Open Access Article
Open Access ArticleIsolation, expression, and biochemical characterization: nitrite reductase from Bacillus cereus LJ01†
Yan-yan Huanga,
Ming-hua Lianga,
Shan Zhaoa,
Si-min Chena,
Jin-song Liu b,
Dong-mei Liu
b,
Dong-mei Liu *a and
Yong-zhi Lu*b
*a and
Yong-zhi Lu*b
aSchool of Food Science and Engineering, South China University of Technology, 381 Wushan Road, Guangzhou, Guangdong 510640, People's Republic of China. E-mail: liudm@scut.edu.cn
bState Key Laboratory of Respiratory Disease, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 190 Kaiyuan Avenue, Science Park, Huangpu District, Guangzhou 510530, People's Republic of China. E-mail: lu_yongzhi@gibh.ac.cn
First published on 14th October 2020
Abstract
Biological remediation of toxic oxygen-containing anions such as nitrate that are common in the environment is of great significance. Therefore, it is necessary to understand the specific role of nitrate and nitrite reductase in the bioremediation process. Bacillus cereus LJ01, which was isolated from traditional Chinese soybean paste, effectively degraded nitrite (such as NaNO2) at 0–15 mmol L−1 in LB medium. Moreover, the nitrite-degrading active substance (ASDN) was isolated and purified from B. cereus LJ01. The nitrite-degrading activity of nitrite reductase (named LJ01-NiR) was 4004.89 U mg−1. The gene encoding the assimilation of nitrite reductase in B. cereus LJ01 was cloned and overexpressed in E. coli. The purified recombinant LJ01-NiR has a wide range of activities under temperature (20–60 °C), pH (6.5–8.0) and metal ions (Fe3+, Fe2+, Cu2+, Mn2+, and Al3+). Kinetic parameters of LJ01-NiR, including the values of Km and Vmax were 1.38 mM and 2.00 μmol g−1 min−1, respectively. The results showed that LJ01-NiR could degrade nitrite with or without an electron donor. In addition, sequence analysis revealed that LJ01-NiR was a ferredoxin-dependent nitrite reductase given the presence of conserved [Fe4–S4] cluster and heme-binding domain. The nitrite ion binds to the LJ01-NiR active site by forming three hydrogen bonds with the residues ASN72, ALA133 and ASN140. Due to its high nitrite-degrading activity, LJ01-NiR could potentially be used for environmental pollution treatment.
1. Introduction
The extensive use of chemical nitrogen fertilizers and nitrogen-based pesticides, as well as the arbitrary discharge of a nitrogen-containing industrial pollutant, have led to excessive nitrate and nitrite effluent discharge into the environment, resulting in extremely severe soil and water pollution. The potential harm that high levels of nitrite pose to the environment has since caught the attention of researchers.1,2 Since nitrite biodegradation is highly efficient and healthy, microorganisms can play significant role in removing pollutants from the environment through bioremediation.Nitrite is a major player in the biological N-cycle. Under anaerobic conditions, nitrite is a key intermediate product in the conversion of nitrate to nitrogen or ammonium in the N-cycle, which can be further utilized by microorganisms.3 Liu et al. reported the degradation of nitrites via the nitrate respiration pathway (NO2− → NO → N2O → N2) in Lactobacillus casei subsp. rhamnosus LCR6013.4 Siroheme was used as electron donor in the assimilation reduction of nitrite in the six electron reduction reaction of sulfur or nitrogen catalyzed by nitrite reductase (NiR, EC 1.7.7.1) from plants cytoplasm of and some microorganisms.5 Thus nitrite can be converted into ammonium salt by bacterial assimilation. Even though the conversion can take place in plants, the yields remain very low.6
The two assimilated nitrite reductases include ferredoxin dependent nitrite reductases (FdNiRs) and NAD(P)H dependent nitrite reductases.7 FdNiRs are found primarily in photosynthetic microorganisms such as algae and cyanobacteria. FdNiRs are globular proteins composed of three domains with two prosthetic groups namely: Fe–S cluster (4Fe–4S) and siroheme.8 FdNiRs is a type of polypeptide chain (60–65 kD) in photosynthetic tissues that can reduce nitrite to ammonia by transferring 6 electrons. Ferredoxin serves as an electron donor and transfers electrons to the siroheme site. Siroheme combines with nitrite ions and reduces nitrite ions to ammonia. The catalytic process is as follows: NiR–NO2− → NiR–NO → NiR–NH2OH → NiR–NH4+. The molecular and structural analysis of NiR has been reported in many denitrifying bacteria, including Lactobacillus sp.,9,10 Bacillus sp.11 and Pseudomonas sp.12 However, there is a shortage in the structural analysis and enzymatic properties of FdNiRs in Gram-positive bacteria. The molecular docking analysis can help explain the structure–activity relationship between an enzyme and the nitrite molecules.
In this study, B. cereus LJ01 was isolated for nitrite degradation from traditional Chinese soybean paste. Moreover, the ASDN in B. cereus LJ01 was purified and identified. This study further describes the expression of LJ01-NiR recombinant protein in E.coil. Homology modeling and molecular docking of LJ01-NiR with nitrite molecule are performed to serve as references for the construction of artificial enzymes in protein engineering research.
2. Materials and methods
2.1 Isolation, identification and nitrite removal capacity of the strain LJ01
The strain LJ01 in this study was extracted from traditional Chinese soybean paste. First, 1 g of traditional soybean paste was suspended in 10 mL of distilled water, and then serially diluted. The final suspension was streaked on Luria-Bertani (LB) medium agar plates containing 9 mmol L−1 nitrite and incubated at 30 °C for 2 days until the formation of bacterial colonies. Several screenings were performed to obtain pure colonies. All bacterial isolates were screened for nitrite degradation on the previously described nitrite medium2 at 30 °C for 2 days. Bacterial growth implied nitrite degradation. LJ01 was identified as one of the main strains isolated from traditional Chinese soybean paste based on its standard morphological and biochemical characteristics.13 For taxonomic identification, the 16S rDNA was amplified from total DNA and cloned into the pMD18-T plasmid, and then sequenced by the Detection Center of Microbiology (Guangdong, China). The Basic Local Alignment Search Tool program (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for sequence alignment of 16S rDNA, and a neighbor-joining tree was constructed using the MEGA7 software.The effect of 0 to 15 mmol L−1 nitrite concentrations on nitrite degradation by LJ01 was investigated in an unsalted LB broth. Cultures were seeded with an inoculum density of 2% (v/v) and incubated at 30 °C for 48 hours. The effect of nitrite concentration on the growth of the strain was observed using an optical microscope (OM) (CX23, Shandong, China) and scanning electron microscopy (SEM) (JEOL 6300F, Tokyo, Japan) at 5.0 kV to establish threshold conditions for further experimentation.
2.2 Purification of ASDN from B. cereus LJ01
According to Liu's method with slight modification, 1.5 mmol L−1 NaNO2 induced LJ01 cells were used to prepare crude ASDN.4 Briefly, the LJ01 cells were completely separated and centrifuged (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C). The supernatant was filtered through a sterilized 0.22 μm membrane to collect ASDN. 40 mL ASDN liquid sample was loaded onto DEAE-Sepharose Fast Flow column (2.6 × 20 cm) pre-equilibrated with the same buffer, and diluted with a linear gradient of 0–0.5 mol L−1 NaCl in 20 mmol L−1 HEPES buffer (pH 7.4) at a flow rate of 1.0 mL min−1, using a fast protein liquid chromatography system (AKTA Purifier Explorer, Pharmacia, Uppsala, Sweden). The fractions with nitrite-degrading activity were pooled, dialyzed against 50 mmol L−1 Tris–HCl buffer (pH 8.0), and loaded onto a Sephadex G-150 column (1.6 × 50 cm), followed by elution with 50 mmol L−1 Tris–HCl buffer (pH 8.0) at a flow rate of 1.0 mL min−1. The separated fractions were collected and probed every 10 min for nitrite-degrading activity. Purity was analyzed by SDS-PAGE.
000g, 4 °C). The supernatant was filtered through a sterilized 0.22 μm membrane to collect ASDN. 40 mL ASDN liquid sample was loaded onto DEAE-Sepharose Fast Flow column (2.6 × 20 cm) pre-equilibrated with the same buffer, and diluted with a linear gradient of 0–0.5 mol L−1 NaCl in 20 mmol L−1 HEPES buffer (pH 7.4) at a flow rate of 1.0 mL min−1, using a fast protein liquid chromatography system (AKTA Purifier Explorer, Pharmacia, Uppsala, Sweden). The fractions with nitrite-degrading activity were pooled, dialyzed against 50 mmol L−1 Tris–HCl buffer (pH 8.0), and loaded onto a Sephadex G-150 column (1.6 × 50 cm), followed by elution with 50 mmol L−1 Tris–HCl buffer (pH 8.0) at a flow rate of 1.0 mL min−1. The separated fractions were collected and probed every 10 min for nitrite-degrading activity. Purity was analyzed by SDS-PAGE.
The purified ASDN band were excised from SDS-PAGE gels. The protein was digested in-gel using trypsin overnight, then the proteolytic peptides were analyzed by a MALDI-TOF/TOF™ 4800 proteomics analyzer from Shenzhen Huada Gene Technology Co., Ltd. (Shenzhen, China).
2.3 In vitro analytical methods for detecting the enzyme activity
Protein concentration was determined by the Bradford method using bovine serum albumin (BSA) as a standard. Gao et al. (2018) described the use of methyl viologen (MV) as an electron donor in determining nitrite reductase activity.14 The reaction was carried out in a 0.5 mL closed vial containing 100 mM potassium phosphate buffer (pH 6.5), 50 mM methyl viologen and 2.5 M sodium nitrite, 1 M sodium chloride and 0.02 mL purified enzyme.14 The reaction was initiated with the addition of 50 μL sodium dithionite (1 M) and the vial gently swirled. After incubation at 37 °C for 10 min, the reaction was terminated by vigorous shaking for 3 min. The nitrite remaining was measured with a spectrophotometer. One unit of activity corresponded to a 1 μmol reduction in nitrite per minute.2.4 Cloning of the cDNA sequence encoding LJ01-NiR and its expression in E. coli
Total RNA was extracted from B. cereus LJ01, and PrimeScript™ first-strand cDNA synthesis kit (Takara, Dalian, China) was used to synthesize cDNA, following the manufacturer's instructions. The full-length coding sequence of the B. cereus LJ01 (LJ01-NiR) nitrite reductase gene was cloned using primers (5′-ATGAGTTATGAAAAAGTATGGGC-3′ and 5′-CTAAGACGCTAT TACTTCTTCTAAC-3′) and submitted to NCBI GenBank (MG839504). The cDNA was used as the template and 5′-CCAGCTAGCATGAGTTATGAAAAAGTATG-3′ and 3′-TGGCTCGAGCTAAGACGCTAT TACTTC-3′ were used as the primers. The LJ01-NiR gene was PCR amplified with the NheI and XhoI sites, without a signal peptide-encoding sequence. The PCR product was connected to the NheI and XhoI sites of the pET-28a (+) vector (Invitrogen, Carlsbad, CA, USA). The resulting plasmid, pET-28a (+)-LJ01-NiR, was then transformed into E. coli DH5α competent cells. The heterogenous expression of recombinant LJ01-NIR in E. coli were purified by Ni2+-NTA affinity chromatography (GE, USA) and Superdex 200 gel filtration chromatography (GE, USA) using a fast protein liquid chromatography system (AKTA Purifier Explorer, Pharmacia, Uppsala, Sweden), referring to Gao's method with slight modification.142.5 Biochemical characterization test
The optimal pH of LJ01-NiR was analyzed using three buffer systems with standard assay conditions including citrate buffer (50 mM, pH 4–6), phosphate buffer (50 mM, pH 6–8), and Tris–HCl buffer (50 mM, pH 8–9). The effect of temperature on nitrite-degrading activity was examined by incubating LJ01-NiR mixture at different temperatures range from 20 °C to 70 °C. Moreover, the effect of metal ions and inhibitory additives was determined by a 10-minute pre-incubation of the enzyme with 10 mM solution containing Fe3+, Fe2+, Cu2+, Zn2+, K+, Mg2+, Ba2+, Mn2+, Al3+, EDTA (ethylenediaminetetraacetic acid) and PMSF (phenylmethanesulfonyl fluoride) for 10 min, followed by the determination of residual activity. To evaluate the impact of the additives, the activity of nitrite reductase with additives was normalized to that without additives. The kinetic parameters of LJ01-NiR were measured in the potassium phosphate buffer (pH 6.5) with different concentrations of sodium nitrite at 35 °C for 10 min. Kinetic parameters (Km and Vmax) were then obtained using nonlinear regression fitting of the Michaelis–Menten equation.2.6 Homology model building of LJ01-NiR
Homology modeling of LJ01-NiR was generated by Discovery Studio using the crystal structure (PDB code: 1ZJ9) of NirA from Mycobacterium tuberculosis (Tb-NirA) as template. The domains of heme and [Fe4–S4] cluster were taken from the template structure.2.7 Enzyme-ligand docking
The AutoDock Vina was used to dock the nitrite molecule (ligand) to the homology model structure of LJ01-NiR. The receptor and ligand molecules were prepared using AutoDock Tools. The docking result analysis and graph preparation were carried out by PyMOL (https://www.pymol.org/).2.8 Statistical analysis
Tests were carried out in triplicate for all experiments. All data were presented as mean ± standard deviation (SD) and analyzed by one-way analysis of variance using SPSS 19.0 (SPSS Inc., Chicago, USA) followed by a least significant difference (LSD) test on a 95% confidence interval.3. Results
3.1 Identification of B. cereus LJ01
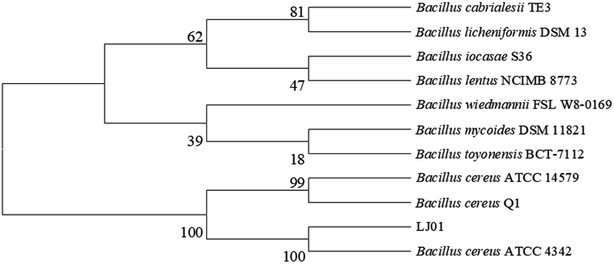
The 16S rDNA cloned from the strain LJ01 was sequenced and submitted to the GenBank with the accession number KM058698. NCBI GenBank BLAST results revealed that the 16S rDNA sequence similarity to Bacillus cereus was up to 99%. The strain LJ01 was initially identified as genus Bacillus. The phylogenetic tree (Fig. 1) indicated a closer relationship between the strain LJ01 and Bacillus cereus ATCC 4342. The physiological and biochemical properties of strain LJ01 are shown in ESI Table S1.† According to the Berger Bacterial Identification Manual, evidence from the phylogenetic tree, morphological characteristics, biochemical characteristics, and 16S rDNA conserved sequence indicated that the strain LJ01 belonged to Bacillus cereus.13 Therefore, the strain LJ01 was named Bacillus cereus LJ01.3.2 Nitrite degradation capacity of B. cereus LJ01
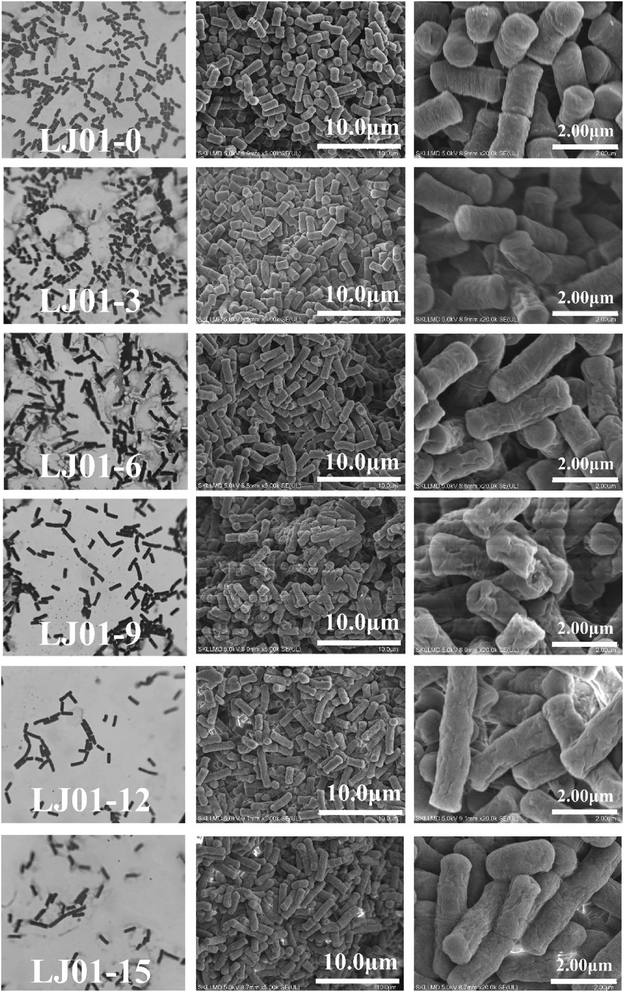
The cell morphologies of B. cereus LJ01 were observed under different culture conditions (0, 3, 6, 9, 12, and 15 mmol L−1 nitrite, 8 h of incubation) by optical microscope (OM) and SEM, as shown in Fig. 2. Compared to the morphology of LJ01 cultured in a nitrite-free medium, the morphology of LJ01 exposed to 6, 9, 12, and 15 mmol L−1 nitrite exhibited rough surfaces with folds based on OM and SEM examination. However, the culture exposed to 3 mmol L−1 nitrite had no significant difference. In addition, with the increase of the concentration of nitrite, the bacteria gradually became longer and some bacteria ruptured to cause cytoplasm leak. Previous studies have found that nitrites can cause serious damage to bacterial DNA. The bacteria resist the damage by evoking SOS response and growing longer.15As shown in Table 1, the nitrite concentrations in the LB broth were measured after 0, 4, 8, 12, 24, 36, and 48 h of incubation to calculate the degradation rate. The degradation rate in the 3 mmol L−1 nitrite group was 100% after 12 hours of incubation. The degradation rate in the 6 mmol L−1 nitrite group was 93.68% which was slightly higher than 9, 12 and 15 mmol L−1 nitrite groups at 12 hours. Subsequently, with the extension of cultivation time and the increase of nitrite concentration, the nitrite degradation rate exhibited a slow downward trend. LJ01 can efficiently degrade 6 mmol L−1 nitrite within 36 hours. The degradation rate of nitrite concentrations in the LJ01 exposed to 9, 12, and 15 mmol L−1 nitrite was significantly lower than in LJ01 exposed to 3 and 6 mmol L−1 nitrite. The maximum nitrite degradation ability exhibited by Lactobacilli from Sauerkraut in the 10-day fermentation broth with an initial content of 2.9 mmol L−1 NaNO2, was 80.0%. Besides, the total number of bacteria in the MRS solid medium was 1.51 × 107 CFU mL−1.16 Therefore, B. cereus LJ01 showed a greater nitrite degradation ability than Lactobacillus.
| a The initial concentrations of sodium nitrite were 3, 6, 9, 12, and 15 mmol L−1 in the reaction system, respectively. The residual nitrites were measured after incubation at 30 °C for different incubation time. Values were expressed as mean ± SD (n = 3). | ||||||
|---|---|---|---|---|---|---|
| Initial concentration of nitrite (mmol L−1) | 3 | 6 | 9 | 12 | 15 | |
| Nitrite degradation rate (%) at different incubation time | 0 h | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 4 h | 77.87 ± 0.57 | 71.60 ± 2.28 | 66.27 ± 0.70 | 65.99 ± 0.08 | 58.00 ± 0.78 | |
| 8 h | 93.83 ± 3.63 | 88.67 ± 0.97 | 74.81 ± 1.13 | 73.31 ± 0.55 | 76.44 ± 4.21 | |
| 12 h | 100.00 ± 1.38 | 93.68 ± 1.06 | 81.57 ± 0.71 | 78.38 ± 0.73 | 78.37 ± 1.71 | |
| 24 h | 100.00 ± 0.16 | 98.45 ± 0.33 | 87.57 ± 2.01 | 82.84 ± 0.58 | 80.19 ± 1.46 | |
| 36 h | 100.00 ± 0.32 | 100.00 ± 1.97 | 92.73 ± 0.70 | 88.07 ± 0.12 | 87.56 ± 0.47 | |
| 48 h | 100.00 ± 0.35 | 100.00 ± 1.85 | 92.21 ± 0.88 | 90.63 ± 0.45 | 88.79 ± 1.80 | |
3.3 Purification and identification of NiR enzyme from B. cereus LJ01
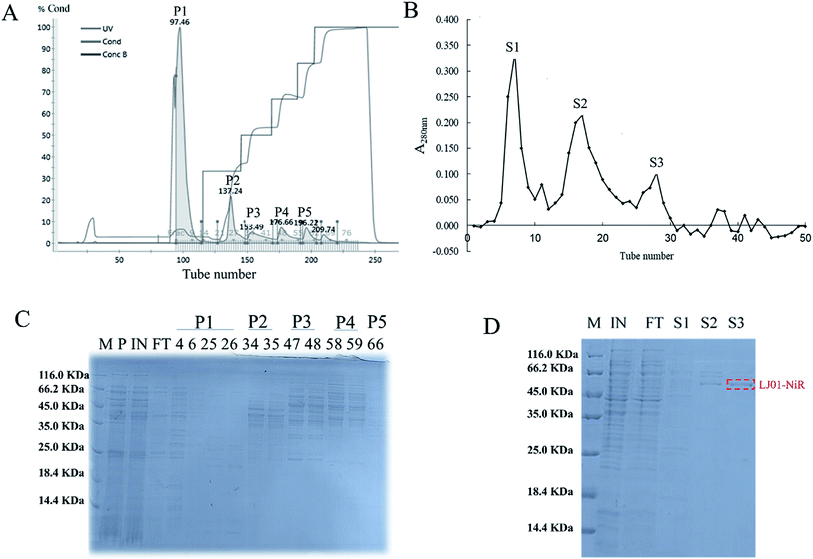
The ASDN from B. cereus LJ01 was purified to homogeneity by a two-step purification procedure (Fig. 3). The protein elution curves of the crude enzyme in DEAE-Sepharose Fast Flow chromatography are shown in Fig. 3A. The peaks P1, P2, P3, P4 and P5 were eluted by an elution buffer with 0, 150, 250, 400 and 500 mmol L−1 NaCl, respectively. Notably, unlike peaks P1 and P2 that did not show any activity of nitrite degradation, peaks P3 (4251.09 ± 20.51 U mg−1), P4 (4561.90 ± 74.09 U mg−1) and P5 (5347.12 ± 56.16 U mg−1) showed relatively strong activity without additional electron donor. Although different, the SDS-PAGE band patterns of peaks P3, P4 and P5 show a similar unit activity of nitrite degradation. Peak P3 was further purified by gel filtration in the subsequent analysis. The active fractions were concentrated and stored at 4 °C for further experiments. As shown in Fig. 3B, the P3 can be separated into 3 different peaks (S1, S2 and S3), of which S3 exerted the highest unit activity of nitrite degradation (4004.89 U mg−1) (Table 2).| Samples | Specific activity (U mg−1) |
|---|---|
| a P1, P2, P3, P4 and P5 represented the five peaks fractions purified by DEAE-Sepharose fast flow anion chromatography, respectively. S1, S2, and S3 represented the crude NiR enzyme purified by Sephadex G-150 gel chromatography, respectively. | |
| P1 | 50.95 |
| P2 | 45.37 |
| P3 | 4251.09 |
| P4 | 4561.90 |
| P5 | 5347.12 |
| S1 | 227.93 |
| S2 | 2540.67 |
| S3 | 4004.89 |
The purified ASDN of B. cereus LJ01 was digested with trypsin and analyzed by mass spectrometry (MALDI-TOF/TOF). Seven internal peptides of the purified ASDN matched the peptide sequences of ATKEIDGEKKVGFHIKVGGGL, GQIRTCNSQ, NKLKKDGLEIFNDIPY, GVQDQKQDGLKYVGFN, IHMVGCPNSCGQRQ, LDGGAYNQKLKGK, and KENKLPAETFY from a putative nitrite reductase from B. cereus ATCC 14579. The characteristics of this enzyme are very similar to NiR from Pseudomonas nautica 617,17 Alcaligenes xylosoxidans subsp. (deposited in N.C.I.M.B.)18 and Haloferax mediterranei.19 Based on the above data (especially mass spectrometry (MALDI-TOF/TOF)), we confirmed that the ASDN purified from B. cereus LJ01 is a NiR, and named it LJ01-NiR.
The molecular weight of LJ01-NiR is approximately 60 kDa. The NiR enzyme activity test showed that the purification rate of enzymes increased 17.57 times, and the enzyme yield and specific activity were 2.37% and 4004.89 U mg−1, respectively. Spirillum 5175 was isolated from an anoxic enrichment culture for Desulfuromonas, and its NiR specific activity (10476.74 U mg−1) was 2.62 times of LJ01-NiR's (4004.89 U mg−1).20 However, the specific activity of NiR in several common denitrifying bacteria was lower than that of LJ01-NiR (4004.89 U mg−1). Hydrogenobacter thermophilus TK-6 represented an inorganically autotrophic thermophile, whose NiR specific activity was only 2.93 U mg−1.21 Clostridium perfringens was screened from gas gangrene muscle, and its NiR specific activity was 81.15 U mg−1.22
3.4 Cloning of LJ01-NiR gene and its expression in E. coli
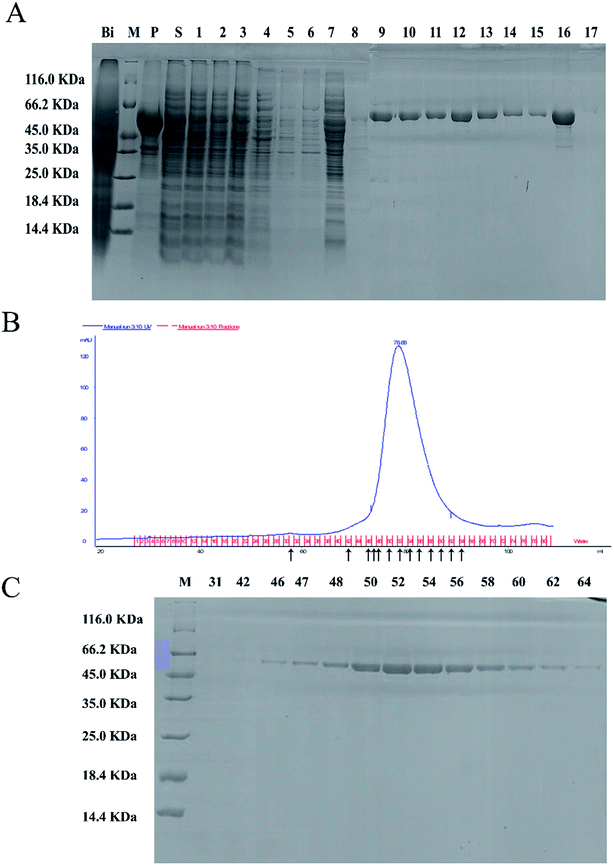
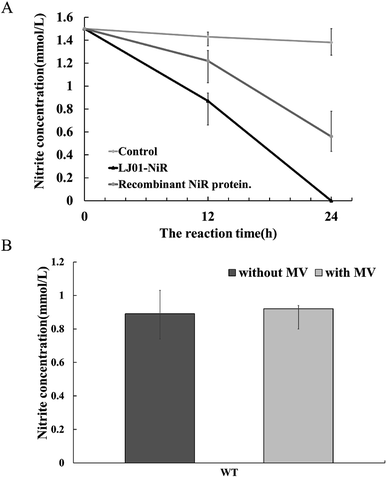
The full-length coding sequence of LJ01-NiR was 1623 bp, encoding 540 amino acid residues. The theoretical molecular weight of LJ01-NiR protein was 60.38 kDa, and the isoelectric point pI 5.47, as analyzed by the Protparam tool (https://web.expasy.org/protparam/). The LJ01-NiR was successfully expressed heterologously in E. coli BL21 (DE3), and the recombinant protein was purified by Ni2+-NTA affinity chromatography. The purified proteins were analyzed by SDS-PAGE (Fig. 4A). The approximately 60 kDa band shown in Fig. 4B and C implied that LJ01-NiR is a monomer, which is consistent with the theoretical mass of LJ01-NiR.Available B. cereus genome sequences were investigated for the presence of the gene. Three strains were selected for comparison with the LJ01 strain on the differences of nucleotide sequences and NiR amino acids (ESI Fig. S1 and S2†). Compared to Bacillus thuringiensis strain XL6 (GenBank: CP013000.1), there was a G to T base substitution at positon 1610 position of the NiR gene nucleotide sequences in the LJ01 strain. This resulted in an amino acid substitution from Glycine to Valine at codon 537. However, their nucleotide sequences similarity reached up to 99%. As for the nucleotide sequences of NiR gene, Bacillus cereus 03BB102 (GenBank: CP009318.1) and Bacillus anthracis A16R (GenBank: CP001974.2) displayed a 97% similarity (with 51 different bases) with B. cereus LJ01. The nitrite-degrading activity of the LJ01-NiR was lower than other denitrifying bacteria, but higher than that of C. perfringens. In terms of specific activity, NiR activity from B. cereus LJ01 was higher than that of several other bacteria, lower than that of Spirillum 5175, and equivalent to that of A. xylosoxidans. As shown in Fig. 5A, LJ01-NiR and recombinant NiR were revealed to similar nitrite degradation tendences. Fig. 5B demonstrates that the specific activity was determined with or without the additional non-physiological electron donor methyl viologen (MV). The nitrite degradation activity was similar in both cases, indicating that artificial electron donors are not essential for the enzymatic nitrite degradation reaction by LJ01-NiR.
3.5 Characterization of LJ01-NiR
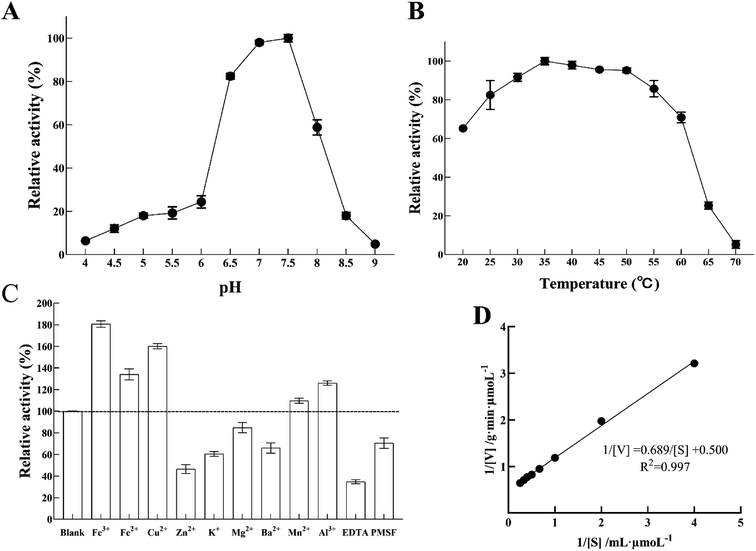
Nitrite reductase activity was measured using MV and sodium nitrite as an electron donor and a substrate, respectively. Fig. 6A shows the enzyme pH dependence at room temperature from pH 4 to pH 9. The optimum pH of LJ01-NiR was pH 7.5, and the retention activities were 98.03% and 58.78% at pH 7.0 and 8.0, respectively (100% at pH 7.5). However, when the pH increased to 8.5, only 17.99% of enzyme activity was retained. Purified LJ01-NiR showed broad activity range in a temperature range from 20 to 70 °C (Fig. 6B). The enzyme showed maximum activity at 35 °C and retained 65.23 and 5.19% activity at 20 °C and 70 °C, respectively. As shown in Fig. 6C, the metal ions Fe3+, Fe2+, Cu2+, Mn2+, and Al3+ can increase the LJ01-NiR activity by 9.76–80.68%. However, in the presence of Zn2+, K+, Mg2+, and Ba2+, the relative activities were 46.47%, 60.47%, 84.77% and 65.96% of the control, respectively. Moreover, inhibitory additives EDTA and PMSF can inhibit the activity of LJ01-NiR by 34.80% and 70.495%, respectively (Fig. 6C). The kinetic parameters Km and Vmax of LJ01-NiR were studied using standard method and in pH 7.5 and 35 °C conditions. The values of Km and Vmax were 1.38 mM and 2.00 μmoL g−1 min−1, respectively (Fig. 6D). Besides, this Km value was similar to Bacillus firmus GY-49 green-type of NirK.143.6 Sequence alignment and homology modelling of LJ01-NiR
Homology modeling of NiR from B. cereus LJ01 is shown in Fig. 7. Like Tb-NirA, LJ01-NiR consisted of three alpha/beta domains namely: the parachute domain, the middle domain, and the C-terminal [Fe4–S4] cluster binding domain (Fig. 7A). All these domains participate in the heme-binding pocket formation while the [Fe4–S4] cluster is located on the other side of the heme-binding domain. As shown in Fig. 7B, the heme-binding domain was coordinated by T101, T102, N140, R179, K180, K182, and S22 6 residues. Four highly conserved cysteine residues including C395, C401, C436, and C440 (Fig. 7C) coordinated the [Fe4–S4] cluster. As for heme-binding domain residues, only 2 alkaline residues (R179 and K180) and S226 were fully conserved (Fig. 7C). In Tb-NirA, an unusual covalent bond was found in the active site of NirA between the side chains of Y69 and C161,23 which promoted the catalytic reaction, but was not required for the enzymatic reaction of artificial electron donors. Interestingly, these 2 residues were conserved in Tb-NirA and LJ01-NiR. While glycine is the residue of other enzymes, the corresponding residues in Tb-NirA and LJ01-NiR are C161, C134, respectively (Fig. 7C). Based on the above analysis, we speculate that LJ01-NiR has a similar catalytic mechanism as Tb-NirA.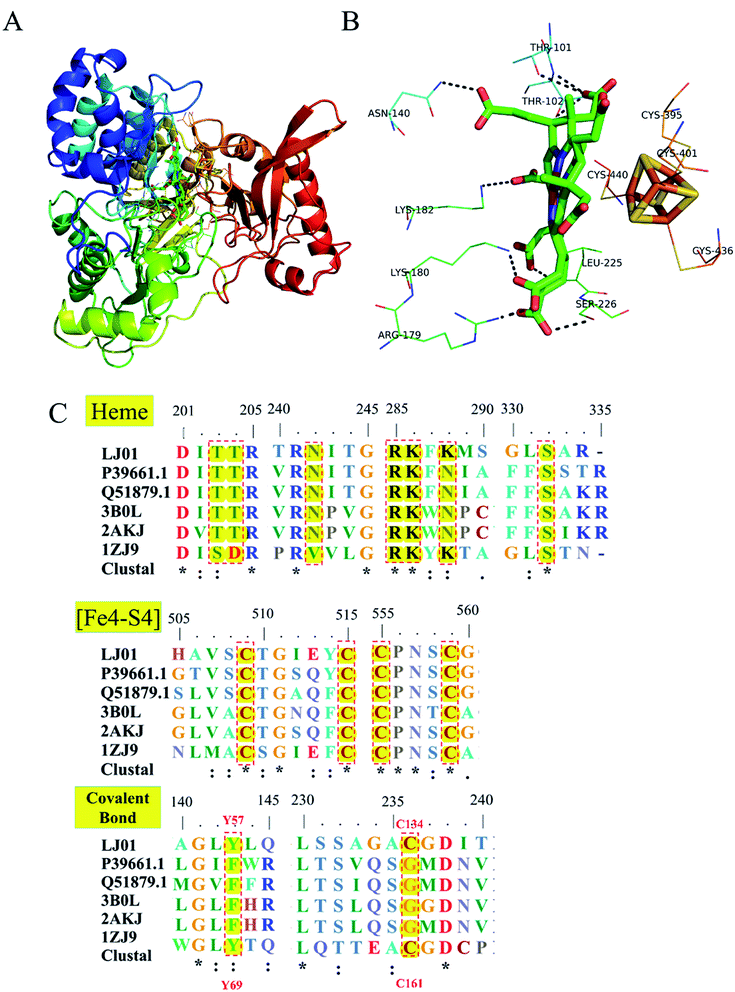 | ||
| Fig. 7 Homology modeling and conserved domains of the LJ01-NiR. (A) The whole structure of LJ01-NiR homology model, including colorful display from N to C and the C-terminal [Fe4–S4] cluster (red). The [Fe4–S4] cluster and the heme-binding residues were shown as sticks; (B) potential interaction between heme-binding domain and [Fe4–S4] cluster. The amino acid residues of this interaction were labelled and shown as lines; (C) alignment of conserved domains (heme-binding domain, [Fe4–S4] cluster, and covalent bonds) of nitrite reductases. LJ01: this study (GenBank: MG839504); P39661.1: Synechococcus elongatus PCC 7942 (UniProtKB/Swiss-Prot: P39661.1); Q51879.1: Phormidium laminosum (UniProtKB/Swiss-Prot: Q51879.1); 3B0L: Nicotiana tabacum (common tobacco) (PDB: 3B0L_A); 2AKJ: Spinacia oleracea (PDB: 2AKJ_A); 1ZJ9: Mycobacterium tuberculosis H37Rv (PDB: 1ZJ9_B). | ||
3.7 Analysis of molecular docking
PyMOL generates docking poses by loading docking programs directly through plugins. The configuration/score relationship is directly analyzed in a small text box containing a rating table of docking scores and their respective combined poses. Nitrite ion forms network of three hydrogen bonds with ASN72 (3.4 Å), ALA133 (3.2 Å) and ASN140 (3.5 Å). The nitrite ion formed a special interaction with the active sites of LJ01-NiR (binding energy: −2.1 kcal mol−1). Fig. 8 displays the hydrogen bonding of the docking ligand and its corresponding binding pose. This stable complex formation offers a model for how LJ01-NiR interacts with nitrite.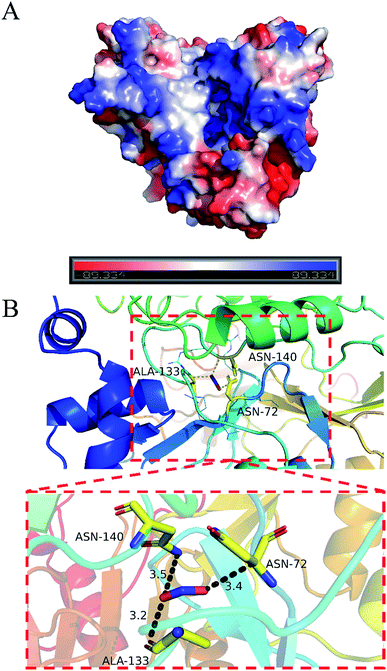 | ||
| Fig. 8 Predicted interaction pattern between LJ01-NiR and nitrite by docking analysis. (A) Surface potential graph. (B) Entire graph. | ||
4. Discussion
So far, NiR generated by B. cereus, which belongs to Gram-positive bacteria, has never been reported. The literature showed that only Tb-NirA from Mycobacterium tuberculosis23 had been functionally verified, and showed a 36% similarity with nitrite reductase of B. cereus. B. cereus LJ01 exhibited greater nitrite degradation ability than Lactobacillus.16 Nitrite reductase in B. cereus LJ01 was then purified by DEAE-Sepharose fast flow ion-exchange chromatography and Sephadex G-150 gel filtration. The specific activity of LJ01-NiR was 4004.89 U mg−1 and its monomer mass was approximately 60 kDa. However, after Sephadex G-150 gel filtration, the nitrite-reducing activity of the LJ01-NiR was considerably worse than the activities of the p3, p4 and p5 components purified by DEAE-Sepharose fast flow ion-exchange chromatography. This could be due to degradation of the LJ01-NiR after Sephadex G-150 gel filtration, resulting in a reduction of the nitrite-reducing activity. The previous study of our laboratory used inductively coupled plasma mass spectrometry (ICP-MS) to determine copper ions in the NiR. It was found that the NIR protein still retains copper ions after NTA purification, and the copper ion content is 184.5 mg kg−1, indicating that copper ions remain after purification.24 Several common denitrifying bacteria were selected for comparison including: Pseudomonas nautica 617 which is a marine denitrifying bacterium;17 Alcaligenes xylosoxidans subsp. (deposited in N.C.I.M.B.)18 and Haloferax mediterranei19 which were classical examples of extreme halophilic archaea; Spirillum 5175 isolated from an anoxic enrichment culture of Desulfuromonas;20 Hydrogenobacter thermophilus TK-6 inorganically autotrophic thermophiles.21 Clostridium perfringens of gas gangrene22 was isolated from the muscle. The nitrite degrading activity of LJ01-NiR from B. cereus LJ01 was lower than other denitrifying bacteria, but higher than that of C. perfringens. NiR activity from B. cereus LJ01 was higher than several other bacteria, equivalent to that of A. xylosoxidans, but lower than that of Spirillum 5175.There are currently two types of nitrite reductase assimilation namely: NAD(P)H NiRs and FdNiRs, whose prosthetic groups are FAD, [Fe4–S4] clusters and siroheme.7 Moreover, the LJ01-NiR sequence alignment and homology modeling from B. cereus LJ01 shows that the [Fe4–S4] cluster and heme-binding residues are highly conserved (Fig. 7). It is speculated that LJ01-NiR influences the assimilation of nitrite reductase. FdNiRs combined siroheme and [Fe4–S4] centers as repair groups, while NAD(P)H NiRs had FAD bound to the extended N-terminus.25 Although it was not possible to establish the physiological electron donor LJ01-NiR from Bacillus cereus LJ01, it was worth noting that both LJ01-NiR and recombinant NiR could degrade nitrite (Fig. 5A). The specific activity was determined with or without the addition of the non-physiological electron donor methyl viologen (MV) as shown in Fig. 5B. It is speculated that [Fe4–S4] cluster serves as a molecular battery to promote the transfer of electrons to heme and to achieve continuous electron reduction.26 Interestingly, Y69 and C161 residues were conserved between Tb-NirA and LJ01-NiR. Although these residues may promote the catalytic reaction, they were not essential for the enzymatic reaction of the artificial electron donors. This study also performed molecular docking of LJ01-NiR with nitrite molecules. The nitrite specifically interacts with the active sites of LJ01-NiR (binding energy: −2.1 kcal mol−1) to form a stable complex that justifies the interaction between LJ01-NiR and nitrite ion. To confirm the above statement, further studies on the mechanism of action of LJ01-NiR are required.
5. Conclusion
In summary, this is the first study to purify, clone, overexpress and characterize Bacillus cereus LJ01, isolated from Chinese traditional soybean paste, as a novel assimilatory nitrite reductase. Bacillus cereus LJ01 is capable of effectively degrading 0–15 mmol L−1 NaNO2 in LB medium. The purified ASDN from B. cereus LJ01 had a specific activity of 4004.89 U mg−1. This study further reveals that LJ01-NiR and recombinant NiR have similar trends for nitrite degradation. Both homology modeling analysis and activity tests indicate that LJ01-NiR does not need the additional electron donors for the enzymatic reaction of nitrite degradation. The docking of LJ01-NiR with nitrite ion offers a model for the nitrite reductase potential modes of action. These findings have great value for the application of nitrite reduction in the contaminated environment and construction of artificial biosensors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (No. 31771908) and the Science and Technology Research Project of Foundation of Guangdong Province (201903010015).References
- W. Lijinsky and S. S. Epstein, N-Nitrosamines as Environmental Carcinogens, Nature, 1970, 225, 21–23 CrossRef CAS.
- Y. T. Fei, D. M. Liu, T. H. Luo, G. Chen, H. Wu, L. Li and Y. G. Yu, Molecular characterization of Lactobacillus plantarum DMDL 9010, a strain with efficient nitrite degradation capacity, PLoS One, 2014, 9, e113792 CrossRef.
- J. Simon, Enzymology and bioenergetics of respiratory nitrite ammonification, FEMS Microbiol. Rev., 2002, 26, 285–309 CrossRef CAS.
- D. M. Liu, P. Wang, X. Y. Zhang, X. L. Xu, H. Wu and L. Li, Characterization of Nitrite Degradation by Lactobacillus casei subsp. rhamnosus LCR 6013, PLoS One, 2014, 9, e93308 CrossRef.
- M. J. Murphy, L. M. Siegel, S. R. Tove and H. Kamin, Siroheme: A new prosthetic group participating in six-electron reduction reactions catalyzed by both sulfite and nitrite reductases, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 612–616 CrossRef CAS.
- E. Cabrera, R. Gonzalezmontelongo, T. Giraldez, D. A. La Rosa and J. M. Siverio, Molecular components of nitrate and nitrite efflux in yeast, Eukaryotic Cell, 2014, 13, 267–278 CrossRef.
- Q. Song, B. Wang, F. Zhao, Y. Han and Z. Zhou, Expression, characterization and molecular docking of the assimilatory NaDH-nitrite reductase from Acidovorax wautersii QZ-4, Biochem. Eng. J., 2020, 107589 CrossRef CAS.
- D. Hira, M. Nojiri, K. Yamaguchi and S. Suzuki, Identification of a blue copper protein from hyphomicrobium denitrificans and its functions in the periplasm, J. Biochem., 2007, 142, 335–341 CrossRef CAS.
- A. A. Hugo, I. S. Rolny, D. Romanin and P. F. Pérez, Lactobacillus delbrueckii subsp. lactis (strain CIDCA 133) stimulates murine macrophages infected with Citrobacter rodentium, World J. Microbiol. Biotechnol., 2017, 33, 48 CrossRef.
- H. Hhm, M. Ma, G. Jin, Y. Jin, I. Khalifa, Q. Zeng and Y. Liu, Nitroso-Hemoglobin increased the color stability and inhibited the pathogenic bacteria in a minced beef model: A combined low-field NMR study, Korean J. FOOD Sci. An., 2019, 39, 704–724 CrossRef.
- I. Verbaendert, N. Boon, V. P. De and K. Heylen, Denitrification is a common feature among members of the genus Bacillus, Syst. Appl. Microbiol., 2011, 34, 385–391 CrossRef CAS.
- J. Zhang, P. Wu, B. Hao and Z. Yu, Heterotrophic nitrification and aerobic denitrification by the bacterium Pseudomonas stutzeri YZN-001, Syst. Appl. Microbiol., 2011, 102, 9866–9869 CAS.
- T. M. Barbosa, C. R. Serra, R. M. L. Ragione, M. J. Woodward and A. O. Henriques, Screening for Bacillus Isolates in the Broiler Gastrointestinal Tract, Appl. Environ. Microbiol., 2005, 71, 968–978 CrossRef CAS.
- H. Gao, C. Li, B. Ramesh and N. Hu, Cloning, purification and characterization of novel Cu-containing nitrite reductase from the Bacillus firmus GY-49, World J. Microbiol. Biotechnol., 2018, 34, 1 CrossRef CAS.
- K. Poole, Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria, Trends Microbiol., 2012, 20, 227–234 CrossRef CAS.
- D. Ren, P. Chen, W. Li, X. Su, K. Bao, Y. Wang, J. Wang and H. Liu, Screening, Mutagenesis of Nitrite-Degrading Lactobacilli in Chinese Traditional Fermented Sauerkraut and its Application in the Production of Sauerkraut, J. Food Saf., 2016, 36, 474–481 CrossRef CAS.
- J. M. Dias, T. Alves, C. Bonifacio, A. S. Pereira, J. Trincao, D. Bourgeois, I. Moura and M. J. Romao, Structural Basis for the Mechanism of Ca2+ Activation of the Di-Heme Cytochrome c Peroxidase from Pseudomonas nautica 617, Structure, 2004, 12, 961–973 CrossRef CAS.
- Z. H. Abraham, D. J. Lowe and B. E. Smith, Purification and characterization of the dissimilatory nitrite reductase from Alcaligenes xylosoxidans subsp. xylosoxidans (N.C.I.M.B. 11015): evidence for the presence of both type 1 and type 2 copper centres, Biochem. J., 1993, 295, 587–593 CrossRef CAS.
- R. M. Martínezespinosa, F. C. Marhuendaegea and M. J. Bonete, Purification and characterisation of a possible assimilatory nitrite reductase from the halophile archaeon Haloferax mediterranei, FEMS Microbiol. Lett., 2001, 196, 113–118 CrossRef.
- A. Zöphel, M. C. Kennedy, H. Beinert and P. M. Kroneck, Investigations on microbial sulfur respiration isolation, purification, and characterization of cellular components from Spirillum 5175, Eur. J. Biochem., 1991, 195, 849–856 CrossRef.
- M. Suzuki, T. Hirai, H. Arai, M. Ishii and Y. Igarashi, Purification, characterization, and gene cloning of thermophilic cytochrome cd1 nitrite reductase from Hydrogenobacter thermophilus TK-6, J. Biomed. Bioeng., 2006, 101, 391–397 CAS.
- S. Sekiguchi, S. Seki and M. Ishimoto, Purification and some properties of nitrite reductase from Clostridium perfringens, J. Biochem., 1983, 94, 1053–1059 CrossRef CAS.
- R. Schnell, T. Sandalova, U. Hellman, Y. Lindqvist and G. Schneider, Siroheme-and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis is a sulfite reductase with a covalent Cys-Tyr bond in the active site, J. Biol. Chem., 2005, 280, 27319–27328 CrossRef CAS.
- S. M. Chen, T. H. Luo, Y. T. Fei, J. F. Wu and D. M. Liu, Cloning, expression and purification of the nitrite reductase gene from bacillus cereus LJ01, Food Sci., 2018, 39, 69–74 Search PubMed.
- W. H. Campbell and J. R. Kinghorn, Functional domains of assimilatory nitrate reductases and nitrite reductases, Trends Biochem. Sci., 1990, 15, 315–319 CrossRef CAS.
- M. Surducan, S. V. Makarov and R. Silaghi-Dumitrescu, O–S bond activation in structures isoelectronic with ferric peroxide species known in O–O-activating enzymes: relevance for sulfide activation and sulfite reductases, Eur. J. Inorg. Chem., 2014, 23, 5827–5837 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06129h |
| This journal is © The Royal Society of Chemistry 2020 |