Open Access Article
Open Access ArticleExploring the broad nucleotide triphosphate and sugar-1-phosphate specificity of thymidylyltransferase Cps23FL from Streptococcus pneumonia serotype 23F†
Siqiang Li‡
 ab,
Hong Wang‡a,
Guoxia Jinc,
Zonggang Chena and
Guofeng Gu
ab,
Hong Wang‡a,
Guoxia Jinc,
Zonggang Chena and
Guofeng Gu *a
*a
aNational Glycoengineering Research Center, Shandong Key Laboratory of Carbohydrate Chemistry and Glycobiology, Shandong University, 72 Binhai Road, Qingdao 266237, China. E-mail: guofenggu@sdu.edu.cn
bSchool of Biological and Food Processing Engineering, Huanghuai University, 76 Kaiyuan Road, Zhumadian 463000, China
cCollege of Chemistry, Chemical Engineering and Materials Science, Shandong Normal University, 88 Wenhua Dong Lu, Jinan 250014, China
First published on 14th August 2020
Abstract
Glucose-1-phosphate thymidylyltransferase (Cps23FL) from Streptococcus pneumonia serotype 23F is the initial enzyme that catalyses the thymidylyl transfer reaction in prokaryotic deoxythymidine diphosphate-L-rhamnose (dTDP-Rha) biosynthetic pathway. In this study, the broad substrate specificity of Cps23FL towards six glucose-1-phosphates and nine nucleoside triphosphates as substrates was systematically explored, eventually providing access to nineteen sugar nucleotide analogs.
1. Introduction
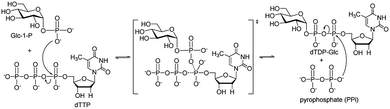
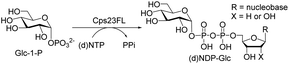
In past decades, many carbohydrates have been developed as drug targets due to their important pharmaceutical and pharmacological bioactivities.1 To generate novel therapeutic molecules, glycorandomization is one of the most popular strategies to produce natural and unnatural glycosylated metabolites,2 which takes advantage of the glycosyltransferases to generate diverse libraries of glycosylated biomolecules.3–5 Although several examples of glycorandomization have been reported,6,7 the deficiency of sugar precursors (usually sugar nucleotides, (d)NDP-sugars) greatly limits the application of glycorandomization. Thus, the efficient synthesis of (d)NDP-sugars has been becoming an attractive and hot topic.8–15Although chemical synthesis of sugar nucleotides has been developed,11 the fastidious manipulations and low yields hindered its broad application. In contrast, enzymatic synthesis of sugar nucleotides is particularly attractive owing to its high synthetic efficiency and high regio-/stereo-specificity.14,15 Numerous enzymes that involved into the biosynthetic pathways of sugar nucleotides have been discovered and applied to synthesize sugar nucleotides in vitro.14,15 Among of them, a thymidylyltransferase (RmlA) has been found to catalyse the first sequential reaction in the biosynthetic pathway of deoxythymidine diphosphate L-rhamnose (dTDP-Rha),16 in which RmlA converts α-D-glucose-1-phosphate (Glc-1-P) and deoxythymidine triphosphate (dTTP) into dTDP-D-glucose (dTDP-Glc) and pyrophosphate (PPi) (Fig. 1). This enzyme reaction presumably proceeded via a bi–bi ordered catalytic mechanism that formed a trigonal bipyrimidal phosphoryl ternary complex.17–19 RmlA was attracted more interest because it displayed unusual promiscuity toward both dozens of sugar phosphates and eight natural nucleotide triphosphates.20,21 RmlA and its mutant have been applied to prepare up to one hundred of (d)NDP-sugars in vitro.19,22–24 In our recent report, Cps23FL, a homolog of RmlA that derived from Streptococcus pneumonia serotype 23F, exhibited the excellent activity on conversion of Glc-1-P and dTTP into dTDP-Glc and PPi.25 Thus, its potential to synthesize various sugar nucleotides is worthwhile for exploitation. In this work, the substrate specificity of Cps23FL was systematically investigated with six sugar-1-phosphates (sugar-1-Ps) and nine nucleotide triphosphates (NTPs) in order to explore the potential ability of recombinant wildtype Cps23FL for synthesizing those important and rare sugar nucleotides in vitro.
2. Results and discussion
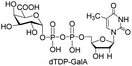
We first investigated the substrate specificity of Csp23FL towards various (d)NTPs using Glc-1-P as the sugar-1-P substrate. Nine (d)NTPs were examined into the reaction systems, respectively (Table 1). Briefly, enzymatic reactions were commenced with the addition of purified Cps23FL (6 or 60 μM) to a solution containing HEPES buffer (30 mM, pH 7.5), MgCl2 (5 mM), YIPP (2 U mL−1), (d)NTPs (3 mM) and Glc-1-P (5 mM) in a final volume of 100 μL. Then, the resulting reactions were incubated at 37 °C for 1 h and quenched in boiling water bath for 30 s followed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 rpm for 10 min. The (d)NDP-Glc product of each enzymatic reaction was detected and isolated by HPLC (DionexCarboPac™ PA-100, 0–1.0 M ammonium acetate eluent) (Table S1†). The yields of (d)NDP-Glcs were depicted in Table 1. In 6 μM enzymatic reaction system, Cps23FL exhibited broad substrate specificities towards deoxy-nucleoside triphosphate (dTTP, dUTP, dCTP, and dGTP) (Table 1, entries 1, 2, 4, 6) and afforded good to high isolate yields of dNDP-Glcs (>50%) except that of dATP (only 27%) (Table 1, entry 8). However, Cps23FL showed only low or none activities toward UTP, CTP, GTP and ATP substrates (Table 1, entries 3, 5, 7 and 9) under this enzyme concentration even with prolonging reaction time to 48 h (for GTP). Accordingly, the influence of different concentration of NTP (0, 1.5, 3.0, 6.0, 12.0, 24.0 mM) and Csp23FL (0, 0.06, 0.6, 6.0, 30, 60 μM) on the product conversion was investigated using ATP as a model substrate (Fig. 2). It was disclosed that the high conversion of ADP-Glc within 6 h was achieved at 3.0 mM concentration of ATP (Fig. 2A), and also efficiently improved with the increased Cps23FL concentration (Fig. 2B). As shown in Table 1, under 60 μM enzyme concentration, all tested nucleoside triphosphates could be commendably recognized by Cps23FL and completely converted into their corresponding (d)NDP-Glcs in high yields (73–95%). Our results indicated Cps23FL had a notable substrate tolerance and the changes on both nucleobase and substituent group on the C-2 position of ribose residue could not influence its enzymatic activity. Therefore, Cps23FL preferred both pyrimidine nucleoside and purine nucleoside whether in nucleotide form or their deoxy form under high enzyme concentration, which was consistent with those results reported by RmlA.20,21 All the prepared (d)NDP-Glcs were well characterized by MS and NMR spectrometry, and the characterization data of dTDP-Glc,25 dUDP-Glc,26,27 UDP-Glc,10,26 CDP-Glc,10,26 GDP-Glc,26 and ADP-Glc26,27 were in good agreement with those previously reported. More remarkably, dCDP-Glc, dGDP-Glc, and dADP-Glc were characterized first time by NMR spectrometry in this study.28
300 rpm for 10 min. The (d)NDP-Glc product of each enzymatic reaction was detected and isolated by HPLC (DionexCarboPac™ PA-100, 0–1.0 M ammonium acetate eluent) (Table S1†). The yields of (d)NDP-Glcs were depicted in Table 1. In 6 μM enzymatic reaction system, Cps23FL exhibited broad substrate specificities towards deoxy-nucleoside triphosphate (dTTP, dUTP, dCTP, and dGTP) (Table 1, entries 1, 2, 4, 6) and afforded good to high isolate yields of dNDP-Glcs (>50%) except that of dATP (only 27%) (Table 1, entry 8). However, Cps23FL showed only low or none activities toward UTP, CTP, GTP and ATP substrates (Table 1, entries 3, 5, 7 and 9) under this enzyme concentration even with prolonging reaction time to 48 h (for GTP). Accordingly, the influence of different concentration of NTP (0, 1.5, 3.0, 6.0, 12.0, 24.0 mM) and Csp23FL (0, 0.06, 0.6, 6.0, 30, 60 μM) on the product conversion was investigated using ATP as a model substrate (Fig. 2). It was disclosed that the high conversion of ADP-Glc within 6 h was achieved at 3.0 mM concentration of ATP (Fig. 2A), and also efficiently improved with the increased Cps23FL concentration (Fig. 2B). As shown in Table 1, under 60 μM enzyme concentration, all tested nucleoside triphosphates could be commendably recognized by Cps23FL and completely converted into their corresponding (d)NDP-Glcs in high yields (73–95%). Our results indicated Cps23FL had a notable substrate tolerance and the changes on both nucleobase and substituent group on the C-2 position of ribose residue could not influence its enzymatic activity. Therefore, Cps23FL preferred both pyrimidine nucleoside and purine nucleoside whether in nucleotide form or their deoxy form under high enzyme concentration, which was consistent with those results reported by RmlA.20,21 All the prepared (d)NDP-Glcs were well characterized by MS and NMR spectrometry, and the characterization data of dTDP-Glc,25 dUDP-Glc,26,27 UDP-Glc,10,26 CDP-Glc,10,26 GDP-Glc,26 and ADP-Glc26,27 were in good agreement with those previously reported. More remarkably, dCDP-Glc, dGDP-Glc, and dADP-Glc were characterized first time by NMR spectrometry in this study.28
| Entry | (d)NTP | (d)NDP-Glc | Yielda (yield)b |
|---|---|---|---|
| a Yields obtained in 6 μM of Cps23FL.b Yields obtained in 60 μM of Cps23FL.c No product detected by HPLC within 48 h.d Enzymatic reaction time was 6 h. | |||
| 1 | dTTP | 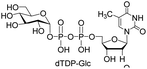 |
86% (93%) |
| 2 | dUTP | 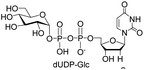 |
88% (95%) |
| 3 | UTP | 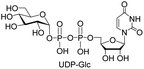 |
40% (94%) |
| 4 | dCTP | 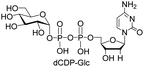 |
75% (93%) |
| 5 | CTP | 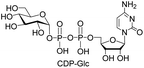 |
35% (87%) |
| 6 | dGTP | 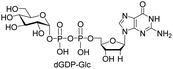 |
53% (89%) |
| 7 | GTP | 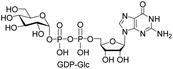 |
NPc (91%)d |
| 8 | dATP | 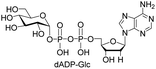 |
27% (73%)d |
| 9 | ATP | 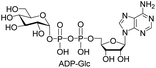 |
13%d (78%)d |
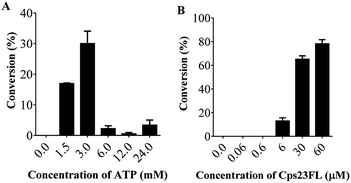 | ||
| Fig. 2 The influence of different concentration of (A) ATP and (B) Cps23FL on the enzymatic reaction of Cps23FL with Glc-1-P and ATP within 6 h. | ||
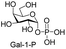
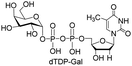
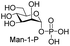
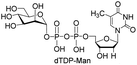
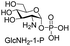
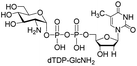
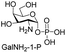
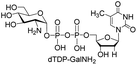
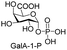
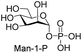
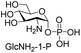
Next, the substrate tolerance of Cps23FL for sugar-1-Ps was investigated with dTTP as pyrophosphate donor. The sugar-1-Ps tested in this study were the native substrate Glc-1-P, galactose-1-phosphate (Gal-1-P), mannose-1-phosphate (Man-1-P), glucosamine-1-phosphate (GlcNH2-1-P), galactosamine-1-phosphate (GalNH2-1-P), and galacturonic acid-1-phosphate (GalA-1-P), respectively. The enzymatic reactions were performed in 60 μM of Cps23FL with the same reaction condition described above. The reaction results shown in Table 2 indicated that Cps23FL exerted different activities for sugar-1-Ps in the following order: Glc-1-P ≈ Gal-1-P > Man-1-P > GlcNH2-1-P ≫ GalNH2-1-P. Cps23FL could efficiently catalyse both Glc-1-P and Gal-1-P to form dTDP-Glc/Gal in excellent yields (Table 2, entries 1 and 2). Furthermore, Man-1-P could be also well tolerated by Cps23FL to yield dTDP-Man in a high yield of 88% (Table 2, entry 3). Nevertheless, GlcNH2-1-P was transformed into dTDP-GlcNH2 by Cps23FL in good conversion rate (71%) whereas only 5% for GalNH2-1-P and no activity for GalA-1-P (Table 2, entries 4, 5 and 6). All these results indicated that the configuration change at C-2 and C-4 positions (Man-1-P, Gal-1-P) or amino group substitution at C-2 position (GlcNH2-1-P) could not affect their recognition by Cps23FL, while the configuration change at both the configuration change at C-4 position and substitution at C-2 or C-6 positions (GalNH2-1-P, GalA-1-P) might greatly decrease their enzymatic activities. Our findings were commendably supported by profile of the crystal structure of RmlA and Glc-1-P complex (PDB entry 1G23), in which the hydroxyl groups on C-2, C-4 and C-6 positions of glucose residue formed hydrogen bonds and van der Waals interacting with RmlA.6,17 Similarly, all the synthetic dTDP-sugars were characterized by MS and NMR spectrometry. The spectra data of dTDP-Gal, and dTDP-Man were well agreed with those previously reported.10 dTDP-GlcNH2 was characterized by NMR spectrometry,28 whilst dTDP-GalNH2 was only acquired by ESI-HRMS spectrum due to only trace amount of product obtained in reaction.
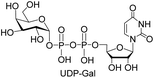
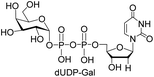
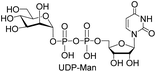
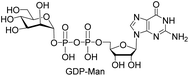
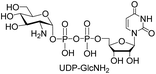
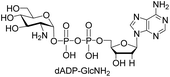
We further investigated the potential synthetic ability of Cps23FL for preparation of other (d)NDP-sugars with both non-native sugar-1-P analogues and (d)NTPs as substrates. Six (d)NDP-sugar analogues were synthesized with diverse yields (Table 3). Surprisingly, Cps23FL exhibited good activity toward Gal-1-P and (d)UTP substrates within 48 h, providing UDP-Gal and dUDP-Gal in an 86% and 83% isolated yield, respectively (Table 3, entries 1 and 2). For Man-1-P and UTP or GTP substrate, however, different conversion yields of UDP-Man (78%) and GDP-Man (13%) were observed after incubation reaction time of 48 h (Table 3, entries 3 and 4). Previous study has suggested that only RmlA mutant (Q24S) could catalyse the reactions of Man-1-P and GTP to synthesize GDP-Man.19 This study was the first time to report the synthesis of GDP-Man by recombinant wildtype Cps23FL. Similarly, Cps23FL could also catalyse GlcNH2-1-P with UTP or dATP to produce UDP-GlcNH2 or dADP-GlcNH2 in low yields (<21%) with the incubation time to 48 h (Table 3, entries 5 and 6). These results indicated its great potential on synthesis of diverse (d)NDP-sugars using non-native substrates. The synthetic UDP-Gal and UDP-Man were well confirmed by NMR and ESI-HRMS spectrometry,23,26 and dUDP-Gal was characterized first time by NMR spectrometry.28
3. Conclusion
In summary, the broad substrate specificity of the recombinant thymidylyltransferase Cps23FL cloned from S. pneumonia serotype 23F against six sugar-1-Ps and nine (d)NTPs was investigated in detail in this study. It was disclosed that Cps23FL exhibited much better recognition towards (d)NTP substrate than sugar-1-P substrate. As a result, nineteen sugar nucleotides were eventually constructed by Cps23FL-catalysed transformation reaction, and fifteen of them were availably isolated by semi-preparation HPLC technique in milligram scales in high yields of 70–95%. However, four of them including GDP-Man, dTDP-GalNH2, UDP-GlcNH2, and dADP-GlcNH2 were obtained in very low yields of less 22%. This can presumably be attributed to the significant structural alternation of both nucleoside triphosphate and sugar 1-phosphate, which is beyond the recognizable substrate tolerance of Cps23FL enzyme and thus likely weakens their binding ability at the active site.19 Moreover, the synthetic (d)NDP-sugars were proved to be stable enough within 48 h that evidenced by the HPLC observation of no more than 5% degradation of (d)NDP-sugars in reaction system. Furthermore, it was reported first time that wildtype Cps23FL could catalyse transformation of GDP-Man (even in low yield of 13%), which could only be achieved by RmlA mutant in previous study.19 This finding would allow us to broaden Cps23FL activity through enzyme engineering technique in case of improving synthetic efficacy of GDP-Man. Overall, this study provides a convenient route for the efficient preparation of a library of sugar nucleotides by wildtype Cps23FL, which will greatly facilitate glycosyltransferase-based glycorandomization synthesis of complex glycosylated metabolites.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (21672129), the Open Projects Fund of Shandong Key Laboratory of Carbohydrate Chemistry and Glycobiology, Shandong University (2019CCG06), and the Project of Education Department of Henan Province (19A180022).Notes and references
- A. C. Weymouth-Wilson, Nat. Prod. Rep., 1997, 14, 99–110 RSC.
- C. J. Thibodeaux, H. W. Liu and J. S. Thorson, Complementary routes to natural product glycodiversification: Pathway engineering and glycorandomization, in Comprehensive Glycoscience, ed. J. Kamerling, G.-J. Boons, Y. Lee, A. Suzuki, N. Taniguchi and A. G. J. Voragen, Elsevier, 2007, vol. 3, pp. 373–396 Search PubMed.
- J. B. McArthur and X. Chen, Biochem. Soc. Trans., 2016, 44, 129–142 CrossRef CAS PubMed.
- H. C. Losey, M. W. Peczuh, Z. Chen, U. S. Eggert, S. D. Dong, I. Pelczer, D. Kahne and C. T. Walsh, Biochemistry, 2001, 40, 4745–4755 CrossRef CAS PubMed.
- P. J. Solenberg, P. Matsushima, D. R. Stack, S. C. Wilkie, R. C. Thompson and R. H. Baltz, Chem. Biol., 1997, 4, 195–202 CrossRef CAS PubMed.
- W. A. Barton, J. Lesniak, J. B. Biggins, P. D. Jeffrey, J. Jiang, K. R. Rajashankar, J. S. Thorson and D. B. Nikolov, Nat. Struct. Biol., 2001, 8, 545–551 CrossRef CAS PubMed.
- G. J. Williams, J. Yang, C. Zhang and J. S. Thorson, ACS Chem. Biol., 2010, 6, 95–100 CrossRef PubMed.
- S. C. Timmons and D. J. Jakeman, Carbohydr. Res., 2008, 343, 865–874 CrossRef CAS PubMed.
- H. Tanaka, Y. Yoshimura, M. R. Jørgensen, J. A. Cuesta-Seijo and O. Hindsgaul, Angew. Chem., Int. Ed., 2012, 51, 11531–11534 CrossRef CAS PubMed.
- S. Wolf, T. Zismann, N. Lunau and C. Meier, Chem.–Eur. J., 2009, 15, 7656–7664 CrossRef CAS PubMed.
- S. Ahmadipour and G. J. Miller, Carbohydr. Res., 2017, 451, 95–109 CrossRef CAS PubMed.
- J. Fang, W. Guan, L. Cai, G. Gu, X. Liu and P. G. Wang, Bioorg. Med. Chem. Lett., 2009, 19, 6429–6432 CrossRef CAS PubMed.
- B. A. Wagstaff, M. Rejzek, T. Pesnot, L. M. Tedaldi, L. Caputi, E. C. O'Neil, S. Benini, G. K. Wagner and R. A. Field, Carbohydr. Res., 2015, 404, 17–25 CrossRef CAS PubMed.
- L. Cai, J. Carbohydr. Chem., 2012, 31, 535–552 CrossRef CAS.
- S. Ahmadipour, L. Beswick and G. J. Miller, Carbohydr. Res., 2018, 469, 38–47 CrossRef CAS PubMed.
- W. Blankenfeldt, M. F. Giraud, G. Leonard, R. Rahim, C. Creuzenet, J. S. Lam and J. H. Naismith, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2000, 56, 1501–1504 CrossRef CAS PubMed.
- W. Blankenfeldt, M. Asuncion, J. S. Lam and J. H. Naismith, EMBO J., 2000, 19, 6652–6663 CrossRef CAS PubMed.
- S. Zuccotti, D. Zanardi, C. Rosano, L. Sturla, M. Tonetti and M. Bolognesi, J. Mol. Biol., 2001, 313, 831–843 CrossRef CAS PubMed.
- D. L. Jakeman, J. L. Young, M. P. Huestis, P. Peltier, R. Daniellou, C. Nugier-Chauvin and V. Ferrières, Biochemistry, 2008, 47, 8719–8725 CrossRef CAS PubMed.
- J. Jiang, J. B. Biggins and J. S. Thorson, J. Am. Chem. Soc., 2000, 122, 6803–6804 CrossRef CAS.
- S. C. Timmons, R. H. Mosher, S. A. Knowles and D. L. Jakeman, Org. Lett., 2007, 9, 857–860 CrossRef CAS PubMed.
- R. Moretti and J. S. Thorson, J. Biol. Chem., 2007, 282, 16942–16947 CrossRef CAS PubMed.
- S. A. Beaton, M. P. Huestis, A. Sadeghi-Khomami, N. R. Thomas and D. L. Jakeman, Chem. Commun., 2009, 45, 238–240 RSC.
- W.-T. Chien, C.-F. Liang, C.-C. Yu, J. H. Lin, H.-T. Wu and C.-C. Lin, Adv. Synth. Catal., 2012, 354, 123–132 CrossRef CAS.
- S. Li, H. Wang, J. Ma, G. Gu, Z. Chen and Z. Guo, Chem. Commun., 2016, 52, 13995–13998 RSC.
- J. Bae, K. Kim, D. Kim, Y. Choi, J. S. Kim, S. Koh, S. Hong and D. Lee, ChemBioChem, 2005, 6, 1963–1966 CrossRef CAS PubMed.
- A. Zervosen, A. Stein, H. Adria and L. Elling, Tetrahedron, 1996, 52, 2395–2404 CrossRef CAS.
- Spectroscopic data of dCDP-Glc: 1H NMR (600 MHz, D2O): δ 7.91 (d, J = 7.8 Hz, 1H, –CH
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.13 (t, J = 6.6 Hz, 1H, H-1), 6.03 (d, J = 7.8 Hz, 1H, –CH
), 6.13 (t, J = 6.6 Hz, 1H, H-1), 6.03 (d, J = 7.8 Hz, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.42 (dd, J = 7.2, 3.0 Hz, 1H, H-1′), 4.43 (br s, H-3), 4.09–3.97 (m, 3H, H-4, H-5a,b), 3.72 (br d, J = 9.6 Hz, 1H, H-5′), 3.68 (d, J = 12.6 Hz, 1H, H-6a′), 3.63-3.56 (m, 2H, H-3′, H-6b′), 3.35 (br d, J = 9.6 Hz, 1H, H-2′), 3.29 (t, J = 9.6 Hz, 1H, H-4′), 2.32–2.25 (m, 1H, H-2), 2.20–2.13 (m, 1H, H-2); 13C NMR (150 MHz, D2O): δ 162.83, 142.76, 95.77, 95.46 (d, J = 6.6 Hz, C-1′), 86.19 (C-1), 85.64 (d, J = 9.0 Hz, C-4), 72.75 (C-3′), 72.68 (C-5′), 71.51 (d, J = 8.4 Hz, C-2′), 70.59 (C-3), 69.05 (C-4′), 65.11 (C-5), 60.18 (C-6′), 39.41 (C-2); 31P NMR (243 MHz, D2O): δ −11.34 (d, J = 20.8 Hz), −12.99 (d, J = 20.8 Hz); ESI-TOF HRMS m/z calcd for C15H24N3O15P2 [M − H]−1 548.0688, found 548.0616. dGDP-Glc: 1H NMR (600 MHz, D2O): δ 7.92 (s, 1H, –CH
), 5.42 (dd, J = 7.2, 3.0 Hz, 1H, H-1′), 4.43 (br s, H-3), 4.09–3.97 (m, 3H, H-4, H-5a,b), 3.72 (br d, J = 9.6 Hz, 1H, H-5′), 3.68 (d, J = 12.6 Hz, 1H, H-6a′), 3.63-3.56 (m, 2H, H-3′, H-6b′), 3.35 (br d, J = 9.6 Hz, 1H, H-2′), 3.29 (t, J = 9.6 Hz, 1H, H-4′), 2.32–2.25 (m, 1H, H-2), 2.20–2.13 (m, 1H, H-2); 13C NMR (150 MHz, D2O): δ 162.83, 142.76, 95.77, 95.46 (d, J = 6.6 Hz, C-1′), 86.19 (C-1), 85.64 (d, J = 9.0 Hz, C-4), 72.75 (C-3′), 72.68 (C-5′), 71.51 (d, J = 8.4 Hz, C-2′), 70.59 (C-3), 69.05 (C-4′), 65.11 (C-5), 60.18 (C-6′), 39.41 (C-2); 31P NMR (243 MHz, D2O): δ −11.34 (d, J = 20.8 Hz), −12.99 (d, J = 20.8 Hz); ESI-TOF HRMS m/z calcd for C15H24N3O15P2 [M − H]−1 548.0688, found 548.0616. dGDP-Glc: 1H NMR (600 MHz, D2O): δ 7.92 (s, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.15 (t, J = 7.2 Hz, 1H, H-1), 5.40 (dd, J = 6.6, 3.0 Hz, 1H, H-1′), 4.58 (br s, 1H, H-3), 4.08 (br s, 1H, H-4), 4.01–3.96 (m, 2H, H-5a,b), 3.74–3.69 (m, 1H, H-5′), 3.67 (d, J = 12.6 Hz, 1H, H-6a′), 3.62–3.54 (m, 2H, H-3′, H-6b′), 3.36–3.31 (m, 1H, H-2′), 3.27 (t, J = 9.6 Hz, 1H, H-4′), 2.68–2.60 (m, 1H, H-2a), 2.38–2.30 (m, 1H, H-2b); 13C NMR (150 MHz, D2O): δ 158.89, 153.75, 151.34, 137.52, 116.11, 95.45 (d, J = 7.5 Hz, C-1′), 85.46 (d, J = 9.0 Hz, C-4), 83.49 (C-1), 72.70 (C-3′), 72.66 (C-5′), 71.49 (d, J = 9.0 Hz, C-2′), 71.22 (C-3), 69.07 (C-4′), 65.37 (C-5), 60.19 (C-6′), 38.37 (C-2); 31P NMR (243 MHz, D2O): δ −11.22 (d, J = 20.6 Hz), −12.99 (d, J = 20.6 Hz); ESI-TOF HRMS m/z calcd for C16H24N5O15P2 [M − H]−1 588.0750, found 588.0680. dADP-Glc: 1H NMR (600 MHz, D2O): δ 8.32 (s, 1H), 8.09 (s, 1H), 6.36 (t, J = 6.6 Hz, 1H, H-1), 5.42-5.39 (m, 1H, H-1′), 4.59 (s, 1H, H-3), 4.14 (s, 1H, H-4), 4.05–3.95 (m, 2H, H-5a,b), 3.71 (d, J = 9.6 Hz, 1H, H-5′), 3.66 (d, J = 12.6 Hz, 1H, H-6a′), 3.60 (t, J = 9.6 Hz, 1H, H-3′), 3.56 (dd, J = 12.6, 4.2 Hz, 1H, H-6b′), 3.33 (d, J = 9.6 Hz, 1H, H-2′), 3.26 (t, J = 9.6 Hz, 1H, H-4′), 2.70-2.64 (m, 1H, H-2), 2.47–2.41 (m, 1H, H-2); 13C NMR (150 MHz, D2O): δ 155.11, 152.05, 148.63, 139.97, 118.55, 95.45 (d, J = 6.6 Hz, C-1′), 85.65 (d, J = 9.0 Hz, C-4), 83.67 (C-1), 72.73 (C-5′), 72.67 (C-3′), 71.52 (d, J = 8.7 Hz, C-2′), 71.14 (C-3), 69.10 (C-4′), 65.35 (d, J = 6.0 Hz, C-5), 60.22 (C-6′), 38.97 (C-2); 31P NMR (243 MHz, D2O): δ −11.22 (d, J = 20.7 Hz), −12.97 (d, J = 20.7 Hz); ESI-TOF HRMS m/z calcd for C16H24N5O14P2 [M − H]−1 572.0800, found 572.0728. dTDP-GlcNH2: 1H NMR (600 MHz, D2O): δ 7.55 (s, 1H, –CH
), 6.15 (t, J = 7.2 Hz, 1H, H-1), 5.40 (dd, J = 6.6, 3.0 Hz, 1H, H-1′), 4.58 (br s, 1H, H-3), 4.08 (br s, 1H, H-4), 4.01–3.96 (m, 2H, H-5a,b), 3.74–3.69 (m, 1H, H-5′), 3.67 (d, J = 12.6 Hz, 1H, H-6a′), 3.62–3.54 (m, 2H, H-3′, H-6b′), 3.36–3.31 (m, 1H, H-2′), 3.27 (t, J = 9.6 Hz, 1H, H-4′), 2.68–2.60 (m, 1H, H-2a), 2.38–2.30 (m, 1H, H-2b); 13C NMR (150 MHz, D2O): δ 158.89, 153.75, 151.34, 137.52, 116.11, 95.45 (d, J = 7.5 Hz, C-1′), 85.46 (d, J = 9.0 Hz, C-4), 83.49 (C-1), 72.70 (C-3′), 72.66 (C-5′), 71.49 (d, J = 9.0 Hz, C-2′), 71.22 (C-3), 69.07 (C-4′), 65.37 (C-5), 60.19 (C-6′), 38.37 (C-2); 31P NMR (243 MHz, D2O): δ −11.22 (d, J = 20.6 Hz), −12.99 (d, J = 20.6 Hz); ESI-TOF HRMS m/z calcd for C16H24N5O15P2 [M − H]−1 588.0750, found 588.0680. dADP-Glc: 1H NMR (600 MHz, D2O): δ 8.32 (s, 1H), 8.09 (s, 1H), 6.36 (t, J = 6.6 Hz, 1H, H-1), 5.42-5.39 (m, 1H, H-1′), 4.59 (s, 1H, H-3), 4.14 (s, 1H, H-4), 4.05–3.95 (m, 2H, H-5a,b), 3.71 (d, J = 9.6 Hz, 1H, H-5′), 3.66 (d, J = 12.6 Hz, 1H, H-6a′), 3.60 (t, J = 9.6 Hz, 1H, H-3′), 3.56 (dd, J = 12.6, 4.2 Hz, 1H, H-6b′), 3.33 (d, J = 9.6 Hz, 1H, H-2′), 3.26 (t, J = 9.6 Hz, 1H, H-4′), 2.70-2.64 (m, 1H, H-2), 2.47–2.41 (m, 1H, H-2); 13C NMR (150 MHz, D2O): δ 155.11, 152.05, 148.63, 139.97, 118.55, 95.45 (d, J = 6.6 Hz, C-1′), 85.65 (d, J = 9.0 Hz, C-4), 83.67 (C-1), 72.73 (C-5′), 72.67 (C-3′), 71.52 (d, J = 8.7 Hz, C-2′), 71.14 (C-3), 69.10 (C-4′), 65.35 (d, J = 6.0 Hz, C-5), 60.22 (C-6′), 38.97 (C-2); 31P NMR (243 MHz, D2O): δ −11.22 (d, J = 20.7 Hz), −12.97 (d, J = 20.7 Hz); ESI-TOF HRMS m/z calcd for C16H24N5O14P2 [M − H]−1 572.0800, found 572.0728. dTDP-GlcNH2: 1H NMR (600 MHz, D2O): δ 7.55 (s, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.17 (t, J = 7.2 Hz, 1H, H-1), 5.66 (dd, J = 6.6, 3.0 Hz, 1H, H-1′), 4.44 (br s, 1H, H-3), 4.05–3.98 (m, 3H, H-4, H-5a,b), 3.78–3.72 (m, 2H, H-3′, H-5′), 3.70 (d, J = 12.6 Hz, 1H, H-6a′), 3.64 (dd, J = 12.6, 3.6 Hz, 1H, H-6b′), 3.38 (t, J = 9.6 Hz, 1H, H-4′), 3.18 (br d, J = 10.2 Hz, 1H, H-2′), 2.25–2.16 (m, 2H, H-2a,b), 1.75 (s, 3H, –CH3); 13C NMR (150 MHz, D2O): δ 166.49, 151.64, 137.21, 111.62, 92.72 (d, J = 4.5 Hz, C-1′), 85.13 (d, J = 9.0 Hz, C-4), 84.94 (C-1), 73.11 (C-5′), 70.82 (C-3), 69.74 (C-3′), 68.94 (C-4′), 65.51 (d, J = 4.5 Hz, C-5), 59.86 (C-6), 54.04 (d, J = 9.0 Hz, C-2′), 38.49 (C-2), 11.54 (–CH3); 31P NMR (243 MHz, D2O): δ −11.34 (d, J = 21.0 Hz), −13.75 (d, J = 21.0 Hz); ESI-TOF HRMS m/z calcd for C16H26N3O15P2 [M − H]−1 562.0850, found 562.0776. dUDP-Gal: δ 7.80 (d, J = 7.8 Hz, 1H, –CH
), 6.17 (t, J = 7.2 Hz, 1H, H-1), 5.66 (dd, J = 6.6, 3.0 Hz, 1H, H-1′), 4.44 (br s, 1H, H-3), 4.05–3.98 (m, 3H, H-4, H-5a,b), 3.78–3.72 (m, 2H, H-3′, H-5′), 3.70 (d, J = 12.6 Hz, 1H, H-6a′), 3.64 (dd, J = 12.6, 3.6 Hz, 1H, H-6b′), 3.38 (t, J = 9.6 Hz, 1H, H-4′), 3.18 (br d, J = 10.2 Hz, 1H, H-2′), 2.25–2.16 (m, 2H, H-2a,b), 1.75 (s, 3H, –CH3); 13C NMR (150 MHz, D2O): δ 166.49, 151.64, 137.21, 111.62, 92.72 (d, J = 4.5 Hz, C-1′), 85.13 (d, J = 9.0 Hz, C-4), 84.94 (C-1), 73.11 (C-5′), 70.82 (C-3), 69.74 (C-3′), 68.94 (C-4′), 65.51 (d, J = 4.5 Hz, C-5), 59.86 (C-6), 54.04 (d, J = 9.0 Hz, C-2′), 38.49 (C-2), 11.54 (–CH3); 31P NMR (243 MHz, D2O): δ −11.34 (d, J = 21.0 Hz), −13.75 (d, J = 21.0 Hz); ESI-TOF HRMS m/z calcd for C16H26N3O15P2 [M − H]−1 562.0850, found 562.0776. dUDP-Gal: δ 7.80 (d, J = 7.8 Hz, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.18 (t, J = 6.6 Hz, 1H, H-1), 5.81 (d, J = 7.8 Hz, 1H, –CH
), 6.18 (t, J = 6.6 Hz, 1H, H-1), 5.81 (d, J = 7.8 Hz, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.45 (dd, J = 7.2, 3.6 Hz, 1H, H-1′), 4.50–4.44 (m, 1H, H-3), 4.08–3.98 (m, 4H, H-4, H-5a,b, H-5′), 3.88 (d, J = 3.0 Hz, 1H, H-4′), 3.76 (dd, J = 10.2, 3.0 Hz, 1H, H-3′), 3.67–3.63 (m, 1H, H-2′), 3.63–3.55 (m, 2H, H-6a,b′), 2.28–2.19 (m, 2H, H-2a,b); 13C NMR (150 MHz, D2O): δ 166.23, 151.55, 141.81, 102.40, 95.74 (d, J = 7.5 Hz, C-1′), 85.42 (d, J = 9.0 Hz, H-4), 85.32 (C-1), 71.81 (C-5′), 70.79 (C-3), 69.23 (C-3′), 69.01 (C-4′), 68.33 (d, J = 9.0 Hz, C-2′), 65.29 (d, J = 6.0 Hz, C-5), 60.91 (C-6′), 38.76 (C-2); 31P NMR (243 MHz, D2O): δ −11.20 (d, J = 19.4 Hz), −12.77 (d, J = 19.4 Hz); ESI-TOF HRMS m/z calcd for C15H24N2O16P2 [M − H]−1 549.0528, found 565.0449.
), 5.45 (dd, J = 7.2, 3.6 Hz, 1H, H-1′), 4.50–4.44 (m, 1H, H-3), 4.08–3.98 (m, 4H, H-4, H-5a,b, H-5′), 3.88 (d, J = 3.0 Hz, 1H, H-4′), 3.76 (dd, J = 10.2, 3.0 Hz, 1H, H-3′), 3.67–3.63 (m, 1H, H-2′), 3.63–3.55 (m, 2H, H-6a,b′), 2.28–2.19 (m, 2H, H-2a,b); 13C NMR (150 MHz, D2O): δ 166.23, 151.55, 141.81, 102.40, 95.74 (d, J = 7.5 Hz, C-1′), 85.42 (d, J = 9.0 Hz, H-4), 85.32 (C-1), 71.81 (C-5′), 70.79 (C-3), 69.23 (C-3′), 69.01 (C-4′), 68.33 (d, J = 9.0 Hz, C-2′), 65.29 (d, J = 6.0 Hz, C-5), 60.91 (C-6′), 38.76 (C-2); 31P NMR (243 MHz, D2O): δ −11.20 (d, J = 19.4 Hz), −12.77 (d, J = 19.4 Hz); ESI-TOF HRMS m/z calcd for C15H24N2O16P2 [M − H]−1 549.0528, found 565.0449.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, MS and NMR data and spectra of the synthetic (d)NDP-sugars. See DOI: 10.1039/d0ra05799a |
| ‡ These authors contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2020 |