Open Access Article
Open Access ArticlePurification and characterization of a thaumatin-like protein-1 with polyphenol oxidase activity found in Prunus mume†
Yuan Ma‡
 *,
Ming Luo‡,
Yingping Xu‡,
Yingjia Liu,
Xiaocui Liu,
Xiufang Bi
*,
Ming Luo‡,
Yingping Xu‡,
Yingjia Liu,
Xiaocui Liu,
Xiufang Bi ,
Yiping Yuan,
Fan Su and
Xiaocui Yin
,
Yiping Yuan,
Fan Su and
Xiaocui Yin
School of Food and Bioengineering, Xihua University, Chengdu 610039, P. R. China. E-mail: ymxhu@mail.xhu.edu.cn
First published on 4th August 2020
Abstract
Thaumatin-like protein-1 (TLP-1), a protein displaying high polyphenol oxidase (PPO) action and a member of the pathogenesis-related (PR) protein family, has a considerable influence on the enzymatic browning of Prunus mume (Chinese plum). In this assay, TLP-1 was identified and extracted from Prunus mume to investigate the protein's properties and better understand its contribution to the fruit's browning during storage or processing. The extracted TLP-1 was purified to apparent homogeneity using a procedure involving citrate phosphate buffer solution (CPBS) extraction, (NH4)2SO4 precipitation, dialysis in a cellulose bag, and ion exchange chromatography using a DEAE Sepharose Fast Flow column, while a Sephadex G-75 column was employed to facilitate gel filtration chromatography. Moreover, the enzyme was characterized in terms of its optimal pH and stability, isoelectric point (pI), molecular weight, optimal temperature and stability, enzyme kinetic parameters and substrate specificity, as well as inhibitor stability. This study indicated that the pI and molecular weight of TLP-1 was approximately 4.4 and 28 kDa, respectively, while 30 °C and 7.5 represented the respective optimal temperature and pH level for PPO catalysis. TLP-1 showed high affinity to catechol and pyrogallol, with Km values of 24.40 mM and 26.23 mM, respectively. Sodium bisulfite significantly inhibited TLP-1 activity. These findings on the properties of TLP-1 can contribute significantly to the search for ways to minimize the losses caused by fruit browning during the storage and processing of Prunus mume.
1. Introduction
Prunus mume Sieb. et Zucc., popularly known as Chinese plum, is one of the most important seasonal fruits in southern China. The fruit is small (reaching an average weight of approximately 18 g) and has a single round seed surrounded by light yellow or green flesh. With its extremely abundant nutrients, including phenolic compounds, carotenoids, organic acid, flavonoids and vitamin C,1 Prunus mume have anti-mutagenic, anti-carcinogenic and antioxidant activities,2 and offer significant medical and health-protection value. However, whether in storage or during processing, the post-harvest physiology of Prunus mumes consistently undergoes undesirable enzymatic browning, which may affect the fruit's flavor, color,3 nutritional and commercial values.4,5 Enzymatic browning occurs when phenolic compounds are catalyzed to quinones through the catalytic reaction of enzymes with polyphenol oxidase (PPO) active proteins in the presence of oxygen.6 PPO is the principal enzyme involved in enzymatic browning and has been previously identified in studies of numerous fruits and vegetables, including those with close genetic relationships to Prunus mumes, such as Prunus armeniaca L. (apricot)1 and Prunus armeniaca L. (Turkish Alyanak apricot),7 and other plants, such as the Napoleon grape8 and apple.9 However, limited information is available regarding the properties of the proteins related to enzymatic browning, and the significance of their defense functionality.The thaumatin-like protein (TLP-1), a protein displaying substantial PPO activity in Prunus mume. The name thaumatin defines this protein as the first delineated part of the Thaumatococcus daniellii Benth. group of pathogenesis-related (PR) proteins.10 Previous studies suggested that all organs contain PR-proteins throughout their formative phases, while substantially contributing to various developmental and defensive plant processes.11 Therefore, the goal of this work is the purification and characterization of TLP-1 from Prunus mume, and to characterize acidic/alkaline stability, thermal stability, as well as inhibitors effect were also performed. These findings are crucial in comprehending the foundation of a functional protein and will provide a better understanding of the TLP-1 in Prunus mume and may contribute to practical applications in the food industry.
2. Materials and methods
2.1 Materials and reagents
Prunus mume Sieb. et Zucc. were purchased at commercial maturity in Dazhou, Sichuan province in June. The pulps were cut from the fruits at 4 °C and frozen at −78 °C until used. Polyvinylpyrrolidone (PVPP, USP), catechol, polyethylene glycol (PEG 15000), and cellulose membranes (76 × 49 mm) were purchased from Yuanye (Shanghai, China). L-Cysteine (Biological reagent, BR) was purchased from Sigma-Aldrich (USA). Tris–HCl was purchased from Amresco (USA). 4-Hexylresorcinol (BR) was purchased from Aladdin (USA). SDS-PAGE Gel Kit and Tris were purchased from Solarbio (Beijing, China). Glycine (BR) was purchased from Biofroxx (Germany). Markers were purchased from Tiangen (China). Bovine albumin serum (BSA, BR) was purchased from Regal (Shanghai, China). All other reagents were purchased from KESHI (Chengdu, China), including the brilliant blue G250 (BR). All reagents and chemicals used in this study were of analytical grade, except where indicated otherwise.2.2 Extraction and purification of the enzymes
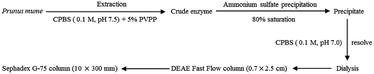
TLP-1 was extracted as described by Unal et al.7 with some modifications. The flow diagram of enzyme extraction and purification is shown in Fig. 1. Pre-cooled citrate-phosphate buffer solution (CPBS, 0.1 M, pH 7.5) with 5% PVPP (400 mL) was mixed with 200 g of frozen plum samples. The homogenate was centrifuged at 8000 × g for 20 min using an Eppendorf Centrifuge 5810R (Germany) at 4 °C. The centrifugal sedimentation procedure was repeated once and the two batches were then mixed. The crude enzyme displaying PPO activity was precipitated with solid ammonium sulfate at 80% saturation. Moreover, 0.01 M CPBS buffer at pH 7.0 was used to resolve the precipitate, including the residual ammonium sulfate, through dialysis in the same buffer for 36 h.The dialysis process was followed by loading the protein fraction onto a DEAE Fast Flow column (0.7 × 2.5 cm), using 0.01 M CPBS buffer at pH 7.0 for pre-equilibration. A linear gradient of the buffer comprising 0–2.0 M NaCl was employed at a 0.5 mL min−1 flow rate to facilitate elution of the protein. An ultrafiltration tube (Millipore Co., USA) with a 10 kDa pore size was used to concentrate the fractions displaying PPO action. The subsequent was then loaded onto a Sephadex G-75 column (10 × 300 mm) using 0.05 M CPBS buffer at pH 7.0 for pre-equilibration. The same elution conditions were used for protein gel filtration chromatography. Furthermore, fractions exhibiting the highest PPO activity were centrifuged using an ultrafiltration tube (Millipore Co., USA) with a 10 kDa pore size to achieve the required concentration, then they were freeze-dried and stored at −18 °C.
2.3 Protein content
The Bradford method with BSA (bovine albumin serum) as the standard was used to measure the protein concentrate.122.4 PPO activity assay
A UV 2800 spectrophotometer (Shunyu Hengping Co., China) was employed to evaluate the PPO activity by observing the absorbance change at 410 nm. Furthermore, the standard reaction solution comprised 0.5 mL of the protein solution (5 μg mL−1) dissolved in 0.01 M CPBS buffer (pH 7.0), 0.5 mL of 0.05 M CPBS buffer (pH 7.0) and 2.0 mL of 0.1 M catechol dissolved in 0.01 M CPBS buffer (pH 7.0). One unit of enzyme activity was defined as the amount of enzyme that increases 0.001 of absorbance per minute per milliliter of protein solution. The PPO activity assay was performed in triplicate at 25 °C.132.5 Polyacrylamide gel electrophoresis (SDS-PAGE)
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method delineated by Laemmli et al.,14 which included a discontinuous buffer system containing 12% separating gel and 5% stacking gel. A 5× protein loading dye mixture (Biosharp, China) and a heating process with boiling water for 4 min were employed to facilitate the denaturation of the enzyme solutions. This process was followed with placing the enzyme markers (5 μL) and samples (20 μL) into the stacking gel wells and performing electrophoresis at 160 V in the separating gel and 80 V in the stacking gel. Subsequently, Coomassie Brilliant Blue R-250 was used to stain the gels, after which distaining in a 10% acetic acid solution. The respective markers (Tiangen, China) denoting the molecular weight calibration were represented by 94.0 kDa, 66.2 kDa, 45.0 kDa, 33.0 kDa, 26.0 kDa, 20.0 kDa, and 14.4 kDa.2.6 Determination of isoelectric point
The isoelectric point (pI) of Prunus mume enzymes was determined according to the turbidimetry method by Lu et al.,15 with minor modifications. The elution of the sample was monitored by A280 nm readings. The eluted sample comprised 1 mL protein mixed with 25 mL distilled water, to which 0.1 M acetic acid was gradually added, buffer solutions with a pH range of 3.6–5.8 added to test tubes, respectively. The light absorbencies of each sample at the wavelengths of 280 nm were detected with a spectrometer. The pH of the solution with the largest absorbency is the isoelectric point.2.7 Liquid chromatography-mass spectrometry (LC-MS/MS) measurement
An LC-MS/MS instrument was employed to identify and analyze the protein displaying PPO activity. Trypsin was used to digest the protein band removed from the gel, which was placed into an ionization chamber, and evaluated as a different peptide sequence. The ExPASy website (http://expasy.org/) was used to approximate the secondary structure arrangement of the protein exhibiting PPO activity. This estimation was established according to the amino acid sequence of the identified protein.102.8 Fluorescence spectral measurement
Fluorescence spectral measurements were performed with a FluoroMax-4 sluorescence spectrophotometer (HORIBA, Japan). To accomplish this, 0.3 mg mL−1 protein solutions were prepared with 10 mM sodium citrate buffer at pH 7, and measured at an emission wavelength of 350 nm to determine the maximum excitation wavelength. This calculation was used during the scanning process to document the emission spectra. The following operating conditions pertaining to the excitation spectrum were used: kex 280 nm, and kem 300–400 nm; the Em slit and Ex slit were both set at 5 nm; the scan speed was set at 200 nm min−1.2.9 Determining optimal pH and pH stability
Combining 2.0 mL of 0.1 M catechol with 0.5 mL of the appropriate buffers (50 mM CPBS buffer for pH 2.0–9.0, 50 mM sodium citrate buffer (SCB) for pH 2.0–7.0 and 50 mM phosphate buffer solution (PBS) for pH 6.0–9.0), and 0.5 mL protein (5 μg mL−1), were examined at 25 °C to identify the optimum pH levels required for maximum PPO activity. The protein in the buffers indicated above was incubated for 24 h at 4 °C to ascertain the pH stability of the enzyme.2.10 Determining optimal temperature and temperature stability
The addition of the protein to preheated reaction buffer at different temperatures ranging from 10 to 80 °C to evaluate the PPO activity. Immediate spectrophotometric measurements were performed on the mixture at 410 nm. The tubes containing 0.5 mL protein (5 μg mL−1) and 0.5 mL of 50 mM CPBS buffer (pH 7.0), were incubated at various temperatures of 40 to 80 °C in a water bath for 10–60 min, respectively, to determine the thermal stability of the PPO activity. The samples were then rapidly cooled to room temperature, followed by the addition of 2.0 mL of 0.1 M catechol, and measurement of the remaining activity in the samples.2.11 Substrate specificity and kinetic parameters
The Michaelis–Menten constant (Km) was determined as follows: catechol, pyrogallol, 4-methylcatechol, phenol and p-hydroxybenzoic acid were dissolved as substrates in 50 mM CPBS buffer (pH 7.0) at concentrations of 0.01 to 0.1 M, respectively. While gallic acid and 5-sulfosalicylic acid that was dissolved as a substrate in 50 mM phosphate buffer (pH 7.0) at concentrations of 0.01 to 0.05 M, respectively. Furthermore, ferulic acid was dissolved as a substrate in 50 mM CPBS buffer (pH 7.0) at a concentration of 0.01 M. The reciprocal lines presented by a Lineweaver–Burk plot was used to determine the Km and Vmax values. With 1/[S] as the abscissa and 1/V denoting the vertical axis, the kinetic data were drawn as 1/substrate concentration (1/[S]) versus 1/specific activity (1/V). Therefore, 1/Km represented the abscissa value, and 1/Vmax denoted the vertical axis value, while the optimum protein substrate was acquired from the constant Km.2.12 The effect of inhibitors on PPO activity
The level of PPO activity was determined in the presence of seven distinct inhibitors (L-glutamine, L-cysteine, sodium sulfite, sodium bisulfite, ethylene diamine tetraacetic acid (EDTA), 4-hexylresorcinol and ascorbic acid) at four concentrations (0.1 mM, 1 mM, 5 mM, and 10 mM) resolved in 0.05 M CPBS buffer (pH 7.0). The activity of the control (the PPO activity of the enzyme without inhibitor) is considered as 100% and the PPO activity is expressed as relative activity. The PPO activity was assayed as 0.2 mL inhibitors (0.1 mM, 1 mM, 5 mM, and 10 mM), 0.2 mL protein (5 μg mL−1), and 2.6 mL of 0.1 M catechol as substrate.2.13 Statistical analysis
The statistical tests were analyzed using Microcal Origin8.6 software (OriginLab Corporation, MA, USA). One-way analysis of variance (ANOVA) was used to evaluate significant (P < 0.05) of differences between samples using IBM SPSS version 17.0 (IBM/SPSS Inc., USA). Every experiment was repeated three times.3. Results and discussion
3.1 Extraction and purification
Fig. 2a indicates that two major protein peaks were eluted with the increase of conductivity, with the second peak much higher than the first. The 10th and 11th tubes with the highest activity under the second peak were collected for gel filtration chromatography on Sephadex G-75 using the AKTA system. Fig. 2b presents the elution profiles of TLP-1, separated only by one significantly high and wide peak. The enzyme activity peak plotted had a high coincidence with the first half protein peak, so the 11th tube was collected for the characterization studies described below. Finally, as shown in Table 1, TLP-1 was successfully purified 122.22-fold with a 57.15% yield and distinct activity in 100 mM catechol of 404![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 033.02 U mg−1.
033.02 U mg−1.
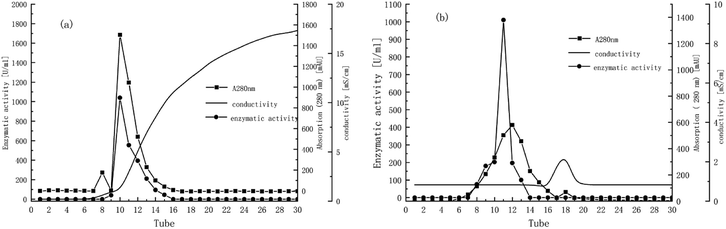 | ||
| Fig. 2 (a) Elution profile of Prunus mume TLP-1 from DEAE-FF Fast Flow. (b) Elution profile of Prunus mume TLP-1 from Superdex G-75. | ||
| Purification steps | Total activity (U) | Total protein (mg) | Specific activity (U mg−1 protein) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 942.40 942.40 |
22.67 | 3305.80 | 1.0 | 100.0 |
| 80% (NH4)2SO4 | 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859.69 859.69 |
10.57 | 6798.46 | 2.06 | 95.89 |
| DEAE Sepharose Fast Flow | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 678.50 678.50 |
0.38 | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 043.19 043.19 |
12.42 | 20.92 |
| Sephadex G-75 | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 827.50 827.50 |
0.11 | 404![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 033.02 033.02 |
122.22 | 57.15 |
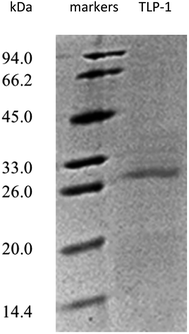
Following the final purification, SDS-PAGE was employed to evaluate the purity of the protein displaying PPO activity. Fig. 3 demonstrates only one visible band containing gel that was stained with Coomassie Brilliant Blue R-250, denoting protein homogeneity. The SDS-PAGE results suggested that the molecular weight of the purified protein with the PPO activity approximately 28 kDa.
3.2 Isoelectric point determination
As shown in Fig. S1†, the isoelectric point of TLP-1 in Prunus mume was found to be 4.4. This concurs with the findings of Hu,16 who reported the isoelectric point of TLP and PR-8 in apple as 5.03 and 4.03, respectively, and Cheng et al.,11 who isolated and purified germin-like protein (GLP) exhibiting PPO activity from tangerine and found the isoelectric point to be 4.48, which is similar to the measured isoelectric point of TLP-1. The measured results suggest that the TLP-1 with PPO activity isolated possesses a similar content of acidic side chains to the enzyme with PPO activity previously reported generally.3.3 LC-MS/MS determination
LC-MS/MS was utilized to evaluate the band on the SDA-PAGE gel. Consequently, the protein was identified as TLP-1 in Prunus mume with 198 points (P < 0.05) as the highest corresponding score, a protein number of gi|645246258, a molecular weight of 28 kDa. This similar to the TLP-1 previously reported in many other plant sources, such as banana (22.1 kDa),17 Pinus sylvestris seedlings (24.6 kDa),18 banana pulp (20 kDa).19 All TLP-1s isolated to date including isolated in this study possess molecular weights, existing a consistent trend, are above 20 kDa. The differences between these results might be due to the different of plant materials. And a predicted pI of 4.94, which is close to the above-measured 4.4. The difference between the isoelectric point value measured by the test and that predicted by the software may be related to the detect conditions and methods, which would affect aggregation morphology of proteins.Fig. 4 indicates that some TLP-1 peptides in Prunus mume display high correspondence to those found in Prunus persica (gi|595941388), Prunus avium (gi|1220087497), and Malus domestica (gi|83853953). It is evident that TLP-1 from different sources share a high degree of identity, which suggests that they may also share similar response attributes to plant defense elicitors. TLP-1 has been termed one of the pathogenesis-related (PR) proteins, which included those that impair the growth of fungal hyphae,20 peroxidases involved in cell wall fortification, disease resistance21 those that participate in enzymatic browning, as well as proteins with as yet unidentified functions.
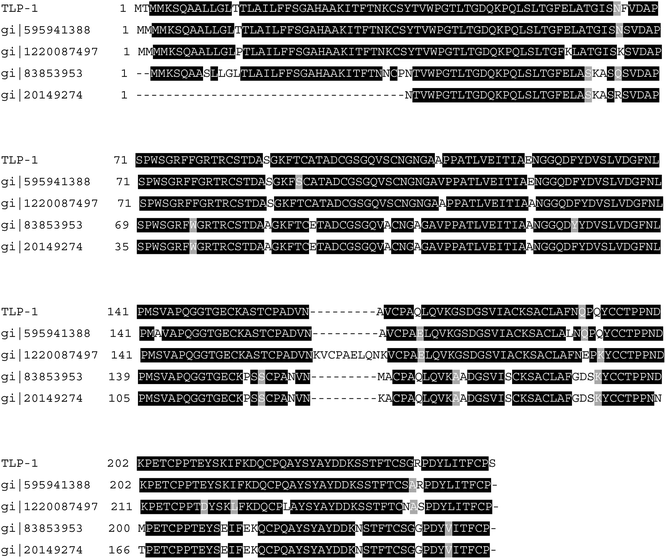 | ||
| Fig. 4 Alignment of sequenced peptides of the TLP-1 gained from Prunus mume onto the TLP-1 of Prunus persica, Prunus avium and Malus domestica. | ||
3.4 Structural analysis of TLP-1
Fig. S2† shows that the λmax of TLP-1 is 348 nm with a high intensity of 348![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 522, suggesting that residual Trp in the TLP-1 reside in a hydrophobic non-polar environment, embedded within the protein interior. This is consistent with the reports of Cheng et al.,11 who measured the maximum emission wavelength of PPO active proteins TLP and PR-8 in apples as 350.84 nm and 345.84 nm, respectively, and the maximum emission wavelength of PPO active GLP in honey oranges as 314 nm.11 Marzban et al.22 found that TLP-1 of MALD2 in apple has two maximum fluorescence emission peaks at room temperature, respectively located at 336 nm and 350 nm, indicating that TLP-1 in MALD2 is located in at least two different environments, one in the non-polar (336 nm) and the other in the polar (350 nm) area.
522, suggesting that residual Trp in the TLP-1 reside in a hydrophobic non-polar environment, embedded within the protein interior. This is consistent with the reports of Cheng et al.,11 who measured the maximum emission wavelength of PPO active proteins TLP and PR-8 in apples as 350.84 nm and 345.84 nm, respectively, and the maximum emission wavelength of PPO active GLP in honey oranges as 314 nm.11 Marzban et al.22 found that TLP-1 of MALD2 in apple has two maximum fluorescence emission peaks at room temperature, respectively located at 336 nm and 350 nm, indicating that TLP-1 in MALD2 is located in at least two different environments, one in the non-polar (336 nm) and the other in the polar (350 nm) area.
3.5 Optimal pH and pH stability
As shown in Fig. 5a, the enzyme presented its highest activity at pH 7.5 (91.06 ± 0.28%). The relative PPO enzyme activity from Prunus mume displayed a gradual increase in the pH range from 2.0 to 7.5, followed by this decrease in the pH range from 7.5 to 9.0. PPO activity in the CPBS system was higher than that in the other buffer systems. In addition, TLP-1 maintained a higher level of PPO activity, in both the neutral and alkaline environments. The least significant values of the relative PPO enzyme activity in CPBS and PBS were obtained at pH 2.0 and pH 6.0, respectively. Cheng et al.11 indicated that a pH value of 6.5 was optimal for GLP from satsuma mandarin with catechol as a substrate, which is below the values obtained in this study. The reason might due to the optimum pH for an enzyme with PPO activity is dependent on the source of the enzyme, as well as experimental elements such as extraction techniques, the nature of the phenolic substrates, and the buffer system used during identification. Derardja et al.1 reported an optimum pH at acidity for PPOs from Prunus armeniaca L. (apricot). Kumar et al.23 reported that a neutral pH was optimal for PPOs from Malpighia glabra L. (Barbados cherry). This suggests that the enzymatic characteristics of PPO may be varied, even in fruits and vegetables with a close genetic relationship.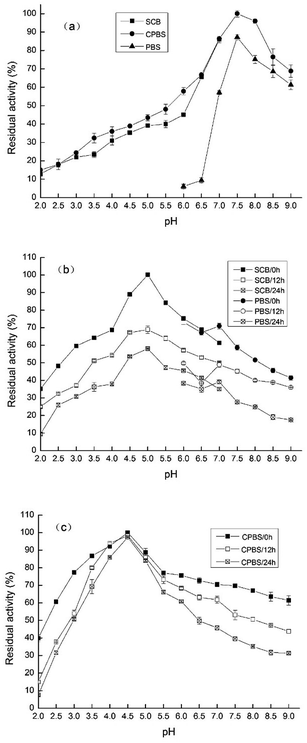 | ||
| Fig. 5 (a) PPO activity of TLP-1 at different pH. (b) PPO activity of TLP-1 in SCB-PBS incubated for 0, 12, 24 h. (c) PPO activity of TLP-1 in CPBS incubated for 0, 12, 24 h. | ||
In both systems, the PPO activity of TLP-1 increased initially, then declined with the increase of pH value, but remained consistently active between pH 2 to 9, even when incubated for 24 h. The relative enzymatic activity value with the highest significance was obtained at pH 4.5 in CPBS (Fig. 5b) and at pH 5.0 in SCB-PBS (Fig. 5c). TLP-1 from Prunus mume maintained a higher pH stability in the CPBS system, when incubated for 24 h at pH 4.0 to 5.0. In strongly acidic or alkaline conditions, peptide hydrolysis, deamination, β-elimination, racemization and other amino acid changes may decrease enzyme activity. Moreover, Maria et al.24 indicated that the pH range of an enzyme was probably close to its pI, and partially denatured TLP-1 transfers its embedded active center to the surface or exposes the hydrophobic group to enhance its ability to bind to the substrate, which are similar to those findings of this study.
3.6 Optimal temperature and temperature stability
As shown in Fig. 6a, the enzyme presented its optimum temperature as 30 °C. PPO from Prunus mume maintained high activity levels at low temperatures, ranging from 10 to 40 °C. However, at higher temperatures, the relative enzyme activity was decreased.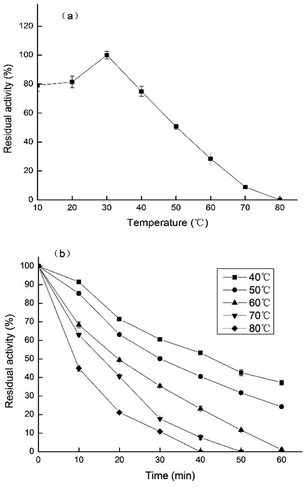 | ||
| Fig. 6 (a) Effects of different temperatures on PPO activity of TLP-1. (b) PPO activity of TLP-1 incubated at different temperatures. | ||
The temperature stability of PPO extracted from Prunus mume, was established by incubating the enzyme solutions at five different temperatures (40–80 °C) for 60 min, and measuring the enzyme activity every 10 min. The results, shown in Fig. 6b, indicate that the highest values of enzymatic activity was obtained at 40 °C, after being treated for 60 min. TLP-1 extracted from Prunus mume exhibited relatively high stability below 50 °C, while the PPO activity decreased significantly between 70 °C and 80 °C, which similar to the research results of Ionita et al.25 The differences on the enzymatic activity induced by these different temperatures could be explained by changes in the ionization of the amino acid residues in the active site of the enzyme or its texture.26
3.7 Substrate specificity and kinetic enzyme parameters
As shown in Fig. 7a, the enzyme showed the highest activity towards catechol considered as 100% in comparing its activity with other substrates. 4-Methylcatechol and pyrogallol as substrates presented lower enzymatic activity, however, the PPO exhibited no activity towards phenol, p-hydroxybenzoic, or gallic acid. Therefore, this study ascertained that TLP-1 could oxidize diphenolic and triphenolic substrates, but was unable to oxidize monophenolic substrates, which consistent with the research results of Siddiq et al.27 and Liu et al.28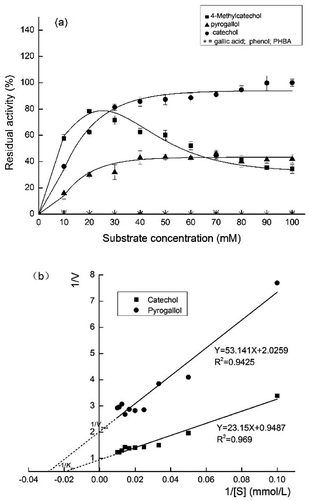 | ||
| Fig. 7 (a) Effects of different substrate concentrations on PPO activity of TLP-1. (b) Lineweaver–Burk plots of catechol and pyrogallol oxidation by PPO activity of TLP-1. | ||
As shown in Fig. 7b, the Km and Vmax of the reactions using catechol and pyrogallol were determined by plotting the activities at optimum pH and temperature as a function of the substrate concentration. The highest kinetic parameter value was provided by catechol (Km 24.40 mmol L−1 and Vmax 1.05 Δ 410 nm min−1), while pyrogallol (Km 26.23 mmol L−1 and Vmax 0.49 Δ 410 nm min−1) produced the lowest values. PPO extracted from Prunus mume had a stronger affinity for diphenol than for triphenol. Other researchers have reported the same result as Ioniţă et al.,25 whose study of the PPO substrate specificity obtained from Prunus domestica (plum) indicated its highest activity for catechol (Vmax 1193.4 ± 68.05 OD per min and Km 26.3 ± 4.4 mmol L−1) followed by 4-methyl catechol (Vmax 792.7 ± 46.85 OD per min and Km 15.5 ± 2.85 mmol L−1). Kumar et al.23 recorded the highest values for the PPO kinetic parameters from Barbados cherry for catechol and 4-methyl catechol, while the least significant values were produced by pyrogallol.
3.8 The effect of inhibitors on PPO activity
As shown in Table 2, the inhibitors employed for investigative purposes in this study, and placed in relative order of inhibition: sodium bisulfite > ascorbic acid > L-cysteine > sodium sulfite > 4-hexylresorcinol > EDTA > L-glutamine. The enzyme activity was complete inhibited with sodium bisulfite, ascorbic acid and L-cysteine at concentrations above 1 mM. The inhibitory effect of ascorbic acid on PPO can be mainly attributed to reducing agent that acts as an antioxidant by reducing quinones to form stable and colorless products.29 The inhibition effect of sodium sulfite on PPO activity was relatively slightly weaker. Sodium sulfite can form colorless substances with quinones, therefore, widely used as a color protection agent in fruit and vegetable processing.30 L-Cysteine, a thiol compound, strongly inhibited the enzyme, which can form stable colorless products by addition reactions with quinones or by binding to the active center of enzyme.31 Here, the lowest inhibitory effect was observed for 4-hexylresorcinol, EDTA and L-glutamine, at the highest concentration of 10 mM. The inhibition effect of 4-hexylresorcinol observed here is at variance with the research result of Derardja et al.,1 who determined the effects of ten inhibitors for Prunus armeniaca L. (apricot). However, EDTA is well-known as a metal ion chelator but displayed low specificity for copper, which indicates TLP-1 is a metalloprotease but may has a different active site from PPO, or has multiple active sites. Similar results have been reported for a purified enzyme obtained from Serratia marcescens.32| Inhibitors | Relative activity (%) | |||
|---|---|---|---|---|
| 0.1 (mM) | 1 (mM) | 5 (mM) | 10 (mM) | |
| a Mean value ± standard deviation (SD) (%). a, b, c, d Values per column without common superscripts were significantly different (P < 0.05). | ||||
| Sodium bisulfite | 77.93 ± 0.81a | 0.00 ± 0.00b | 0.00 ± 0.00b | 0.00 ± 0.00b |
| Ascorbic acid | 84.03 ± 0.44a | 0.00 ± 0.00b | 0.00 ± 0.00b | 0.00 ± 0.00b |
| L-Cysteine | 91.75 ± 0.42a | 0.00 ± 0.00b | 0.00 ± 0.00b | 0.00 ± 0.00b |
| Sodium sulfite | 91.86 ± 0.68a | 63.37 ± 0.38b | 0.00 ± 0.00c | 0.00 ± 0.00c |
| 4-Hexylresorcinol | 86.26 ± 1.13a | 69.97 ± 1.08b | 31.65 ± 1.06c | 15.57 ± 0.44d |
| EDTA | 87.66 ± 0.87a | 84.39 ± 1.37b | 77.04 ± 0.06c | 55.94 ± 0.65d |
| L-Glutamine | 90.64 ± 0.65a | 87.59 ± 1.02b | 86.72 ± 0.73b | 83.46 ± 2.24c |
4. Conclusions
This study isolated a protein with polyphenol oxidase activity, identified as thaumatin-like protein (TLP-1). Characterization indicates that the purified enzyme has the character of a metalloprotease and a high activity at physiological conditions of appropriate pH and temperature, which follows a Michaelis–Menten kinetic.In conclusion, this study demonstrates that purified TLP-1 from Prunus mume exhibits polyphenol oxidase activity with a defining enzymatic characteristic. This finding contributes to improve understanding of the TLP-1, revealing greater scope for the development of methodologies to inactivate enzymes, which may be applied to food processing. Future work will investigate the effects of temperature, ultrasound and pressure on TLP-1 in its activity, structure and the enzymatic browning mechanism.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Sichuan Science and Technology Program [grant number 2018ZR0283]; Sichuan Science and Technology Poverty Alleviation Project [grant number 2018NFP0007]; Sichuan Science and Technology Innovation Miaozi Project [grant number 2019060]; and Innovation Fund of Postgraduate, Xihua University [grant number ycjj2019074].Notes and references
- A. E. Derardja, M. Pretzler, I. Kampatsikas, M. Barkat and A. Rompel, Purification and Characterization of Latent Polyphenol Oxidase from Apricot (Prunus armeniaca L.), J. Agric. Food Chem., 2017, 65, 8203–8212 CrossRef CAS PubMed.
- A. D. Caro, A. Piga, V. Vacca and M. Agabbio, Changes of flavonoids, vitamin C and antioxidant capacity in minimally processed citrus segments and juices during storage, Food Chem., 2004, 84, 99–105 CrossRef.
- Z. Q. Gong, D. J. Li, C. Q. Liu, C. Q. Liu, A. W. Cheng and W. L. Wang, Partial purification and characterization of polyphenol oxidase and peroxidase from chestnut kernel, LWT--Food Sci. Technol., 2015, 60, 1095–1099 CrossRef CAS.
- J. J. Wu, J. Y. Gao, H. B. Chen, X. Liu, W. Cheng, X. J. Ma and P. Tong, Purification and Characterization of Polyphenol Oxidase From Agaricus bisporus, Int. J. Food Prop., 2013, 16, 1483–1493 CrossRef CAS.
- J. Y. Shi, J. X. Li, S. H. Zhu and J. Zhou, Browning inhibition on fresh-cut chestnut kernel by exogenous nitric oxide, Int. J. Food Sci. Technol., 2011, 46, 944–950 CrossRef CAS.
- G. P. Orozco, N. A. M. Hernandez, J. G. Sampedro and H. Najera, Purification and partial biochemical characterization of polyphenol oxidase from mango (Mangifera indica cv. Manila), J. Agric. Food Chem., 2014, 62, 9832–9840 CrossRef PubMed.
- M. U. Unal and A. Sener, Two-year comparison of the biochemical properties of polyphenol oxidase from Turkish Alyanak apricot (Prunus armenica L.), Food Chem., 2016, 190, 741–747 CrossRef CAS PubMed.
- E. N. Delicado, M. S. Megias, A. J. P. Lopez and J. M. L. Nicolas, Characterization of polyphenol oxidase from Napoleon grape, Food Chem., 2007, 100, 108–114 CrossRef.
- M. Deirdre, N. Eidhin, E. Murphy and D. O'Beirne, Polyphenol Oxidase from Apple (Malus domestica Borkh. cv Bramley's Seedling): Purification Strategies and Characterization, J. Food Sci., 2006, 71, C51–C58 CrossRef.
- J. Hejgaard, S. Jacobsen and I. Svendsen, Two antifungal thaumatin-like proteins from barley grain, Fed. Eur. Biochem. Soc., Lett., 1991, 291, 127–131 CrossRef CAS.
- X. Cheng, X. J. Huang, S. Y. Liu, M. Tang, W. F. Hu and S. Y. Pan, Characterization of germin-like protein with polyphenol oxidase activity from Satsuma mandarine, Biochem. Biophys. Res. Commun., 2014, 449, 313–318 CrossRef CAS PubMed.
- R. A. Asryants, I. V. Duszenkova and N. K. Nagradova, Determination of Sepharose-bound protein with Coomassie brilliant blue G-250, Anal. Biochem., 1985, 151, 571–574 CrossRef CAS PubMed.
- A. Sener, M. U. Unal and S. Aksay, Purification and Characterization of Polyphenol Oxidase from Goldnugget Loquat (Eriobotrya Japonica Cv. Goldnugget), J. Food Biochem., 2011, 35, 1568–1575 CrossRef CAS.
- U. K. Laemmli, F. Beguin and G. G. Kellenberger, A factor preventing the major head protein of bacteriophage T4 from random aggregation, J. Mol. Biol., 1970, 47, 69–85 CrossRef CAS PubMed.
- L. Yang and P. B. Xian, Comparative Study of Determining Iep and Iep Distribution of Gelatin with Turbidimetry_Spectrometry and Capillary Iep Focusing Methods, J. Grad. Sch. Chin. Acad. Sci., 2006, 24, 114–118 Search PubMed.
- W. F. Hu, Purification and characterization of pathogenesis-related proteins in apple (Malus domestica) and effect of high pressure carbon dioxide on their activity and structure, J. China Agric. Univ., 2011, 47–48 Search PubMed.
- W. X. Jiao, X. X. Li, H. D. Zhao, J. K. Cao and W. B. Jiang, Antifungal Activity of an Abundant Thaumatin-Like Protein from Banana against Penicillium expansum, and Its Possible Mechanisms of Action, Molecules, 2018, 23, 1442–1458 CrossRef PubMed.
- I. Šņepste, V. Šķipars, B. Krivmane, L. Brūna and D. Ruņgis, Characterization of a Pinus sylvestris thaumatin-like protein gene and determination of antimicrobial activity of the in vitro expressed protein, Tree Genet. Genomes, 2018, 14, 58–71 CrossRef.
- N. Yasmin, M. Saleem, M. Naz, R. Gul and H. M. Rehman, Molecular Characterization, Structural Modeling, and Evaluation of Antimicrobial Activity of Basrai Thaumatin-Like Protein against Fungal Infection, BioMed Res. Int., 2017, 2017, 1–9 CrossRef PubMed.
- V. S. M. Ho, J. H. Wong and T. B. Ng, A thaumatin-like antifungal protein from the emperor banana, Peptides, 2007, 28, 760–766 CrossRef CAS PubMed.
- X. X. Yan, H. B. Qiao, X. M. Zhang, C. L. Guo, M. N. Wang, Y. J. Wang and X. P. Wang, Analysis of the grape (Vitis vinifera L.) thaumatin-like protein (TLP) gene family and demonstration that TLP29 contributes to disease resistance, Sci. Rep., 2017, 7, 4269–4283 CrossRef PubMed.
- G. Marzban, A. Herndl, S. Pietrozotto, S. Banerjee, C. Obinger, F. Maghuly, R. Hahn, D. Boscia, H. Katinger and M. Laimer, Conformational changes of Mal d 2, a thaumatin-like apple allergen, induced by food processing, Food Chem., 2009, 112, 803–811 CrossRef CAS.
- V. B. A. Kumar, T. C. K. Mohan and K. Murugan, Purification and kinetic characterization of polyphenol oxidase from Barbados cherry (Malpighia glabra L.), Food Chem., 2008, 110, 328–333 CrossRef CAS PubMed.
- G. Maria, Quick identification of a simple enzyme deactivation model for an extended-Michaelis-Menten reaction type. Exemplification for the D-glucose oxidation with a complex enzyme deactivation kinetics, Comput. Chem. Eng., 2016, 93, 323–330 CrossRef CAS.
- E. Ionita, L. Gurgu, I. Aprodu, N. Stanciuc, I. Dalmadi, G. Bahrim and G. Rapeanu, Characterization, purification, and temperature/pressure stability of polyphenol oxidase extracted from plums (Prunus domestica), Process Biochem., 2017, 56, 177–185 CrossRef CAS.
- H. P. Modarres, M. R. Mofrad and A. Sanati-Nezhad, Protein thermostability engineering, RSC Adv., 2016, 6, 115252–115270 RSC.
- M. Siddiq and K. D. Dolan, Characterization of polyphenol oxidase from blueberry (Vaccinium corymbosum L.), Food Chem., 2017, 218, 216–220 CrossRef CAS PubMed.
- N. N. Liu, W. Liu, D. J. Wang, Y. B. Zhou, X. J. Lin, X. Wang and S. B. Li, Purification and partial characterization of polyphenol oxidase from the flower buds of Lonicera japonica Thunb, Food Chem., 2013, 138, 478–483 CrossRef CAS PubMed.
- M. Ü. Ünal and A. Şener, Determination of some biochemical properties of polyphenol oxidase from Emir grape (Vitis vinifera L. cv. Emir), J. Sci. Food Agric., 2006, 86, 2374–2379 CrossRef.
- K. Bravo and E. Osorio, Characterization of polyphenol oxidase from Cape gooseberry (Physalis peruviana L.) fruit, Food Chem., 2016, 197, 185–190 CrossRef CAS PubMed.
- K. N. Waliszewski, O. Márquez and V. T. Pardio, Quantification and characterisation of polyphenol oxidase from vanilla bean, Food Chem., 2009, 117, 196–203 CrossRef CAS.
- A. Krishnamurthy and P. D. Belur, A novel fibrinolytic serine metalloprotease from the marine Serratia marcescens subsp. sakuensis: purification and characterization, Int. J. Biol. Macromol., 2018, 112, 110–118 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05659f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |