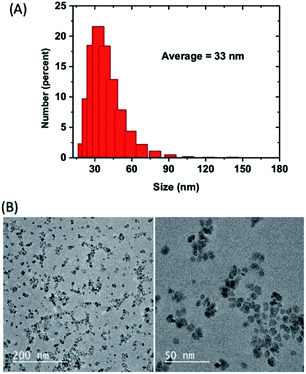
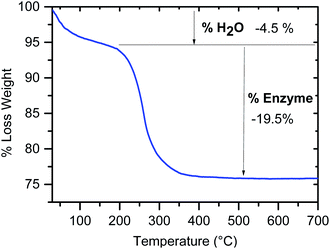
Open Access Article
Open Access ArticleSuperparamagnetic iron oxide nanoparticles (SPIONs) conjugated with lipase Candida antarctica A for biodiesel synthesis†
Luis Fernando Peffi Ferreira *a,
Thayná Mazzi de Oliveiraa,
Sergio Hiroshi Toma
*a,
Thayná Mazzi de Oliveiraa,
Sergio Hiroshi Toma b,
Marcos Makoto Toyama
b,
Marcos Makoto Toyama b,
Koiti Araki
b,
Koiti Araki b and
Luis Humberto Avanzi
b and
Luis Humberto Avanzi c
c
aChemical Engineering Department, FEI University Center, São Bernardo do Campo, SP, 09850-901, Brazil. E-mail: lpeffi@fei.edu.br
bInstitute of Chemistry, University of São Paulo, SP, 05508-000, Brazil
cPhysics Department, FEI University Center, São Bernardo do Campo, SP, 09850-901, Brazil
First published on 20th October 2020
Abstract
Biodiesel is an alternative biodegradable and non-toxic fuel, with a low emission profile and capable of reducing significantly the level of carcinogenic pollutants released into the atmosphere. A newly designed nano-biocatalyst prepared by conjugation of lipase A on superparamagnetic iron oxide nanoparticles (SPIONs) demonstrated high efficiency for production of biodiesel by the reaction of soybean oil with anhydrous methanol. The nanomaterial was characterized by FTIR, TGA and XRD, and its enzymatic activity compared with Lipozyme 435, a commercial gold standard from Novozyme™, which presented average enzymatic activity of 4559 ± 75 only twice as large as that of the SPION-CAL-A catalyst (2283 ± 249 PLU g−1), whereas Lipozyme TLIM showed a much lower activity of 588 ± 16 PLU g−1. These results were confirmed in the transesterification reaction for production of biodiesel where a yield of 11.4% was achieved with Lipozyme 435 and 4.6 ± 0.5% with the nano-biocatalyst. Such an improved performance associated with easy magnetic recovery and reuse make the material potentially interesting for production of biodiesel from used cooking oil, adding value to this abundant resource.
Introduction
Biodiesel is a more sustainable alternative to petroleum diesel since it has the advantage of being produced from strategic renewable agricultural crop sources.1–3 Also, enzymatic conversion of vegetable oils to biodiesel offers a more efficient and lower waste option than conventional processes.4–8 In fact, hydrolytic enzymes such as lipases can be used as biocatalysts, although the industrial application of a biotechnological process has not been found in the literature. The transesterification reaction9,10 that takes place, using lipase as a catalyst in the process, is the same as using chemical catalysts, but with a much higher selectivity than the reaction based on alkalis as catalyst, and under milder conditions than using acids.11Enzymes, such as lipase, can be used to transform the available biomass into greener, more environmentally friendly and sustainable value-added products. However, the difficulties in separating such catalysts from the reaction products, has led to the development of supported heterogeneous catalysts that have as a significant advantage the possibility of recovery and reuse, while preserving the efficiency and the nanostructure. Thus, these materials exhibit a much larger surface to volume ratio, i.e. a larger specific catalytic active area, in addition to enable the development of more efficient and selective materials. Properties similar to those found in molecular systems can be incorporated by conjugating molecular catalysts on the surface of nanoparticles, whereas the superparamagnetic iron oxide nanoparticles (SPIONs) incorporate the possibility of easy recovery using magnets, a very relevant advantage considering the quite high cost of enzymes. However, other supporting materials such as silica, carbon, alumina and polymer nanoparticles, have also been used for preparation of nano catalysts for specific applications conferring greater structural rigidity12 and stability to the biomolecules,13,14 while keeping the enzymatic activity.15 Additional advantages such as improved activity and selectivity, reduced inhibition, as well as ease recovery and reuse of biological material16–19 upon immobilization have been reported.
There are two main distinct methods to immobilize enzymes on nanoparticles surface: (a) by physically trapping (physical method) and (b) by covalent attachment (chemical method). In the first method, enzymes are entrapped in microvolumes/voids, or within semipermeable wall membranes. In this case, there is no bonds formed between the support and enzyme, nevertheless molecules such as substrates and products are able to diffuse in and out to the reaction medium and come into contact with the enzyme's active sites, whereas more complex molecules, such as proteins, are entrapped in the support and are not able to diffuse through the wall.20 In the chemical method, proteins are bond to the surface of the support in three different ways: (a) by ionic, (b) van der Walls interactions, and (c) covalent bonds with the amino acid residues in the enzyme and reactive groups grafted on the support.20
The choice of the type of support and the method of immobilization employed depends on the characteristics of the biological material and the conditions of use of the immobilized catalyst. Therefore, there are no single specific method and no universal supporting material that can be considered in all cases.21 Among the support materials, magnetite (Fe3O4) and maghemite (γ-Fe2O3) nanoparticles22 are considered ideal since are cheap, easy to prepare, chemically stable, and the resultant enzymatic catalyst can be easily separated by applying a magnetic field. On the other hand, among the enzymes supported on SPIONs,23–25 one of the most commonly used ones for trans-esterification reaction are lipases.26–29
The immobilization of enzymes such as lipases on magnetic nanoparticles or other similar supports, where reported to enhance their stability in a wide range of pHs and solvents, at mild temperatures, as compared with the free enzyme. In addition, they showed a reasonable recyclability allowing five up to ten cycles of recovery and reuse.30–33 Interestingly, CAL-A presented much higher enzymatic activity in the transesterification of waste cooking oil34 than PcL (Pseudomonas cepacian) and TLIM (Thermomyces lanuginosus) in a comparative study, demonstrating to be a suitable biocatalyst for that process leading to the formation of higher amounts of product at a lower activation energy. Those findings provided a better understanding of the biocatalytic activity and showed that CAL-A can be promising for real applications.
However, realizing a hybrid nanomaterial preserving or enhancing the enzymatic activity is not a trivial task, especially considering the several possible conformations and anchoring sites that would block the enzyme active site. Accordingly, hereon described is a simple method to prepare a lipase-based nanocatalyst with good activity as compared with Lipozyme 435 for esterification and trans-esterification reaction producing biodiesel from soybean oil and anhydrous methanol as raw materials. For this purpose, a glutaraldehyde free nanocatalyst was developed by using the unusual linker APC (3,4-dihydroxyaldehyde or protocatechuic aldehyde) for lipase immobilization on SPIONs; and the enzymatic activity and biodiesel production performance compared with a gold standard supported commercial enzyme (Lipozyme 435). The magnetic nano-biocatalyst was easily and rapidly recovered from the reaction medium using a NdBFe magnet. Another key point of this work is the preparation of a catalyst by immobilization of a lower cost CAL-A lipase instead of CAL-B lipase onto SPION surface, and a non-ordinary choice for biodiesel production.
Results and discussion
This section is divided in two parts. First, the raw materials were characterized to ensure the quantification of biodiesel production assays and the quality of the oil itself. Secondly, the characterization of the catalyst based on enzyme conjugated magnetic nanoparticles and its application, enzymatic activity and performance in biodiesel synthesis.Characterization of raw materials
The average molar mass of the soybean oil, its acidity (IA) and saponification (IS) index were determined according to the procedure described in “methodology of characterization of raw materials”, and presented as the average of at least two assays.The acidity index represents the fatty acid content while the saponification index is related to the amount of base necessary for saponification of the oil. Both chemical parameters are used by ANVISA35 to determine the quality of oils and fats, which directly affects the quality of the biodiesel obtained, especially in base catalyzed reactions. The measured average acidity index was 0.1554 ± 0.036 mg KOH per g, and the saponification index 198.28 ± 3.06 mg KOH per g of oil sample. These parameters were used to determine the average molar mass of soybean oil of 849.56 ± 12.96 g mol−1 based on eqn (3), a value consistent with those found in the literature ranging around 900 g mol−1.
Characterization of superparamagnetic nanoparticles conjugated with Candida antarctica enzyme
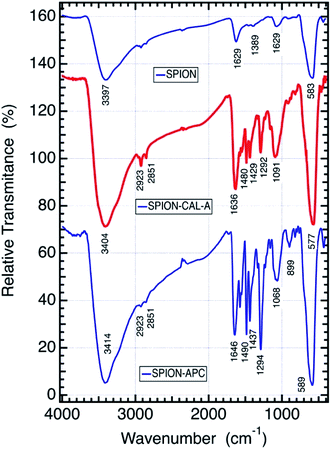
The infrared spectrum of the as prepared SPION, SPION-APC and SPION-CAL-A hybrid enzyme can be compared in Fig. 3. The as prepared SPION exhibits a broad band at 3397 and 1629 cm−1 that can be assigned to symmetric and antisymmetric O–H stretching modes of water molecules, whereas the strong band at 583 cm−1 can be assigned to Fe–O stretching modes of magnetite (Fe3O4), and the low intensity sharp peaks at 2800-2950 cm−1 and at 1389 cm−1 to organic species formed in the thermal decomposition process.
The conjugation of 3,4-dihydroxybenzaldehyde or protocatechuic aldehyde (APC),36.37,38 increased dramatically the number of characteristic peaks in the 1650 to 400 cm−1 range, as expected for the anchoring of that molecule on the nanoparticle surface through the catechol group. The IR spectrum bands in (cm−1) were assigned as follow: 3414 (νOH), 1646 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O aldehyde), 1564 and 1490 (νC
O aldehyde), 1564 and 1490 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic), 1294 (νC–H aldehyde), 1068 (O–H), 899 (aromatic ring) and 589 and 424 (out-of-plane angular deformation of the ring C
C aromatic), 1294 (νC–H aldehyde), 1068 (O–H), 899 (aromatic ring) and 589 and 424 (out-of-plane angular deformation of the ring C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds).
C bonds).
The identification of the typical lipase CAL-A modes was carried out by comparison with SPION-CAL-A and SPION-APC FTIR spectra. The precise assignment of the vibrational bands is not feasible because of the overlap of APC linker and lipase spectrum but significant differences can be clearly observed in the 1200 to 1700 cm−1 range upon covalent binding of the enzyme to the nanomaterial (Fig. 3), characteristic of the Antarctic Candida Lipase Enzyme (CAL),21,22 in addition to the magnetite vibrational mode at 577 cm−1. The appearance of two bands at 2851 cm−1 and 2923 cm−1, assigned to asymmetric and symmetric C–H stretching modes is remarkable, confirming the successful conjugation of CAL-A on the SPION surface. However, it is not clear yet if the enzyme is bound with suitable conformation keeping its high catalytic activity. Nevertheless, the magnetic properties of SPION-CAL-A allowing its easy separation from the reaction medium, thus making it environmentally more sustainable, is given by the crystal structure of the magnetite core.
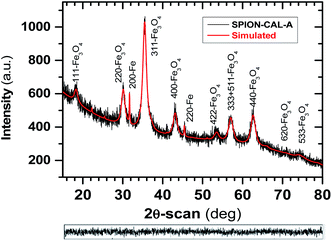
All relevant crystallographic lattice parameters for both crystalline phases, as well as the goodness of fit results obtained from simulation, are shown in Table 1.
| Magnetite (Fe3O4)a | Fe | |
|---|---|---|
| a Magnetite is the main phase with standard lattice parameter at 25 °C of 8.396 Å (JCPDS file 16-0629). | ||
| a (Å) | 8.38820 ± 0.00086 | 5.64474 |
| b (Å) | — | 5.64474 |
| c (Å) | — | 20.8857 |
| % wt | 98.4 | 1.60 ± 1.0 |
| Crystallite size (Å) | 103.9 ± 1.2 | — |
| Crystal lattice symmetry | Cubic | Tetragonal |
| Goodness of fit (gof) | ||
| Rwp = 5.38% | Rexp = 5.16% | σ = 1.05 |
| Enzymes | Activity 1 (PLU g−1) | Activity 2 (PLU g−1) | Average & deviations | Normalized activity (PLU g−1)a |
|---|---|---|---|---|
| a PLU normalized per gram of enzyme supported on SPION, considering the mass of enzyme actually present on catalyst. No catalytic activity was measured for SPION. | ||||
| Lipozyme 435 | 3074.42 | 3003.56 | 3039 ± 50 | 4559 ± 75 |
| Lipozyme TLIM | 192.15 | 199.69 | 196 ± 5 | 588 ± 16 |
| SPION-CAL-A | 631.5 | 541.15 | 586 ± 64 | 2283 ± 249 |
The results listed in Table 2 are comparable since all three assays were carried out in similar experimental conditions. Clearly, Lipozyme TLIM from Thermomyces lanuginosus has a much lower enzymatic activity for esterification reaction than Lipozyme 435 (7.7 times) based on CAL-B and SPION-CAL-A (3.9 times). A typical composition of the biodiesel obtained in the trans-esterification reactions are presented in Fig. S3 and S4 (ESI†).
The lipase used in this commercial product, Candida antarctica lipase B, is not exactly the same as that in SPION-CAL-A conjugate, where CAL-A was used instead. The Lipozyme 435 (CAL-B) presented an enzymatic activity twice as large than SPION-CAL-A (Table 2),43,44 a value similar to those reported in the literature, indicating that APC is a good bridging/anchoring unit and the binding conformation achieved is suitable to preserve the enzymatic activity of variant A of the enzyme itself. For example, the enzymatic activity of CAL-A and CAL-B immobilized on iron oxide nanoparticles described in the literature23,44 were respectively 3499 U g−1 and 6300 U g−1 of enzyme with 84% of immobilization yield, for the hydrolysis of p-NPB (p-nitrophenyl butyrate), i.e. CAL-B showed 1.8 times larger activity than CAL-A indicating that our conjugation method is as good as the commercial one.
The validity of our method for evaluation of the enzymatic activity is corroborated by comparing the activity of the enzyme TLIM, whose value found in the literature is 443 ± 25 PLU g−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 whilst we obtained 588 ± 16 PLU g−1. In addition, the catalytic activity of the CAL-A enzyme supported on SPION was also measured to evaluate its effect. A systematic assay was carried out using 1, 2, 3, 5 and 10% of SPION and molar proportion of soybean oil/methanol of 1
45 whilst we obtained 588 ± 16 PLU g−1. In addition, the catalytic activity of the CAL-A enzyme supported on SPION was also measured to evaluate its effect. A systematic assay was carried out using 1, 2, 3, 5 and 10% of SPION and molar proportion of soybean oil/methanol of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, and 4 to 6 hours of reaction time, at 60 °C, at 300 rpm in a shaker. No esterification reaction product was found demonstrating that SPION is sole acting as support in addition to incorporate magnetic properties to the SPION-CAL-A hybrid, imparting no effect to the catalytic properties of lipase CAL-A enzyme immobilized on the surface. Similar result is expected for SPION-APC as well since APC does not exhibit any catalytic activity.
6, and 4 to 6 hours of reaction time, at 60 °C, at 300 rpm in a shaker. No esterification reaction product was found demonstrating that SPION is sole acting as support in addition to incorporate magnetic properties to the SPION-CAL-A hybrid, imparting no effect to the catalytic properties of lipase CAL-A enzyme immobilized on the surface. Similar result is expected for SPION-APC as well since APC does not exhibit any catalytic activity.
| Enzymes | Glycerin (g) | Initial mass of oil (g) | Yield (%) |
|---|---|---|---|
| Lipozyme 435 | 0.0623 | 5.04 | 11.4 |
| SPION-CAL-A | 0.0252 | 5.05 | 4.60 |
Assays were conducted by adding equal volumes of methanol in three portions of both catalysts. As expected, Lipozyme 435 achieved a better biodiesel yield than SPION-CAL-A, which corroborates the commercial production based on CAL-B lipase enzyme instead of CAL-A lipase. However, the observed difference was not as large as that obtained in the enzymatic activities reported in the literature. In fact, the biodiesel yield obtained in the synthesis was 40%, whereas the catalytic capacity of SPION-CAL-A was about 50% of that obtained with Lipozyme 435, an excellent result considering the performance of CAL-A reported in the literature.46,47
An interesting fact was noted upon washing SPION-CAL-A with pH = 7.0 phosphate buffer solution to check for glycerin adsorption on the nanoparticles surface. The results indicated that 0.9 wt% of glycerin was retained in the material lowering the expected biodiesel yield determined based on that byproduct. Note that a significant fraction produced in the transesterification reaction was not considered in the initial analyses, thus distorting the results by the periodate method. Furthermore, the glycerin molecules may cause steric hindrance slowing down the diffusion of the reagents to the active sites of the enzymes immobilized on the SPION.
Although biodiesel production results are low when compared to those obtained with the commercial catalyst (Lypozime 435), a switch to CAL-B and new assays are warranted due to the better efficiency of this enzyme in trans-esterification reaction and the possibility of cleaner production of biodiesel using magnetically recoverable heterogeneous nano-biocatalysts. Another point to be considered is the simplification of biodiesel and glycerin purification steps due to the easy separation of the catalyst by a magnet and its absence in the final product.
Experimental
Materials
Analytical grade reagents were used to prepare all solutions. Sulfuric acid (98 wt%) was acquired from Vetec®, hydrochloric acid (37 wt%) were purchased from Dinâmica®; isopropyl alcohol (99.5 wt%), sodium periodate (99.8 wt%), monoethylene glycol (99 wt%), sodium hydroxide (>97 wt%) and potassium hydroxide (>97 wt%) were purchased from Neon. Absolute methanol (99.95 wt%), absolute ethanol (99.95 wt%), THF and ammonium hydroxide (99.8 wt%) were acquired from Merck®. lauric acid (99.8 wt%), Candida antarctica Lipase A (CAL-A) recombinant from Aspergillus oryzae, and 4 Å molecular sieves were purchased from Sigma-Aldrich®. Lipozyme 435 and lipozyme TLIM were purchased from Novozyme®, and Soya® soybean oil was acquired at local market.Methods
Characterization of raw material
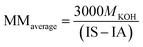 | (1) |
An activity unit (PLU = Propyl Laurate Unit)26,29 is defined as the amount of enzymatic activity capable of generating 1 μmol of isopropyl laurate per minute under defined assay conditions. The enzymatic activity was determined accordingly using the eqn (2):
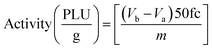 | (2) |
The biodiesel percentage yield was obtained as the ratio of the number of mols of glycerin actually formed in the reaction and the number of mols of glycerin theoretically expected from the reaction stoichiometry, according to eqn (3):
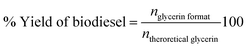 | (3) |
Preparation of SPION-CAL-A catalyst
Characterization of SPION-CAL-A
Infrared spectra (FTIR) were registered in an ALPHA spectrophotometer from Bruker, in the 400 to 4000 cm−1 range, in transmittance mode. The samples were prepared as KBr disks by dispersing 1 mg of material in 100 mg of KBr and pressing. The results presented are the average of at least 16 scans.
Conclusions
The new superparamagnetic iron oxide nanoparticle conjugated to Lipase A (SPION-CAL-A) was synthesized using APC as a linker in the enzyme immobilization process on its surface, a strategy that demonstrated to be simple and very effective for production of such a magnetic nano-biocatalyst. The nanomaterial was characterized primarily by comparing the FTIR pattern of the components, and then by TGA and XRD. These measurements confirmed the successful conjugation of the enzyme on the nanoparticle surface. The effectiveness of this process was verified based on the measurement of the enzymatic activity and performance in the trans-esterification reaction for production of biodiesel from soybean oil.The characterization of soybean oil for reaction with methanol aiming the preparation of biodiesel, was performed by determination of the respective acidity and saponification indexes, which were consistent with an average molar mass of 849.56 ± 12.96 g mol−1.
The tests confirmed that the commercial enzyme Lipozyme 435 has better activity and biodiesel production performance when compared to the SPION-CAL-A enzyme conjugated nanoparticle. This is possibly due to the fact that Lipozyme 435 uses Candida antarctica lipase B, which shows higher enzymatic activity than lipase A used in the preparation of SPION-CAL-A catalyst, that demonstrated a much higher enzymatic activity than expected considering the enzymatic activity reported in the literature for CAL-A relative to CAL-B. Such an enhanced activity associated with the possibility of easy magnetic recovery and reuse make the material potentially interesting for real application as esterification and trans-esterification catalyst, especially for production of biodiesel from used cooking oil, adding value to this abundant resource.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to FAPESP (18/21489-1), Petróleo Brasileiro (Petrobras 0050.0101557.16.9), and CNPq (401581/2016-0) for financial support. This study was financed in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES, Finance Code 001). LFPF and MMT also thank University Center of FEI for supporting the development of this work (IN-1D1R01/17). KA and SHT thanks Conselho Nacional de Desenvolvimento Científico e Tecnológico for Scholarship (CNPq 303137/2016-9 and 305950/2016-9).References
- D. Huang; H. Zhou and L. Lin; Energy Procedia 2012, 16, p. 1874 Search PubMed.
- S. Pinzi, L. Garcia, F. Gimenez, M. D. Castro and M. P. Dorado, Energy Fuels, 2009, 23(5), 2325 CrossRef CAS.
- H. E. Toma, Química de Coordenação, Organometálica e Catálise, Publishing Company: Edgard Blucher, São Paulo, 2013, vol. 4 Search PubMed.
- M. Aarthy, P. Saravana, M. K. Gowthaman, C. Rose and N. R. Kamini, Chem. Eng. Res. Des., 2014, 92, 1591 CrossRef CAS.
- A. Bajaj, et al., J. Mol. Catal. B: Enzym., 2010, 62, 9 CrossRef CAS.
- A. Ghuide, B. Singh, T. Mutanda, K. Permaul and F. Bux, Renew. Sustain. Energy Rev., 2015, 41, 1447 CrossRef.
- G. Knothe, J. Gerpen, J. Krahl and L. P. Ramos, Manual do Biodiesel, Publishing Company: Edgard Blucher, São Paulo, 2006 Search PubMed.
- S. Tang, L. Wang, Y. Zhang, S. Li, S. Tian and B. Wang, Fuel Process. Technol., 2012, 95, 84 CrossRef CAS.
- A. Singh and K. Gaurav, J. Biochem. Technol., 2018, 7(3), 1148 CAS.
- T. Suzuta, M. Toba, Y. Abe and Y. Yoshimura, J. Am. Oil Chem. Soc., 2012, 89(11), 1981 CrossRef CAS.
- H. F. D. E. Castro, A. A. Mendes, C. dos Santos Júlio and C. L. de. Aguiar, Quim. Nova, 2004, 27(1), 146 CrossRef CAS; G. S. Silva, P. C. Oliveira, D. S. Giordani and H. F. de Castro, J. Braz. Chem. Soc., 2011, 22(8), 1407 CrossRef; I. C. R. Costa, S. G. F. Leite, I. C. R. Leal, L. S. M. Miranda and R. O. M. A. de Souza, J. Braz. Chem. Soc., 2011, 22(10), 1993 CrossRef; C. R. Matte, C. Bordinhão, J. K. Poppe, E. V. Benvenutti, T. M. H. Costa, R. C. Rodrigues, P. F. Hertz and M. A. Z. Ayub, J. Braz. Chem. Soc., 2017, 28(8), 1430 Search PubMed.
- J. Tampion and M. D. Tampion, Immobilized cells: principles and applications, Cambridge University Press, 1988, p. 257 Search PubMed.
- P. C. M. Da Rós, G. A. M. Silva, A. A. Mendes, J. C. Santos and H. F. de Castro, Bioresour. Technol., 2010, 101, 5508 CrossRef CAS; A. A. Mendes, R. C. Giordano, R. de L. C. Giordano and H. F. de Castro, J. Mol. Catal. B: Enzym., 2011, 68, 109 CrossRef.
- C. Cantarelli, J. Food Sci., 1989, 3, 3 Search PubMed.
- R. Dalla-Vecchia, M. G. Nascimento and V. Soldi, Quím. Nova, 2004, 27(4), 623 CrossRef CAS.
- M. Vitolo, Biotecnologia, 1988, 11, 2 Search PubMed.
- L. Canilha; W. Carvalho and J. B. Almeida E Silva Biotecnologia Ciência & Desenvolvimento, ano IX, 2006, 36, p. 48 Search PubMed.
- J. C. da Silva and M. da G. Nascimento, J. Braz. Chem. Soc., 2016, 27(12), 2226 Search PubMed.
- M. Mittersteiner, T. M. Machado, P. C. de Jesus, P. B. Brondani, D. R. Scharf and R. Wendhausen Jr, J. Braz. Chem. Soc., 2017, 28(7), 1185 CAS.
- J. C. Quilles Junior, A. L. Ferrarezi, J. P. Borges, R. R. Brito, E. gomes, R. da Silva, J. M. Guisán and M. Boscolo, Bioprocess Biosyst. Eng., 2016, 39, 1933 CrossRef.
- F. A. G. Gonçalves, G. Colen and J. A. Takahashi, Afr. J. Biotechnol., 2013, 12(17), 2270 CrossRef; F. A. G. Gonçalves, G. Colen and J. A. Takahashi, The Scientific World Journal, 2014, 1 CrossRef.
- S. Macala, W. Robertson, C. L. Johnson, Z. B. Day, R. S. Lewis, M. G. White, A. V. Iretskii and P. C. Ford, Catal. Lett., 2008, 122, 205 CrossRef.
- M. R. Mehrasbi, J. Mohammadi, M. Peyda and M. Mohammadi, Renewable Energy, 2017, 101, 593 CrossRef CAS.
- C. G. M. Netto, L. H. Andrade and H. E. Toma, Tetrahedron: Asymmetry, 2009, 20, 2299 CrossRef CAS.
- W. Xie and N. MA, Biomass Bioenergy, 2010, 34, 890 CrossRef CAS.
- A. D. Madalozzo, L. S. Muniz, A. M. Baron, L. Piovan, D. A. Mitchell and N. Krieger, Biocatal. Agric. Biotechnol., 2014, 3(3), 13 CrossRef.
- O. L. Bernardes, J. V. Bevilaqua, M. C. M. Lael, D. M. G. Freire and M. A. P. Langone, Appl. Biochem. Biotechnol., 2007, 136–140, 105 CrossRef.
- V. Gotor-Fernàndez, E. Busto and V. Gotor, Adv. Synth. Catal., 2006, 348, 797 CrossRef.
- T. F. C. Salum, A. M. Baron, E. Zago, V. Turra, J. Baratti, D. A. Mitchell and N. Krieger, Biocatal. Biotransform., 2008, 26(3), 197 CrossRef CAS.
- M. Molina-Gutiérrez, N. L. S. Hakalin, L. Rodríguez-Sánchez, L. Alcaraz, F. A. López, M. J. Martínez and A. Prieto, Molecules, 2019, 24, 1313 CrossRef.
- C. G. Spelmezan, L. C. Bencze, G. Katona, F. D. Irimie, C. Paizs and M. I. Tosa, Molecules, 2020, 25, 350 CrossRef CAS.
- Y. Bi, Z. Wang, Y. Zhang, Bi, Z. Wang, R. Zhang, Y. Diao, Y. Tian and Z. Jin, Processes, 2020, 8, 629 CrossRef CAS.
- R. R. C. Monteiro, D. M. A. Neto, P. B. A. Fechine, A. A. S. Lopes, L. R. B. Gonçalves, J. C. S. dos Santos, M. C. M. de Souza and R. Fernandez-Lafuente, Int. J. Mol. Sci., 2019, 20, 5807 CrossRef CAS.
- A. Gusniah, H. Veny and F. Hamzah, Bull. Chem. React. Eng. Catal., 2020, 15(1), 243 Search PubMed.
- ANVISA. Agência Nacional de Vigilância Sanitária. Resolução no 482, de 23 de setembro de 1999. The United States Pharmacopeia - USP 27; RDC No. 482. Regulamento Técnico para Fixação de Identidade e Qualidade de Óleos e Gorduras Vegetais Search PubMed.
- T. S. Smith, J. R. Lane, M. R. Mucalo and W. Henderson, Transition Met. Chem., 2016, 41(5), 581 CrossRef CAS.
- P. J. Larkin.; Infrared and Raman spectroscopy: principles and spectral interpretation, 2011, Elsevier Inc., USA Search PubMed.
- A. G. Roca, M. P. Morales, K. O'Grady and C. J. Serna, Nanotechnology, 2006, 17, 2783 CrossRef CAS.
- MAUD software can be downloaded from http://maud.radiographema.com/.
- L. Lutterotti, Nucl. Instrum. Methods Phys. Res., Sect. B, 2010, 268, 334 CrossRef CAS.
- L. Lutterotti, D. Chateigner, S. Ferrari and J. Ricote, Thin Solid Films, 2004, 450, 34 CrossRef CAS.
- See http://www.crystallography.net/cod/.
- M. F. C. Andrade, A. L. A. Parussulo, G. C. M. Netto, L. H. Andrade and H. E. Toma, Biofuel Res. J., 2016, 10, 403 CrossRef.
- R. R. C. Monteiro, P. J. M. Lima, B. B. Pinheiro, T. M. freire, L. M. U. Dutra, P. B. A. Fechine, L. R. B. Gonçalves, M. C. M. Souza, J. C. S. dos Santos and R. Fernandez-Lafuente, Int. J. Mol. Sci., 2019, 20, 4018 CrossRef CAS.
- N. Shultz, C. Syldatk, M. Franzreb and T. J. Hobley, J. Biotechnol., 2007, 132, 202 CrossRef.
- C. L. B. Reis, E. Y. Sousa, J. de F. Serpa, R. C. Oliveira and J. C. S. dos Santos, Quím. Nova, 2019, 42, 768 CAS.
- S. J. Pierre, J. C. Thies, A. Dureault, N. R. Cmeron, J. C. M. van Hest, N. Crette, T. Michon and R. Weberskirch, Adv. Mater., 2006, 18, 1822 CrossRef CAS.
- INSTITUTO ADOLFO LUTZ, Óleos e gorduras, in INSTITUTO ADOLFO LUTZ. Métodos físico-químicos para análise de alimentos, 2008, p. 600 Search PubMed.
- M. L. Pisarello, B. O. Dalla Costa, N. S. Veizaga and C. A. Querini, Ind. Eng. Chem. Res., 2010, 49, 8935 CrossRef CAS.
- L. V. Cocks and C. Van Rede, Laboratory handbook for oils and fats analysis, London: Academic Press, 1966 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary data, TEM, DLS, zeta Potential of pure magnetic nanoparticle, and typical biodiesel composition are available. See DOI: 10.1039/d0ra06215d |
| This journal is © The Royal Society of Chemistry 2020 |