Open Access Article
Open Access ArticleDNA adsorption on nanoscale zeolitic imidazolate framework-8 enabling rational design of a DNA-based nanoprobe for gene detection and regulation in living cells†
Shengmei Wang‡
ab,
Linqi Ouyang‡ab,
Guiming Dengb,
Zhenzhen Denga and
Shengfeng Wang *a
*a
aDepartment of Pharmacy, The Third Xiangya Hospital, Central South University, Changsha, Hunan 410013, China. E-mail: sunfeelwang@126.com
bDepartment of Pharmacy, The First Hospital of Hunan University of Chinese Medicine, Changsha, Hunan 410007, China
First published on 21st August 2020
Abstract
DNA functionalized nanomaterials have attracted tremendous attention for bioanalytical applications. Owing to exceptional fluorescence quenching ability, most DNA-based nanoprobes were designed with turn-on signals for target gene detection, while only a few of them could simultaneously achieve gene detection and regulation in one system. In this study, we explored the use of nanoscale zeolitic imidazolate framework-8 (ZIF-8) as a building block to construct a DNA-based nanoprobe. We found ZIF-8 could stably adsorb DNA to resist the dissociation by various biological ligands, enabling potential biological applications. However, ZIF-8 was not a nano-quencher to turn off the fluorophore labeling on the adsorbed DNA. We therefore designed a DNAzyme embedded molecular beacon (DMB) to functionalize ZIF-8. After endocytosis, ZIF-8 was disintegrated to release DMB for target mRNA detection, and the co-released Zn2+ acted as an effective cofactor to activate the embedded DNAzyme for mRNA regulation. This study provides a versatile nano-platform to realize multiple functions inside cells by using functional nucleic acids, which holds great promise for theranostic applications.
Introduction
To improve the overall survival of patients, remarkable efforts have been made in the timely diagnosis and effective treatment of cancer at the early stage.1,2 To this end, it is important to search for valuable biomarkers that can specifically signify cancer at the cellular level,3,4 and various oncogenes, such as survivin,5 Bcl-2,6 have been identified for this purpose. These tumor-related genes play critical roles in tumor progression and metastasis, and thus selective suppression of their expression could effectively inhibit tumor with little side effects.7 Therefore, a ‘‘Trojan horse’’ approach that simultaneously detects and regulates tumor-related genes would be desirable for personalized cancer therapy,8 which is favored over conventional tissue biopsy and anatomical measurement.9,10Up to now, many theranostic systems in cancer therapy have been attempted.11 DNAzymes (DZs) are single-stranded catalytic DNA molecules which can catalyze a broad spectrum of reactions, including RNA/DNA cleavage, ligation and other reactions.12 Due to the cofactor-dependent and sequence-specific properties, RNA-cleaving DZs have been extensively used as diagnostic biosensors,13–15 and also used as therapeutic agents for gene regulation.16,17 However, one barrier for their cellular application is the difficulty to penetrate the lipophilic and negative cell membrane because of the negative charge and hydrophilic nature. Over the past decades, researchers have investigated to conjugate nanomaterial with DZs to realize intracellular delivery.18 For example, fluorophore (FAM) – or chlorin e6 (Ce6) – labeled DZs were adsorbed on graphene oxide (GO) or manganese dioxide (MnO2) to achieve fluorescence quenching, and target mRNA induced DZs desorption for fluorescent sensing as well as cleavage-based gene regulation.19,20 Despite the simple and feasible construction, these single-dye-labeled sensors were prone to false positive/high background signals caused by biological ligands, resulting in compromise sensor performance.21 To solve this problem, attempts were made by co-adsorbing internal reference probes,22 or by covalently conjugating probes,23 but the advantage of simple physisorption loses.
To simultaneously realize mRNA detection and regulation in living cells, we previously constructed a DNAzyme-embedded molecular beacon (DMB) by encoding the catalytic core sequence of DZ into the recognition loop of molecular beacon (MB), and the intracellular delivery was achieved by further tethering a tumor binding aptamer.24 Although this system could improve the sensitivity and selectivity for target mRNA detection, the regulation efficiency was rather poor, due to low catalytic activity of the DZ inside cells. As metal-dependent enzymes, the RNA-cleaving DZs (i.e., 8–17 and 10–23) require specific metal ions as cofactor for activation, while intracellular metals (mainly Mg2+ and Ca2+) are not sufficient enough to activate DZs.12,25 Therefore, a smart nanocarrier is highly desired to provide sufficient cofactors for in situ activation of DZ.
Zeolitic imidazolate framework-8 (ZIF-8) is a type of metal–organic frameworks (MOFs) with high porosity, low cytotoxity, and intrinsic pH-induced biodegradability, which builds via coordination between Zn2+ ions and 2-methylimidazoles (2-MIMs).26,27 We anticipate that ZIF-8 is a suitable nanoplatform for improving the performance of DMB. First, ZIF-8 has been used as a general carrier to load various functional nucleic acids, such as DZ,28 CpG,29 plasmid,30 and DZ-substrate duplex,31 although the adsorption mechanism is still elusive. Meanwhile, ZIF-8 can decompose in response to intracellular endo/lysosome microenvironment to release DNA payloads.29 Importantly, the co-released Zn2+ ions can be used as efficient cofactors for DZ activation as the gene silencing tool.28
In this work, we started by exploring the DNA adsorption on nanoscale ZIF-8 for fundamental understanding, followed by rational design of the DNA-based nanoprobe using ZIF-8 as carrier (Scheme 1). Interestingly, while ZIF-8 could stably adsorb DNA under biological conditions, it cannot quench the labeling fluorophore. We therefore constructed the nanoprobe using DMB as sensor fragment and ZIF-8 as carriers and metal cofactor reservoir. With the aid of ZIF-8, DMB could be delivered into cancer cells effectively, recognize target for turn-on signal, and subsequently cleave the target mRNA for gene regulation.
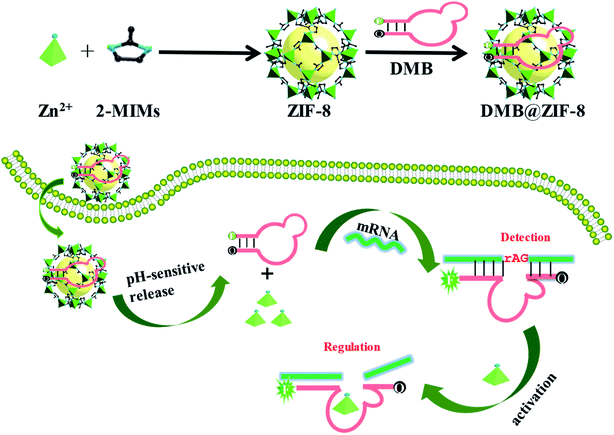 | ||
| Scheme 1 Schematic illustration of a DNAzyme-embedded molecular beacon (DMB) delivered by ZIF-8 for simultaneous detection and regulation of tumor-related genes in living cells. | ||
Experimental section
Materials
All DNA samples (see Table S1† for sequences) with HPLC purification were obtained from Sangon Biotech Co., Ltd (Shanghai, China). Zn(NO3)2·6H2O, 2-methylimidazole (2-MIMs), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), methanol (HPLC grade) and poly(ethylene glycol) (PEG, MW 20k) were bought from Sigma-Aldrich Trading Co., Ltd (Shanghai, China). A RevertAid First Strand cDNA Synthesis Kit and a Lipofectamine 2000 transfection reagent were purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). A real-time PCR kit (SYBR Premix Ex Taq™) was from Takara Biotechnology Co., Ltd (Dalian, China). Survivin rabbit monoclonal antibody was purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA). A bicinchoninic acid (BCA) protein assay kit, horseradish peroxidase (HRP) conjugated goat anti-rabbit IgG (H + L), and radioimmunoprecipitation assay (RIPA) buffer were from Auragene Bioscience Corporation Inc. (Changsha, China). BSA (bovine serum albumin) was purchased from Aladdin Industrial Co., Ltd. (Shanghai, China). Fetal bovine serum (FBS), RPMI-1640 medium, Dulbecco's modified Eagle's medium (DMEM) and 0.25% (w/v) trypsin solution were provided by Gibco BRL (Grand Island, NY, USA). Penicillin-streptomycin solution, 4′,6-diamidino-2-phenylindole (DAPI), and TRIzol™ Reagent were obtained from Solarbio Biotech, Co., Ltd. (Beijing, China).Cell lines and culture conditions
Human A549 lung cancer cells and human embryonic kidney 293 (HEK293) cells were obtained from Xiangya Central Experiment Laboratory (Hunan, China). A549 cancer cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin solution. HEK293 normal cells were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin–streptomycin solution. Both cell lines were cultured in the incubators under a humidified atmosphere containing 5% CO2 at 37 °C. All experiments were performed on cells in the logarithmic phase of growth.Preparation of ZIF-8 nanoparticles
ZIF-8 nanoparticles (NPs) were synthesized in methanol according to a published procedure with some modification.31 Zn(NO3)2·6H2O (150 mg, 0.5 mmol) was dissolved in 7 mL of methanol, and 2-MIMs (330 mg, 4 mmol) was also dissolved in 7 mL of methanol with thorough dissolution. Under magnetic stirring, the two solutions were mixed at room temperature. After 5 min, the solution became milky, indicating the formation of ZIF-8 NPs. Then, solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, and the precipitants were washed three times with 10 mL of methanol. Finally, the obtained ZIF-8 NPs were re-dispersed in 7 mL of 2% PEG20k solution for the subsequent experiments and the concentration of the NPs was about 2.0 mg mL−1.
000 rpm for 10 min, and the precipitants were washed three times with 10 mL of methanol. Finally, the obtained ZIF-8 NPs were re-dispersed in 7 mL of 2% PEG20k solution for the subsequent experiments and the concentration of the NPs was about 2.0 mg mL−1.
Characterization of ZIF-8 NPs
Particle size and zeta potential of ZIF-8 NPs were determined at 25 °C by a Zetasizer Nano-ZS (Malvern Instruments Ltd., UK). The morphology of the NPs was observed by a CM 10 transmission electron microscope (TEM, Philips, USA). With the same preparation of ZIF-8 NPs, the synthesized NPs in methanol were dried at room temperature under vacuum overnight to obtain ZIF-8 powders for the characterization of the chemical bond change. The structure of the ZIF-8 powders was characterized by a Nicolet 6700 Fourier transform infrared spectrometer (FT-IR, Thermo Fisher Scientific, USA) and an inVia Confocal Raman spectrometer (Renishaw, UK).In vitro Zn2+ release
In vitro Zn2+ release was evaluated by the membrane-free dissolution method. Typically, ZIF-8 NPs (1 mg) were dispersed in 1 mL of 50 mM HEPES buffer (pH 7.4) or 50 mM HEPES buffer (pH 5.5) and the mixtures were shaken mechanically at 37 °C. At predetermined intervals, samples were withdrawn and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The concentration of Zn2+ in the supernatant solution was determined after dilution by the atomic absorption spectroscopy (AAS) assay.32 HNO3 solution (5%, 1 mL) was used to dissolve ZIF-8 NPs completely to assay the 100% releasing efficiency.
000 rpm for 10 min. The concentration of Zn2+ in the supernatant solution was determined after dilution by the atomic absorption spectroscopy (AAS) assay.32 HNO3 solution (5%, 1 mL) was used to dissolve ZIF-8 NPs completely to assay the 100% releasing efficiency.
DNA adsorption studies
The FAM-labeled homo-DNAs (A15, T15, C15 or G15, 100 nM) were incubated with ZIF-8 (100 μg mL−1) in HEPES buffer (10 mM, pH 7.4) for 30 min, and the mixtures were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. Fluorescence images of the mixtures were taken with 470 nm LED excitation in a dark room before and after centrifugation. As a control, free FAM fluorophore instead of DNA was used to perform the same experiments.
000 rpm for 10 min. Fluorescence images of the mixtures were taken with 470 nm LED excitation in a dark room before and after centrifugation. As a control, free FAM fluorophore instead of DNA was used to perform the same experiments.
DNA loading capacity
The fluorescence of FAM-labeled homo-DNAs in HEPES buffer (10 mM, pH 7.4) without adding ZIF-8 was measured using an Infinite M200 fluorescence microplate reader (TECAN, Switzerland) with a flat-bottomed black 96-well plate (excitation at 485 nm, emission at 524 nm). To assay the effect of ZIF-8 concentration on adsorption, the loading capacity of DNA was investigated by mixing various concentrations of ZIF-8 (0, 25, 50, 75, or 100 μg mL−1) with FAM-labeled homo-DNAs (100 nM) in HEPES buffer (10 mM, pH 7.4) to a total volume of 100 μL for each sample. To study the effect of DNA concentration on adsorption, FAM-labeled homo-DNAs of two concentrations (500 nM or 1000 nM) were mixed with ZIF-8 (100 μg mL−1) in HEPES buffer (10 mM, pH 7.4). Then the mixtures were incubated for 30 min and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. After centrifugation, the fluorescence intensity of the supernatant solution was detected and the adsorbed amount of DNA was calculated based on the decreased fluorescence.
000 rpm for 10 min. After centrifugation, the fluorescence intensity of the supernatant solution was detected and the adsorbed amount of DNA was calculated based on the decreased fluorescence.
DNA desorption studies
To study the displacement of adsorbed DNA, DNA@ZIF-8 nanoprobe was prepared by incubating FAM-labeled homo-DNAs (25 nM) with ZIF-8 (100 μg mL−1) in HEPES buffer (10 mM, pH 7.4) for 30 min to achieve the sufficient adsorption of DNA. Typically, varying concentrations of competing agents (NaCl, phosphate, or adenosine) or 100 nM of homo-DNAs were added to the prepared A15-FAM@ZIF-8 solution for 1 h. After centrifugation, the fluorescence intensity of the supernatant solution was measured to calculate the percentage of the desorbed DNA. Similarly, the competing agents (150 mM NaCl, 5 mM phosphate, 5 mM nucleosides, 100 nM homo-DNAs, 0.5% FBS or 100 μg mL−1 BSA) were added to the prepared T15-FAM@ZIF-8 or C15-FAM@ZIF-8 solution for 1 h in the same way.The sensor performance of DMB
The detection assay was performed in HEPES buffer (10 mM, pH 7.4) containing 10 mM NaCl and 100 mM KCl. DMB sensor (50 nM) was incubated with a series of concentrations of target DNA (0, 5, 10, 20, 50, 100 nM) for 30 min at 37 °C. The fluorescence peak intensity was measured at 524 nm for quantification by excitation at 485 nm. Target DNA, single-base and three-base mismatch DNA, A15, T15, and C15 (50 nM) were used to test the specificity. To evaluate the applicability of DMB sensor in complex physiological matrix, the detection assay was also performed in 0.5% FBS or 100 μg mL−1 BSA.The RNA-cleavage activity of DMB
To investigate in vitro cleavage activity, DMB was hybridized with the cleavable substrate in 10 mM HEPES buffer (pH 7.4, containing 100 mM KCl), and 0.2 mM ZnCl2 was added to initiate the cleavage reaction. At predetermined time points (0, 1, 2, 4, 8 h), an equal amount of the reaction mixture was withdrawn and the reaction was quenched by mixing with 8 M urea. Then the samples of cleaved substrates were analyzed using denaturing polyacrylamide gel electrophoresis (PAGE, 15%) at 220 V for 50 min. The gel images were recorded and the percentage of cleavage was analyzed by a ChemiDoc XRS+ imaging system (Bio-Rad, USA). To test the activation of DMB by ZIF-8, ZIF-8 was pretreated with acidic buffer (pH 5.5) to mimic the acidic endo/lysosome conditions, and the supernatant was collected for substrate cleavage activity test after 1 h incubation.Adsorption of DMB on ZIF-8
The loading capacity of DMB on ZIF-8 was studied by incubating various concentrations of ZIF-8 NPs (0, 25, 50, 75, or 100 μg mL−1) with DMB-FAM (100 nM) in HEPES buffer (10 mM, pH 7.4) for 30 min. The adsorption isotherm was carried out by mixing various concentrations of DMB-FAM with ZIF-8 NPs (100 μg mL−1) in HEPES buffer (10 mM, pH 7.4) for 30 min. The mixtures were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, and the adsorbed amount of DMB on ZIF-8 was calculated according to the decreased fluorescence of the supernatant solution.
000 rpm for 10 min, and the adsorbed amount of DMB on ZIF-8 was calculated according to the decreased fluorescence of the supernatant solution.
In intro cytotoxicity study of ZIF-8
The in intro cytotoxicity of ZIF-8 against A549 cells was evaluated by MTT assay. A549 cells were seeded in the 96-well plate with a cell density of 1 × 104 per well for 24 h. After washing with PBS, the cells were treated with ZIF-8 NPs with different concentrations of 0, 5, 10, 15, 20 μg mL−1 (diluted with RPMI-1640 complete medium containing 10% FBS) followed by incubation for 24 h. After treatment, MTT solution (0.5 mg mL−1in PBS) was added into the 96-well plate (100 μL per well) and incubated at 37 °C for another 4 h. Then, all media were removed and 100 μL of dimethyl sulfoxide (DMSO) was added to each well to dissolve the insoluble purple formazan product formed in living cells. Finally, the 96-well plate was shaken for 10 min and the absorbance at 490 nm was measured by an Infinite M200 microplate reader (TECAN, Switzerland) to calculate the cell viability.Intracellular bioimaging of DMB@ZIF-8
The DMB@ZIF-8 nanoprobe was prepared by incubating DMB sensor (1 μM) with ZIF-8 (200 μg mL−1) in HEPES buffer (10 mM, pH 7.4) for 30 min to achieve sufficient adsorption. A549 cells and HEK293 cells were seeded in the 96-well culture plate with a cell density of 2 × 104 per well for 24 h and then removed the medium and washed with PBS. DMB@ZIF-8 (20 μg mL−1) was added to FBS-free medium and incubated with the cells for 2 h at 37 °C. Afterwards, the cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min. The cells were stained with DAPI solution (1 μg mL−1) for 5 min and washed with PBS. Finally, the cells were visualized using an operetta high content imaging system (PerkinElmer, USA) with fluorescence imaging.Intracellular gene regulation of DMB@ZIF-8
To evaluate the gene regulation ability of DMB@ZIF-8, we analyzed the survivin expression levels by RT-PCR and western blot after incubating A549 cells with no treatment, DMB, DMB transfected by Lipo2000 (DMB@Lipo) and DMB@ZIF-8. A549 cells were seeded in the 6-well culture plate at the density of 4 × 105 cells per well for 24 h. The DMB@ZIF-8 nanoprobe was prepared as before. DMB@ZIF-8 (20 μg mL−1), DMB (100 nM) or DMB@Lipo (100 nM) was added to FBS-free RPMI-1640 medium and incubated with the cells for 4 h, then the medium was changed with RPMI-1640 complete medium supplemented with 10% FBS, followed by 44 h incubation. After treatment, the procedures of the RT-PCR and western blot analyses were detailed as follows.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), followed by incubating with HRP conjugated goat anti-rabbit IgG (H + L) as a secondary antibody (1
1000), followed by incubating with HRP conjugated goat anti-rabbit IgG (H + L) as a secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500). An anti-β-actin antibody was used as a control. The immunoblots were visualized with chemiluminescence and analyzed using a ChemiDoc XRS+ imaging system (Bio-Rad, USA).
2500). An anti-β-actin antibody was used as a control. The immunoblots were visualized with chemiluminescence and analyzed using a ChemiDoc XRS+ imaging system (Bio-Rad, USA).Statistical analysis
One-way analysis of variance (ANOVA) in SPSS statistics (Version 18.0) was performed to determine the statistical significance of differences among groups. P < 0.05 was considered statistically significant in all cases.Results and discussion
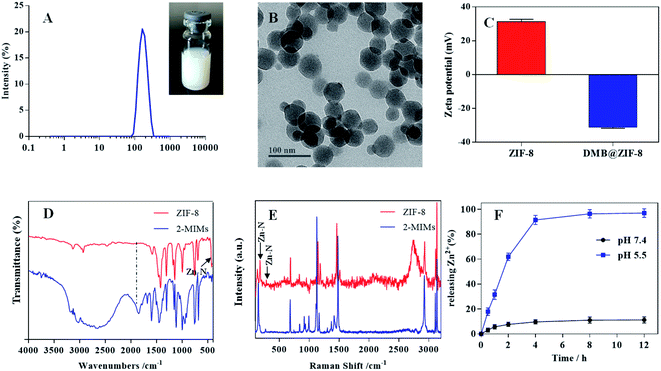
Preparation and characterizations of ZIF-8 NPs
ZIF-8 NPs were prepared via a facile one-pot synthesis method, and the resulting NPs were semi-transparent with light blue opalescence, displaying an average size of ∼160 nm based on dynamic lights scattering (DLS) analysis (Fig. 1A). From TEM, the NPs showed well-defined rhombic polyhedral shape (Fig. 1B). The particle size was estimated to be ∼60 nm by TEM, which was much smaller than that of DLS measurement due to dehydration and shrinkage of NPs before TEM measurement. The surface zeta potential of ZIF-8 was +31.3 mV (Fig. 1C), and such positive charge can be ascribed to the abundance of Zn2+ on the particle surface.The chemical structure of ZIF-8 NPs was then evaluated by FT-IR characterization (Fig. 1D). The peak at 1580 cm−1 assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching mode, while the bands between 1350 cm−1 and 1500 cm−1 was associated with the entire imidazolate ring stretching. For ZIF-8, the absorption peak at 420 cm−1 corresponded to the Zn–N stretching mode, indicating Zn2+ coordination with the nitrogen atom of 2-MIMs to form ZIF-8. Compared with free 2-MIMs, the absorption peak at 1845 cm−1 disappeared and the C–H bending vibration band shifted from 1113 to 1142 cm−1 for ZIF-8, further confirming the formation of ZIF-8.33,34 To complement the FT-IR measurement and probe the microscopic vibrations of chemical bonds, the Raman spectroscopy was also performed (Fig. 1E). ZIF-8 showed a characteristic band at 176 cm−1 and weak band at 281 cm−1, originated from the Zn–N vibrations in the ZnN4 tetrahedrons.35,36 The high-frequency bands at 686 cm−1, 1148 cm−1, and 1460 cm−1 were attributed to imidazolate ring puckering, C–N stretching and C–H bending in the methyl group, respectively.
N stretching mode, while the bands between 1350 cm−1 and 1500 cm−1 was associated with the entire imidazolate ring stretching. For ZIF-8, the absorption peak at 420 cm−1 corresponded to the Zn–N stretching mode, indicating Zn2+ coordination with the nitrogen atom of 2-MIMs to form ZIF-8. Compared with free 2-MIMs, the absorption peak at 1845 cm−1 disappeared and the C–H bending vibration band shifted from 1113 to 1142 cm−1 for ZIF-8, further confirming the formation of ZIF-8.33,34 To complement the FT-IR measurement and probe the microscopic vibrations of chemical bonds, the Raman spectroscopy was also performed (Fig. 1E). ZIF-8 showed a characteristic band at 176 cm−1 and weak band at 281 cm−1, originated from the Zn–N vibrations in the ZnN4 tetrahedrons.35,36 The high-frequency bands at 686 cm−1, 1148 cm−1, and 1460 cm−1 were attributed to imidazolate ring puckering, C–N stretching and C–H bending in the methyl group, respectively.
The pH-responsive release profile of Zn2+ from ZIF-8
In our design, ZIF-8 was employed as Zn2+ reservoir for DZ activation. For this purpose, ZIF-8 would be dissociated inside cells to release Zn2+. To characterize this profile, the release of Zn2+ was studied under various conditions (Fig. 1F).31 We first tested release at pH 7.4 to mimic physiological condition, and minimal Zn2+ was liberated from ZIF-8 (∼10%) after 12 h incubation. Therefore, ZIF-8 was highly stable under biological condition. However, when lowering the pH to 5.5 (which simulates the acidic endo/lysosome condition), a burst and complete Zn2+ release was observed within 4 h, indicating a pH-responsive release profile. The fast release was caused by the effects of protonation and the dissolution of the coordination bonds between Zn2+ and 2-MIMs in acidic microenvironment.37 Therefore, after assimilating by cells and delivering into endo/lysosome, the ZIF-8 could collapse rapidly to release Zn2+, which acts as an effective metal cofactor for DZ activation (vide infra).DNA adsorption by ZIF-8
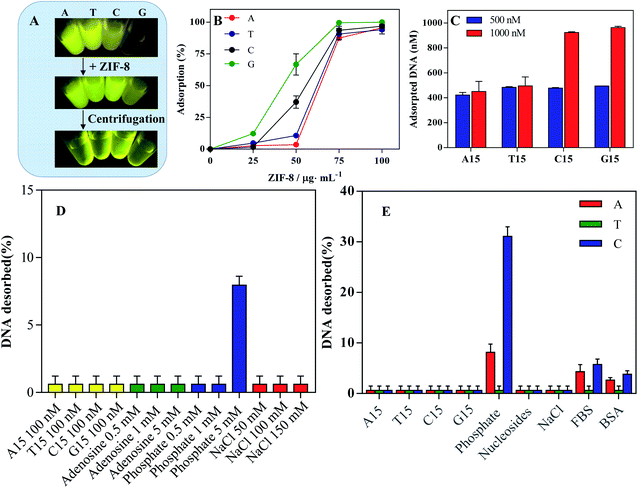
To study the adsorption of DNA on ZIF-8, all DNAs were labeled with a FAM fluorophore. Different homo-DNAs showed distinct fluorescence intensity, due to the photoinduced electron transfer by the nucleobases.38 Interestingly, upon addition of ZIF-8 into DNA solution, no obvious fluorescence quench was observed (Fig. 2A). Most nanomaterials display strong fluorescence quench efficiency due to distance-dependent optical features, which lays the basis for turn-on fluorescence detection by using fluorophore-labeled DNA.39–41 In our case, however, the phenomenon was different. This could be explained by two possibilities: no DNA adsorption, or adsorption without fluorescence quench. To explain the underlining reason, we further performed the centrifugation to precipitate the ZIF-8. Surprisingly, the fluorescence was strongly weakened in solution, while a bright fluorescence was observed in the precipitants (Fig. 2A). Therefore, the FAM-labeled DNA was adsorbed but without fluorescence quench. This conclusion was further confirmed by directly using free FAM fluorophore, which can also be adsorbed on ZIF-8 but not being quenched (Fig. S1†). This may also explain the lack of applications of ZIF-8 for turn-on fluorescent detection.42 Thus, an additional quencher is required to design a fluorescent sensor.43 We also noticed that the fluorescence intensity of C15-FAM and G15-FAM was obviously enhanced after absorption on ZIF-8 (Fig. S2†). This could be explained by the strong electrostatic interaction of FAM fluorophore with ZIF-8 that might disrupt its electron transfer toward DNA nucleobase, thus resulting in the fluorescence recovery.42,44We next studied the loading capacity by fixing FAM-labeled homo-DNAs at 100 nM (Fig. 2B). With increasing ZIF-8 concentration, more DNA was adsorbed, and quantitative DNA adsorption was achieved at 100 μg mL−1 ZIF-8. Then, we fixed ZIF-8 at 100 μg mL−1 and varied the concentration of FAM-labeled DNAs (Fig. 2C). At the concentration of 500 nM, almost all DNA was adsorbed. Further adding DNA to 1000 nM, the amount of DNA adsorption was unchanged for A15 and T15 while doubled for C15 and G15. Therefore, the apparent adsorption capacity was in order of G15 ≈ C15 > T15 ≈ A15. The higher loading capacity of C15 and G15 could be explained by their complex secondary structures occupying smaller footprints on the surface of ZIF-8.45
Adsorption stability probed by competing ligands
Having studied DNA adsorption capability, we continued to explore the binding stability between DNA and ZIF-8 by adding a series of competing agents. First, different homo-DNAs (A15, T15, C15 or G15) were incubated with A15-FAM@ZIF-8 nanoprobe for 1 h, and the supernatant was collected to measure the DNA desorption. Interestingly, none of the homo-DNAs would effectively displace DNA from ZIF-8 surface (Fig. 2D, yellow bar), including the T15 DNA that can hybridize with A15. In this sense, the binding between ZIF-8 and DNA is strong enough to resist the dissociation of complementary DNA, which is different from most other nanomaterials.45,46The nucleobases and phosphate backbones are two main functional building blocks of DNA which are responsible for DNA adsorption.40 We added adenosine into the A15-FAM@ZIF-8 nanoprobe. However, different concentration of adenosine all failed to induce DNA desorbed (Fig. 2D, green bars). In contrast, 5 mM of phosphate began to desorb DNA moderately, suggesting the phosphate backbone plays an important role in DNA adsorption on ZIF-8 (Fig. 2D, blue bars). It should be noted that physiological phosphate level is ∼1 mM, while its level in cytoplasm is ∼30 mM. Therefore, the DNA could be stably attached on ZIF-8 surface under physiological solution while detached in responsive to intracellular high phosphate concentration.
NaCl is often used to probe the electrostatic interaction between DNA and NPs by screen the charge attraction/repulsion. NaCl is usually added to facilitate the DNA adsorption on negatively charged NPs surface,47 whereas the adsorption capacity was decreased by NaCl for some positively charged NPs.48 For ZIF-8, NaCl (150 mM) had little effect on DNA adsorption (Fig. 2D, red bars). Therefore, ZIF-8 could effective adsorb DNA under physiological ionic strength.
To explore the general applicability, we further compared the relative adsorption affinity of different DNA by performing desorption experiments as mentioned above (Fig. 2E). NaCl, nucleosides and homo-DNAs failed to induce DNA desorption. In contrast, phosphate caused much C15, moderate A15 and scarce T15 departing from ZIF-8. Thus, the adsorption affinity under physiological condition followed the order of T15 > A15 > C15. C15 could form a folded configuration with less phosphate backbones binding on ZIF-8 surface,44 which may explain its relatively lower affinity than other DNA homo-polymers. Next, we tested the stability of the DNA@ZIF-8 nanoprobe in a simulative physiological matrix containing FBS or BSA, an only marginal DNA desorption occurred. Taken together, DNA would stably adsorb on ZIF-8 under various physiological conditions, enabling such DNA functionalized material for biological applications.
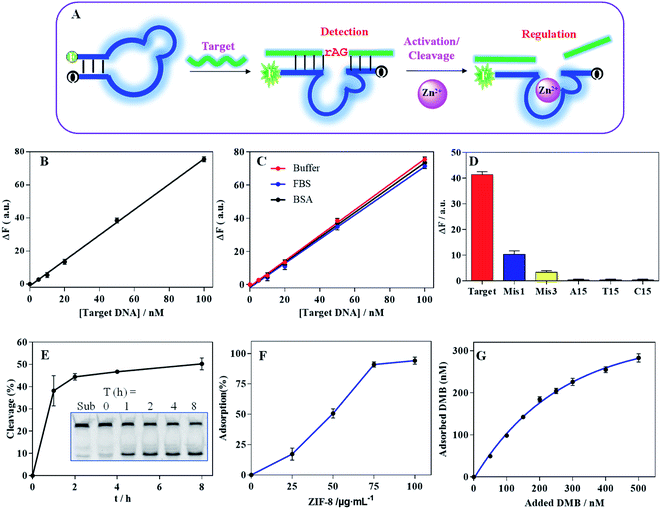
Characterization of DMB@ZIF-8
We next explored the applications of DNA-functionalized ZIF-8. For this purpose, we designed a DMB sensor by embedding the catalytic core sequence of the DZ into MB loop structure (Fig. 3A, and the secondary structure was shown in Fig. S3A†). The hybridization of DMB with its target caused a fluorescence increase for target detection (the secondary structure was shown in Fig. S3B†), and the DZ was activated with the aid of Zn2+ to cleave the binding target.To investigate DMB sensor performance, we monitored the fluorescence recovery upon addition of various concentration of the target DNA, showing a linear dynamic response within 0–100 nM (Fig. 3B). The limit of detection (LOD) was calculated to be 2.1 nM (LOD = 3σ/slope, where σ = standard deviation of background variation of the sensor without target DNA), showing comparable sensitivity to the MBs designed in our previous work and other literatures.24,49 It was suggested that the embedding of the DZ sequence did not influence the MB detection performance. The sensor was also evaluated in various physiological matrix including FBS or BSA, exhibiting similar performance to that in HEPES buffer (Fig. 3C). Therefore, DMB sensor was quite competent for biosensing. Moreover, the specificity of sensor was also investigated (Fig. 3D). The target DNA produced a much stronger signal than the single-base and three-base mismatched DNA (termed Mis1 and Mis3, see Table S1† for detailed DNA sequences) did. None of homo-DNAs produced obvious signal, proving high specificity.
Compared with MB, the advantage of DMB was its property to catalyze the cleavage of mRNA. To prove this, we embedded a single RNA cleavage (rAG) junction in the target DNA sequence with a FAM fluorescence at its 5′-end, and the cleavage of the substrate was visualized using PAGE gel (inset in Fig. 3E). With 0.2 mM Zn2+ ions, ∼40% substrate was cleaved after 1 h (Fig. 3E). Notably, ZIF-8 can be dissociated in response to acidic pH (Fig. S4A†), and the released Zn2+ can activate DZ fragment for substrate cleavage without extra addition of Zn2+ (Fig. S4B†). Therefore, the catalytic activity of the DMB can be effectively activated by Zn2+, which can be provided by the ZIF-8 reservoir.
With above characterization, we next prepared the DMB@ZIF-8 nanoprobe by adsorption of DMB on ZIF-8 surface. With 100 nM DMB, the quantitative DNA adsorption was achieved at 100 μg mL−1 ZIF-8 (Fig. 3F). We then fixed ZIF-8 at 100 μg mL−1, and more DNA was adsorbed with higher concentration of feeding DMB (Fig. 3G). The adsorption behavior was fitted to a Langmuir isotherm,46 indicating DMB wrapped around ZIF-8 with monolayer adsorption, and an equilibrium between adsorption and desorption. With DNA adsorption, the surface charge was revised to negative (Fig. 1C).
Simultaneous gene detection and regulation using DMB@ZIF-8
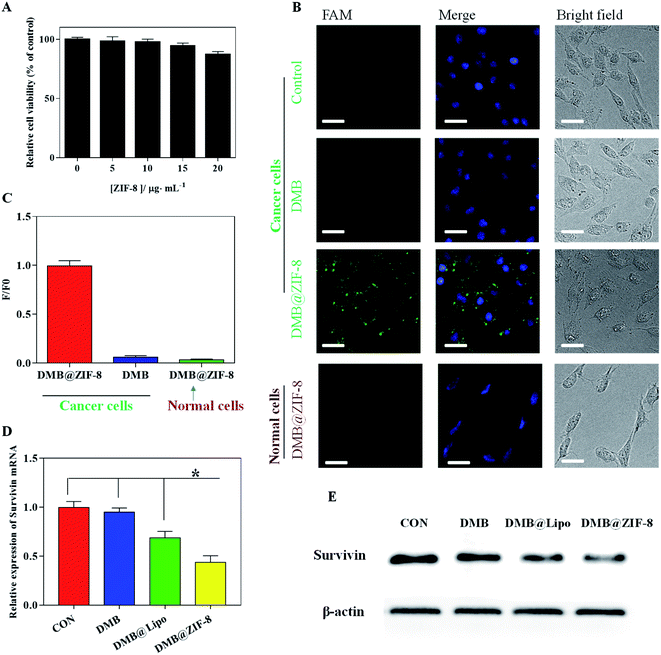
To explore the cellular performance of DMB@ZIF-8, we first evaluated the biocompatibility of ZIF-8 by MTT assay using A549 cells. The cell viability remained ∼90% with 20 μg mL−1 of ZIF-8 for 24 h, suggesting that ZIF-8 had negligible cytotoxicity below this concentration (Fig. 4A).50,51 To have a proof-of-concept demonstration, the survivin mRNA was chosen as target. Survivin is a member of the inhibitor of apoptosis protein (IAP) family, which plays a pivotal role in inhibiting the apoptosis of cancer cells and promoting their survival.52 Survivin is overexpressed in most cancer cells while rarely expressed in normal cells, offering a specific tumor biomarker. In addition, survivin overexpression is strongly associated with poor prognosis, cancer recurrence and drug resistance in chemotherapy,53 which renders it an appropriate theranostic target for cancer therapy.We chose A549 cancer cells since the overexpression of survivin gene was clearly evident, and normal HEK293 cells were used for comparison. The cells were incubated with DMB@ZIF-8 for 2 h, and the cell nuclei was stained blue with DAPI for cell localization (Fig. 4B). Free DMB sensor treated cells did not emit visible fluorescence, due to the minimal internalization of DMB. By comparison, an obvious green fluorescence throughout the cytoplasm was observed in A549 cells after incubation with DMB@ZIF-8, indicating the turn-on signal from DMB sensor after recognition with survivin. Therefore, ZIF-8 could decompose inside cells to release the DMB payloads for target detection. To confirm the fluorescence in A549 cells was not an artifact, we further test DMB@ZIF-8 in HEK293 normal cells, but no detectable fluorescence was produced in this case. This was reasonable that survivin was rarely expressed in HEK293 cells and DMB sensor kept turn-off signal without target.
For quantitative comparison, the relative fluorescence intensity of cells was also presented (Fig. 4C). The fluorescence intensity of DMB@ZIF-8 in A549 cells was obviously stronger than that of DMB sensor, consistent with the fluorescence images. Notably, DMB@ZIF-8 showed fluorescence signal only in cancer cells but not in normal cells, demonstrating that our sensor was useful for differentiating cancer cells from normal cells, which was important for cancer diagnosis.
We next investigated the mRNA regulation ability of DMB@ZIF-8. After treatment, the relative expression of survivin mRNA was determined by RT-PCR and western blot. It is clear that survivin gene was overexpressed in A549 cell without treatment, and free DMB had little effect on the survivin expression. With the aid of transfection reagent (Lipo2000, DMB@Lipo), a moderate downregulation was observed owing to the anti-sense effect of DMB. With DMB@ZIF-8 treatment, by contrast, the survivin mRNA expression was significantly reduced by 56.4% (Fig. 4D), and the survivin protein was also obviously down-regulated (Fig. 4E). Therefore, ZIF-8 not only facilitates the efficient cellular uptake of DMB for target detection, but also supplies Zn2+ ions to the cleavage of the survivin mRNA by DMB, which lays the basis for its potential theranostic applications.
Conclusions
In summary, we have successfully developed a versatile DMB@ZIF-8 nanoprobe which can simultaneously detect and regulate tumor-related genes in living cells. The ZIF-8 can stably adsorb DMB under physiological conditions, facilitate intracellular delivery of DMB, release DMB in response to intracellular stimulus for target mRNA detection, and concomitantly release Zn2+ to activate DZ for gene regulation. It is expected that this versatile nanoplatform could be extended to encapsulate chemotherapeutic drugs and photosensitizers to realize multimodal tumor therapy. Therefore, this work has provided a smart strategy to combine various functional nucleic acids for gene detection and regulation, which holds great potential for disease theranostics.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by Natural Science Foundation of Hunan Province (No. 2019JJ50924, 2020JJ5435).References
- J. Cortes, J. M. Perez-Garcia, A. Llombart-Cussac, G. Curigliano, N. S. El Saghir, F. Cardoso, C. H. Barrios, S. Wagle, J. Roman, N. Harbeck, A. Eniu, P. A. Kaufman, J. Tabernero, L. Garcia-Estevez, P. Schmid and J. Arribas, Ca-Cancer J. Clin., 2020, 70, 105–124 CrossRef PubMed.
- J. B. Li, S. B. Tan, R. Kooger, C. Y. Zhang and Y. Zhang, Chem. Soc. Rev., 2014, 43, 506–517 RSC.
- H. Schwarzenbach, D. S. B. Hoon and K. Pantel, Nat. Rev. Cancer, 2011, 11, 426–437 CrossRef CAS PubMed.
- L. Li, Y. C. Meng, L. Li, S. F. Wang, J. S. Ding and W. H. Zhou, Microchim. Acta, 2019, 186, 824 CrossRef CAS PubMed.
- D. C. Altieri, Nat. Rev. Cancer, 2008, 8, 61–70 CrossRef CAS PubMed.
- S. Cory, D. C. S. Huang and J. M. Adams, Oncogene, 2003, 22, 8590–8607 CrossRef CAS PubMed.
- Y. Jin, Z. H. Li, H. F. Liu, S. Z. Chen, F. Wang, L. Wang, N. Li, K. Ge, X. J. Yang, X. J. Liang and J. C. Zhang, NPG Asia Mater., 2017, 9, e365 CrossRef CAS.
- R. Kumar, W. S. Shin, K. Sunwoo, W. Y. Kim, S. Koo, S. Bhuniya and J. S. Kim, Chem. Soc. Rev., 2015, 44, 6670–6683 RSC.
- L. M. Nie, P. Huang, W. T. Li, X. F. Yan, A. Jin, Z. Wang, Y. X. Tang, S. J. Wang, X. F. Zhang, G. Niu and X. Y. Chen, ACS Nano, 2014, 8, 12141–12150 CrossRef CAS PubMed.
- S. S. Wan, Q. Cheng, X. Zeng and X. Z. Zhang, ACS Nano, 2019, 13, 6561–6571 CrossRef CAS PubMed.
- H. Kim, G. Kwak, K. Kim, H. Y. Yoon and I. C. Kwon, Biomaterials, 2019, 213, 119207 CrossRef CAS PubMed.
- W. H. Zhou, J. S. Ding and J. W. Liu, Theranostics, 2017, 7, 1010–1025 CrossRef CAS PubMed.
- M. Liu, D. R. Chang and Y. F. Li, Acc. Chem. Res., 2017, 50, 2273–2283 CrossRef CAS PubMed.
- S. N. He, L. Qu, Z. F. Shen, Y. Tan, M. Y. Zeng, F. Liu, Y. Y. Jiang and Y. F. Li, Anal. Chem., 2015, 87, 569–577 CrossRef CAS PubMed.
- X. L. Zhu, H. H. Zhang, C. Feng, Z. H. Ye and G. X. Li, RSC Adv., 2014, 4, 2421–2426 RSC.
- H. H. Fan, X. B. Zhang and Y. Lu, Sci. China: Chem., 2017, 60, 591–601 CrossRef CAS.
- A. Mitchell, C. R. Dass, L. Q. Sun and L. M. Khachigian, Nucleic Acids Res., 2004, 32, 3065–3069 CrossRef CAS PubMed.
- J. J. Zhang, Catalysts, 2018, 8, 550 CrossRef.
- S. Kim, S. R. Ryoo, H. K. Na, Y. K. Kim, B. S. Choi, Y. Lee, D. E. Kim and D. H. Min, Chem. Commun., 2013, 49, 8241–8243 RSC.
- H. H. Fan, Z. L. Zhao, G. B. Yan, X. B. Zhang, C. Yang, H. M. Meng, Z. Chen, H. Liu and W. H. Tan, Angew. Chem., Int. Ed., 2015, 54, 4801–4805 CrossRef CAS PubMed.
- Y. J. Yang, J. Huang, X. H. Yang, K. Quan, H. Wang, L. Ying, N. L. Xie, M. Ou and K. M. Wang, J. Am. Chem. Soc., 2015, 137, 8340–8343 CrossRef CAS PubMed.
- X. H. Tan, T. Chen, X. L. Xiong, Y. Mao, G. Z. Zhu, E. Yasun, C. M. Li, Z. Zhu and W. H. Tan, Anal. Chem., 2012, 84, 8622–8627 CrossRef CAS PubMed.
- P. J. J. Huang and J. W. Liu, Anal. Chem., 2012, 84, 4192–4198 CrossRef CAS PubMed.
- S. F. Wang, J. S. Ding and W. H. Zhou, Analyst, 2019, 144, 5098–5107 RSC.
- W. H. Zhou, Y. P. Zhang, J. S. Ding and J. W. Liu, ACS Sens., 2016, 1, 600–606 CrossRef CAS.
- L. H. Gao, Q. Chen, T. T. Gong, J. H. Liu and C. X. Li, Nanoscale, 2019, 11, 21030–21045 RSC.
- I. B. Vasconcelos, T. G. da Silva, G. C. G. Militao, T. A. Soares, N. M. Rodrigues, M. O. Rodrigues, N. B. da Costa, R. O. Freire and S. A. Junior, RSC Adv., 2012, 2, 9437–9442 RSC.
- H. M. Wang, Y. Q. Chen, H. Wang, X. Q. Liu, X. Zhou and F. Wang, Angew. Chem., Int. Ed., 2019, 58, 7380–7384 CrossRef CAS PubMed.
- H. J. Zhang, W. Chen, K. Gong and J. H. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 31519–31525 CrossRef CAS PubMed.
- A. Poddar, J. J. Conesa, K. Liang, S. Dhakal, P. Reineck, G. Bryant, E. Pereiro, R. Ricco, H. Amenitsch, C. Doonan, X. Mulet, C. M. Doherty, P. Falcaro and R. Shukla, Small, 2019, 15, 1902268 CrossRef PubMed.
- J. T. Yi, T. T. Chen, J. Huo and X. Chu, Anal. Chem., 2017, 89, 12351–12359 CrossRef CAS PubMed.
- Z. Y. Song, Y. Wu, Q. Cao, H. J. Wang, X. R. Wang and H. Y. Han, Adv. Funct. Mater., 2018, 28, 1800011 CrossRef.
- E. L. Bustamante, J. L. Fernandez and J. M. Zamaro, J. Colloid Interface Sci., 2014, 424, 37–43 CrossRef CAS PubMed.
- Y. Hu, H. Kazemian, S. Rohani, Y. N. Huang and Y. Song, Chem. Commun., 2011, 47, 12694–12696 RSC.
- G. Kumari, K. Jayaramulu, T. K. Maji and C. Narayana, J. Phys. Chem. A, 2013, 117, 11006–11012 CrossRef CAS PubMed.
- S. L. Chen, X. Li, E. L. Dong, H. Lv, X. B. Yang, R. Liu and B. B. Liu, J. Phys. Chem. C, 2019, 123, 29693–29707 CrossRef CAS.
- H. Q. Zheng, Y. N. Zhang, L. F. Liu, W. Wan, P. Guo, A. M. Nystrom and X. D. Zou, J. Am. Chem. Soc., 2016, 138, 962–968 CrossRef CAS PubMed.
- M. Torimura, S. Kurata, K. Yamada, T. Yokomaku, Y. Kamagata, T. Kanagawa and R. Kurane, Anal. Sci., 2001, 17, 155–160 CrossRef CAS PubMed.
- A. E. Prigodich, D. S. Seferos, M. D. Massich, D. A. Giljohann, B. C. Lane and C. A. Mirkin, ACS Nano, 2009, 3, 2147–2152 CrossRef CAS PubMed.
- B. W. Liu, L. Z. Ma, Z. C. Huang, H. Hu, P. Wu and J. W. Liu, Mater. Horiz., 2018, 5, 65–69 RSC.
- F. L. Gao, J. Wu, Y. Yao, Y. Zhang, X. J. Liao, D. Q. Geng and D. Q. Tang, RSC Adv., 2018, 8, 28161–28171 RSC.
- Y. B. Hao, Z. S. Shao, C. Cheng, X. Y. Xie, J. Zhang, W. J. Song and H. S. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 31755–31762 CrossRef CAS PubMed.
- J. Zheng, R. H. Yang, M. L. Shi, C. C. Wu, X. H. Fang, Y. H. Li, J. H. Li and W. H. Tan, Chem. Soc. Rev., 2015, 44, 3036–3055 RSC.
- A. Lopez, B. W. Liu, Z. C. Huang, F. Zhang and J. W. Liu, Langmuir, 2019, 35, 11932–11939 CrossRef CAS PubMed.
- Y. C. Meng, P. Liu, W. H. Zhou, J. S. Ding and J. W. Liu, ACS Nano, 2018, 12, 9070–9080 CrossRef CAS PubMed.
- B. W. Liu and J. W. Liu, Langmuir, 2015, 31, 371–377 CrossRef CAS PubMed.
- L. Wang, Z. C. Huang, Y. B. Liu, J. Wu and J. W. Liu, Langmuir, 2018, 34, 3094–3101 CrossRef CAS PubMed.
- L. Wang, Z. J. Zhang, B. W. Liu, Y. B. Liu, A. Lopez, J. A. Wu and J. W. Liu, Langmuir, 2018, 34, 14975–14982 CrossRef CAS PubMed.
- P. Zhang, T. Beck and W. H. Tan, Angew. Chem., Int. Ed., 2001, 40, 402–405 CrossRef CAS PubMed.
- J. Zhuang, C. H. Kuo, L. Y. Chou, D. Y. Liu, E. Weerapana and C. K. Tsung, ACS Nano, 2014, 8, 2812–2819 CrossRef CAS PubMed.
- Z. Q. Shi, X. R. Chen, L. Zhang, S. P. Ding, X. Wang, Q. F. Lei and W. J. Fang, Biomater. Sci., 2018, 6, 2582–2590 RSC.
- D. Martinez-Garcia, N. Manero-Ruperez, R. Quesada, L. Korrodi-Gregorio and V. Soto-Cerrato, Med. Res. Rev., 2019, 39, 887–909 CrossRef CAS PubMed.
- N. Singh, S. Krishnakumar, R. K. Kanwar, C. H. A. Cheung and J. R. Kanwar, Drug Discovery Today, 2015, 20, 578–587 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06218a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |