level is shown in Fig. 2A. The occurrence of biotinylation of bacteriophage head was at its lowest level (49.37 biotin per mole protein) when 1 mM biotin was introduced and was doubled when 5–10 mM range was used (95.71–98.15 biotin per mole protein). The highest concentration of incorporated biotin was 121.92 biotin per mole protein with 20 mM biotin. Viability and infectivity remained high even after chemical biotinylation (94.47%). Fig. 2B shows morphological observation after biotinylation by tracking bound streptavidin-coated Qdots to bacteriophage head. Within 5–15 mM biotin concentrations range, no major morphological changes were observed while the thickening of head and denser capsid at 20 mM biotin was obvious. Data showed that the optimum biotin concentration range was within 10–15 mM range; thus 10 mM was selected was in the subsequent procedures.3.2. Characterization and behavioral analysis of unmodified SPCEs
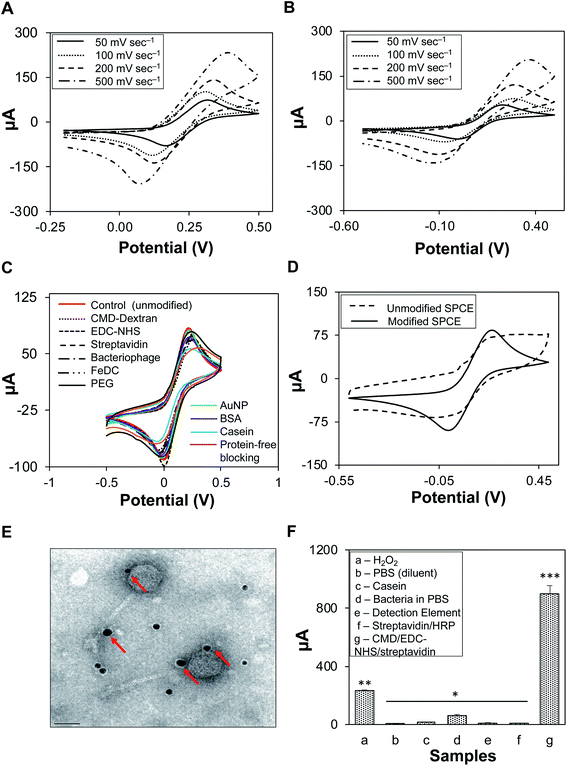
Cyclic voltammetry of K3[Fe(CN6)] was conducted in two supporting electrolytes, 0.1 M H2SO4 (Fig. 3A) and 1× PBS (Fig. 3B), at varying scan rates. The peak potential separation values (ΔEp) increased with scan rate; for 0.1 M H2SO4, ΔEp was 243.2 mV at 200 mV s−1 and 324.3 mV at 500 mV s−1. A similar increasing trend was observed for 1× PBS; ΔEp was 170.6 mV at 200 mV s−1 and 216.2 mV at 500 mV s−1 (Fig. S2, Table S3†). The supporting electrolyte did not participate in any electrode reactions; however, it increased the conductivity of the solution.38 Salt as a supporting electrolyte ensures high ionic strength (0.1 M), thus maintaining a homogenous and near-zero electric field that shields the solution from redox reactions of the target analytes and reduces electrode resistance to a negligible level.39,40 Similarly, sulfuric acid reduces the electric field in the solution as well as the transport of cupric ions.38 1× PBS generated lower ΔEp than H2SO4; therefore, it was used in the subsequent characterization of SPCEs at a scan rate of 100 mV s−1, as low ΔEp values indicated an excellent quality of the electrode surfaces as well as optimal parameter settings (i.e., scan rate). It was also found that 50 mV s−1 scan rate generated the lowest current, irrespective of the supporting electrolyte. However, at 100 mV s−1, the current was consistent and stable during cyclic voltammetry and amperometric tests.3.3. Biofunctionalization of SPCEs
In Fig. 3C unmodified SPCEs showed higher cathodic peaks (Ecp) than chemically modified SPCEs. The blocking features of the chemicals may have affected the electron transfer kinetics of the redox probe. Casein, known as an excellent blocking reagent, had the lowest Ecp, and may have effectively blocked the SPCE surfaces, as observed from the voltammogram produced from SPCEs generated using casein (light blue curve) in comparison with those of SPCEs generated using other blocking reagents. In Fig. 3D, it can be observed that voltammograms generated from unmodified and modified-biofunctionalized SPCEs are clearly distinct with ΔEp values of 201.02 mV and 393.60 mV, respectively. This difference (ΔEp) indicates a successful modification of SPCEs by coating and layering with various components onto the surface of the working electrode while retaining its stability even after several incubating and washing steps. Additional data on the modification of SPCEs are presented in Table S4.†Modified SPCEs generated lower currents than the unmodified counterpart, primarily due to the blocking property of the working electrode surface. A properly ordered self-assembled monolayer of small molecules allows electron transfer from the solution to the electrode. The lack of a peak in the voltammograms of monolayer-modified SPCEs indicated that redox reactions did not occur. Reductions in the voltammetric response were observed in all modification steps, i.e., after the addition of individual reagents and when the actual linkages were assembled. CMD was added onto the SPCE surface to introduce carboxyl groups and was activated by the addition of EDC–NHS. This self-assembled monolayer was terminated with an active amine-carrying streptavidin via carboxyl-to-amine crosslinking, and ultimately covalently bonded with the biotinylated bacteriophages. These surface layers hindered the diffusion of redox probe toward the electrode surface, indicating efficient and successful immobilization and modification of the SPCE surface.41 After functionalization, the stability and viability of immobilized biotinylated bacteriophages (STEC O179) were evaluated by the agar diffusion method (Fig. S3†). STEC O179 bacteriophage showed zones of clearing, indicating stable immobilization, biofunctionality, and biocompatibility with the electrodes.42 The bacteriophages that were included in this study belong to families Siphoviridae and Myoviridae. Both families have been reported to be highly resistant to long-term storage, dry conditions, and large temperature fluctuations.43 Chemical modification was chosen over other techniques for SPCE functionalization because of the ease and simplicity of this approach. Active chemical modification of the SPCEs with streptavidin allowed tethering of biotinylated bacteriophages via the strong streptavidin–biotin interaction. The stability and viability of bacteriophages indicated that the biotinylated bacteriophages were strongly linked to the surface of the working electrode via biotin-streptavidin bonds rather than physical adsorption, as most of the bacteriophages would have been washed away during the washing steps. Previous bacteriophage immobilization techniques involved passive,44 charge-directed oriented immobilization,45,46 and chemical immobilization via streptavidin on quantum dots.33 Passive immobilization resulted in poor cell capture efficiency,34 while chemical immobilization involved genetically-modified bacteriophages, which are not always economical, especially for a comprehensive collection of environmental and genetically uncharacterized bacteriophages.
3.4. Development of bacteriophage-based detection elements with signal amplifiers
Detection element solution containing biotinylated bacteriophage/S-HRP/AuNP complex was viewed under the TEM. Gold nanoparticles as signal amplifiers can be seen as dark spots and were bound to the biotinylated STEC O179 bacteriophages (Fig. 3E). Most of the attached AuNPs were observed along with the heads and tails of the bacteriophages, but not directly to the tail fibers, which could have blocked the specific binding of bacteriophages to host cells. Gold nanoparticles (AuNPs) are widely utilized as signal amplifiers because they are biocompatible, have synergistic effects on catalytic activity, promote electron transfer between mediators and electrodes, and have a large electrochemically active surface.35,47,48 Other nanomaterials and nanosheets such as nickel nanoparticles (NP) and graphitic carbon nitrides (gC3N4) have also attracted attention for potential applications due to their surface area and excellent electron transportability.49,50 The infectivity of biotinylated bacteriophages in the detection element solution was not negatively affected by the binding of AuNPs.3.5. Individual reagents as sources of background noise
Sources of noise and indirect signals are presented in Fig. 3F. Reagents were classified into three major groups based on their functionality. The response currents (μA) for individual reagents that could have indirectly interfered with the signal and ultimately contributed to the background noise of the system were measured. The results showed that response currents from reagents and detection elements were mostly negligible, except for that of H2O2 (230.66 ± 6.32 μA). Modified SPCEs (CMD/EDC–NHS/streptavidin) had the strongest response current detected, 896.51 ± 58.24 μA. Based on these findings, the baseline response value was set at 900 μA, which was applied to subsequent amperometric measurements. The dimension of the working electrode strongly influenced the noise and stability of the sensor, such as the noise was directly proportional to the electrode area.51,52 Though the working electrode had a diameter of only 4 mm, the transducer-induced noise highly likely originated from the thermal motion of ions in the electrolyte–electrode interface where electrode pores created a frictional environment.53 Another possibility was power line interference pickup, which contributed to the overall background noise; therefore, it was recommended to insulate all microelectrode connections.533.6. Electrochemical detection of pure STEC strain cultures
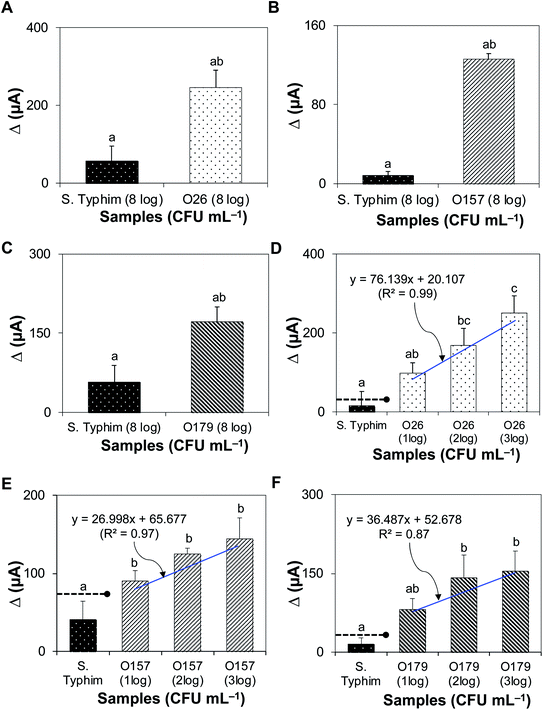
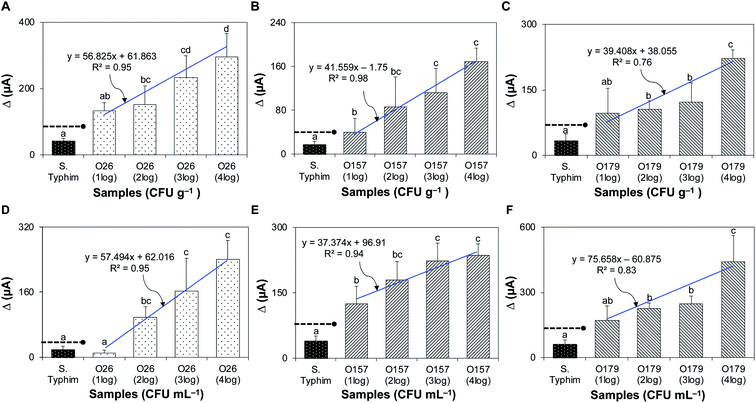
Fig. 4A–C shows that the bacteriophage-based biosensor was highly-specific. Delta (Δ) currents of all target bacteria tested were significantly higher than those of non-target samples. The sensitivity of the assay is presented in Fig. 4D–F; the detection limits for STEC strains O26, O157, and O179 were 102, 10, and 102 CFU mL−1, with R2 values of 0.99, 0.97, and 0.87, respectively.3.7. Detection of STEC strains in artificially inoculated and natural complex matrices
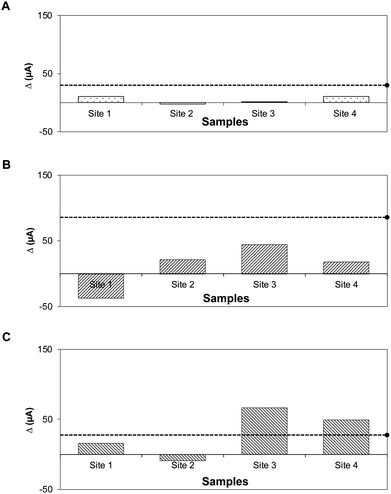
Solid and liquid food samples (fresh ground beef and pasteurized apple juice) were tested without pre-treatment. The device detected STEC strains O26, O157, and O179 in ground beef, with detection limits of 102, 10, and 102 CFU g−1 and R2 values of 0.95, 0.98, and 0.76, respectively (Fig. 5A–C), and in pasteurized apple juice, with detection limits of 102, 10, and 102 CFU mL−1 and R2 values of 0.95, 0.94, and 0.83, respectively (Fig. 5D–F).Fig. 6A–C show the results for the environmental water samples. The Δ current (μA) for each sample was compared to the threshold for positive detection established from pure cultures. Water samples tested negative and had generated Δ current (μA) below the positive threshold for STEC O26 and O157. For STEC O179, Δ currents for samples from sites 3 and 4 were slightly above the positive threshold. However, parallel tests showed negative; the standard plate method using selective media and PCR for stx genes showed that none of the environmental water samples contained the virulence gene-specific for STEC serogroups (Fig. S4 and S5). This difference could be attributed to the generated signals, which were below the reference detection limit in pure culture for STEC O179 (102 CFU mL−1, Δ current = 141 μA).
The results of this study strongly suggested that antibody-free portable detection systems allowed the rapid detection of foodborne pathogens. Viable pathogenic bacterial cells are a primary concern in processed foods, and bacteriophages can facilitate the monitoring and detection of viable pathogens because they can distinguish between viable and dead bacterial cells.54 Bacteriophages possess high specificity and affinity for their hosts.55 The specificity depends on nature and physiological assembly, localization, spatial configuration, and chemical composition of receptors on the bacterial surface. Excellent specificity is advantageous, especially in samples with a high non-target-to-target ratio. In addition, the developed biosensor did not require food or environmental samples to undergo pre-enrichment even for low-level inocula. Direct detection of viable targets and elimination of pre-treatment steps are significant improvements over traditional methods, which will mainly be advantageous for rapid on-site testing of samples. Last, a cocktail of bacteriophages is planned to be used in the future to simultaneously detect multiple targets in a single assay.
4. Conclusions
The detection of STEC strains has been generally based on traditional culture and immunological methods that take several days to complete. The novel, portable, and highly sensitive bacteriophage sandwich detection-based amperometric biosensor that allowed rapid and specific detection of viable STEC addressed the limitations of conventional approaches. In addition, biofunctionalized SPCEs with biotinylated bacteriophages offered tremendous flexibility in terms of performance and cost. Electrochemical analysis using a portable device that was wirelessly connected to a tablet proved the applicability of the sensor in on-site screening. Our sensor requires only a few simple steps prior to sample testing in microvolumes and has a turn-around time of less than 1 h. Further, it showed remarkable detection limits (10–102 CFU mL−1 or g) when applied to complex food samples, without the need for enrichment. The robustness and sensitivity of the biosensor were confirmed in testing natural environmental water samples by comparison with traditional plating and conventional PCR methods. This detection technology has great application potential in routine on-site STEC detection, in both pre- and post-harvest environments.Conflicts of interest
There are no conflicts to declare.Disclosure statement
The authors disclose that accompanying US Patent and Trademark Office (PTO) and Patent Cooperation Treaty (PCT) patent applications were filed. The patent applications USSN 16/115,97 and PCT/US19/47729 both comprised of the described bacteriophage-based biosensor technology.Acknowledgements
The authors would like to thank Dr Yen-Te Liao at Produce Safety and Microbiology Research Unit, USDA ARS WRRC, for the isolation of STEC O179 bacteriophage. This work was supported by the United States Department of Agriculture (USDA NIFA AFRI) food safety grant (award number 2015-69003-23410) and the USDA-ARS CRIS projects 2030-42000-050-00D.References
- E. Vallières, M. Saint-Jean and F. Rallu, J. Clin. Microbiol., 2013, 51, 481–486 CrossRef.
- L. Beutin and P. Fach, Microbiol. Spectrum, 2014, 2, 3 Search PubMed.
- I. A. Quintela, B. G. de los Reyes, C.-S. Lin and V. C. Wu, Nanoscale, 2015, 7, 2417–2426 RSC.
- C. A. Fuller, C. A. Pellino, M. J. Flagler, J. E. Strasser and A. A. Weiss, Infect. Immun., 2011, 79, 1329–1337 CrossRef CAS.
- C. L. Mayer, C. S. Leibowitz, S. Kurosawa and D. J. Stearns-Kurosawa, Toxins, 2012, 4, 1261–1287 CrossRef CAS.
- H. Trachtman, C. Austin, M. Lewinski and R. A. Stahl, Nat. Rev. Nephrol., 2012, 8, 658–669 CrossRef CAS.
- M. Rippa, R. Castagna, M. Pannico, P. Musto, G. Borriello, R. Paradiso, G. Galiero, S. Bolletti Censi, J. Zhou and J. Zyss, ACS Sens., 2017, 2, 947–954 CrossRef CAS.
- F. Mustafa, R. Hassan and S. Andreescu, Sensors, 2017, 17, 2121 CrossRef.
- N. Bhardwaj, S. K. Bhardwaj, M. K. Nayak, J. Mehta, K.-H. Kim and A. Deep, Trends Anal. Chem., 2017, 97, 120–135 CrossRef CAS.
- D. Wang, Q. Chen, H. Huo, S. Bai, G. Cai, W. Lai and J. Lin, Food Control, 2017, 73, 555–561 CrossRef CAS.
- G. Ertürk and R. Lood, Sens. Actuators, B, 2018, 258, 535–543 CrossRef.
- A. K. Trilling, T. Hesselink, A. v. Houwelingen, J. H. G. Cordewener, M. A. Jongsma, S. Schoffelen, J. C. M. v. Hest, H. Zuilhof and J. Beekwilder, Biosens. Bioelectron., 2014, 60, 130–136 CrossRef CAS.
- N. Karoonuthaisiri, R. Charlermroj, U. Uawisetwathana, P. Luxananil, K. Kirtikara and O. Gajanandana, Biosens. Bioelectron., 2009, 24, 1641–1648 CrossRef CAS.
- K. M. Leach, J. M. Stroot and D. V. Lim, Appl. Environ. Microbiol., 2010, 76, 8044–8052 CrossRef CAS.
- Z.-Q. Shen, J.-F. Wang, Z.-G. Qiu, M. Jin, X.-W. Wang, Z.-L. Chen, J.-W. Li and F.-H. Cao, Biosens. Bioelectron., 2011, 26, 3376–3381 CrossRef CAS.
- N. Moll, E. Pascal, D. H. Dinh, J.-P. Pillot, B. Bennetau, D. Rebiere, D. Moynet, Y. Mas, D. Mossalayi and J. Pistré, Biosens. Bioelectron., 2007, 22, 2145–2150 CrossRef CAS.
- A. S. Afonso, B. Pérez-López, R. C. Faria, L. H. C. Mattoso, M. Hernández-Herrero, A. X. Roig-Sagués, M. Maltez-da Costa and A. Merkoçi, Biosens. Bioelectron., 2013, 40, 121–126 CrossRef CAS.
- A. M. Valadez, C. A. Lana, S.-I. Tu, M. T. Morgan and A. K. Bhunia, Sensors, 2009, 9, 5810–5824 CrossRef CAS.
- Y. Zhao, H. Wang, P. Zhang, C. Sun, X. Wang, X. Wang, R. Yang, C. Wang and L. Zhou, Sci. Rep., 2016, 6, 21342 CrossRef CAS.
- B. J. Yakes, E. Papafragkou, S. M. Conrad, J. D. Neill, J. F. Ridpath, W. Burkhardt, M. Kulka and S. L. DeGrasse, Int. J. Food Microbiol., 2013, 162, 152–158 CrossRef.
- A. Ben Aissa, J. J. Jara, R. M. Sebastián, A. Vallribera, S. Campoy and M. I. Pividori, Biosens. Bioelectron., 2017, 88, 265–272 CrossRef CAS.
- A. L. Bole and P. Manesiotis, Adv. Mater., 2016, 28, 5349–5366 CrossRef CAS.
- O. Lazcka, F. J. D. Campo and F. X. Muñoz, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS.
- H. Deng, Y. Xu, Y. Liu, Z. Che, H. Guo, S. Shan, Y. Sun, X. Liu, K. Huang and X. Ma, Anal. Chem., 2012, 84, 1253–1258 CrossRef CAS.
- V. Velusamy, K. Arshak, O. Korostynska, K. Oliwa and C. Adley, Biotechnol. Adv., 2010, 28, 232–254 CrossRef CAS.
- D. C. Vanegas, C. L. Gomes, N. D. Cavallaro, D. Giraldo-Escobar and E. S. McLamore, Compr. Rev. Food Sci. Food Saf., 2017, 16, 1188–1205 CrossRef CAS.
- A. Singh, S. Poshtiban and S. Evoy, Sensors, 2013, 13, 1763–1786 CrossRef CAS.
- W. Sun, L. Brovko and M. Griffiths, J. Ind. Microbiol. Biotechnol., 2000, 25, 273–275 CrossRef CAS.
- E. Fernandes, V. Martins, C. Nóbrega, C. Carvalho, F. Cardoso, S. Cardoso, J. Dias, D. Deng, L. Kluskens and P. Freitas, Biosens. Bioelectron., 2014, 52, 239–246 CrossRef CAS.
- M. J. Loessner, M. Rudolf and S. Scherer, Appl. Environ. Microbiol., 1997, 63, 2961–2965 CrossRef CAS.
- H. M. Byeon, V. J. Vodyanoy, J.-H. Oh, J.-H. Kwon and M.-K. Park, J. Electrochem. Soc., 2015, 162, B230–B235 CrossRef CAS.
- Z. Wang, D. Wang, J. Chen, D. A. Sela and S. R. Nugen, Analyst, 2016, 141, 1009–1016 RSC.
- R. Edgar, M. McKinstry, J. Hwang, A. B. Oppenheim, R. A. Fekete, G. Giulian, C. Merril, K. Nagashima and S. Adhya, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4841–4845 CrossRef CAS.
- L. Gervais, M. Gel, B. Allain, M. Tolba, L. Brovko, M. Zourob, R. Mandeville, M. Griffiths and S. Evoy, Sens. Actuators, B, 2007, 125, 615–621 CrossRef CAS.
- Y.-H. Lin, S.-H. Chen, Y.-C. Chuang, Y.-C. Lu, T. Y. Shen, C. A. Chang and C.-S. Lin, Biosens. Bioelectron., 2008, 23, 1832–1837 CrossRef CAS.
- J. M. Denyes, M. Dunne, S. Steiner, M. Mittelviefhaus, A. Weiss, H. Schmidt, J. Klumpp and M. J. Loessner, Appl. Environ. Microbiol., 2017, 83, e00277-17 CrossRef.
- J. D. Willford, B. Bisha, K. E. Bolenbaugh and L. D. Goodridge, Bacteriophage, 2011, 1, 101–110 CrossRef.
- J. Newman and K. E. Thomas-Alyea, Electrochemical Systems, John Wiley & Sons, 2012 Search PubMed.
- E. J. F. Dickinson, J. G. Limon-Petersen, N. V. Rees and R. G. Compton, J. Phys. Chem. C, 2009, 113, 11157–11171 CrossRef CAS.
- N. m. Elgrishi, K. J. Rountree, B. D. McCarthy, E. S. Rountree, T. T. Eisenhart and J. L. Dempsey, J. Chem. Educ., 2018, 95, 197–206 CrossRef CAS.
- J.-Z. Tsai, C.-J. Chen, K. Settu, Y.-F. Lin, C.-L. Chen and J.-T. Liu, Biosens. Bioelectron., 2016, 77, 1175–1182 CrossRef CAS.
- Z. Hosseinidoust, T. G. M. Van de Ven and N. Tufenkji, Langmuir, 2011, 27, 5472–5480 CrossRef CAS.
- E. Jończyk, M. Kłak, R. Międzybrodzki and A. Górski, Folia Microbiol., 2011, 56, 191–200 CrossRef.
- A. R. Bennett, F. G. C. Davids, S. Vlahodimou, J. G. Banks and R. P. Betts, J. Appl. Microbiol., 1997, 83, 259–265 CrossRef CAS.
- Y. Zhou, A. Marar, P. Kner and R. P. Ramasamy, Anal. Chem., 2017, 89, 5735–5742 Search PubMed.
- R. Cademartiri, H. Anany, I. Gross, R. Bhayani, M. Griffiths and M. A. Brook, Biomaterials, 2010, 31, 1904–1910 CrossRef CAS.
- L. Y. Zhang, R. Yuan, Y. Q. Chai and X. L. Li, Anal. Chim. Acta, 2007, 596, 99–105 CrossRef CAS.
- X. Liu and D. K. Y. Wong, Talanta, 2009, 77, 1437–1443 CrossRef CAS.
- X. Li, P. Zhu, C. Liu and H. Pang, Chem. Commun., 2019, 55, 9160–9163 RSC.
- M. Yuan, X. Guo, N. Li, Q. Li, S. Wang, C.-S. Liu and H. Pang, Sens. Actuators, B, 2019, 297, 126809 CrossRef CAS.
- P. Kuberský, A. Hamáček, S. Nešpůrek, R. Soukup and R. Vik, Sens. Actuators, B, 2013, 187, 546–552 CrossRef.
- J. Yao, X. Liu and K. Gillis, Anal. Methods, 2015, 7, 5760–5766 RSC.
- J. Yao and K. D. Gillis, Analyst, 2012, 137, 2674–2681 RSC.
- M. Zourob and S. Ripp, in Recognition Receptors in Biosensors, Springer, 2010, pp. 415–448 Search PubMed.
- A. Singh, D. Arutyunov, C. M. Szymanski and S. Evoy, Analyst, 2012, 137, 3405–3421 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06223e |
| This journal is © The Royal Society of Chemistry 2020 |