Open Access Article
Open Access ArticleIntegrated glycomics strategy for the evaluation of glycosylation alterations in salivary proteins associated with type 2 diabetes mellitus†
Hanjie Yu‡
 a,
Junhong Wang‡b,
Zhen Tanga,
Xia Lia,
Mengqi Yina,
Fan Zhanga,
Jian Shua,
Wentian Chena,
Shuang Yang
a,
Junhong Wang‡b,
Zhen Tanga,
Xia Lia,
Mengqi Yina,
Fan Zhanga,
Jian Shua,
Wentian Chena,
Shuang Yang *c and
Zheng Li*a
*c and
Zheng Li*a
aLaboratory for Functional Glycomics, College of Life Sciences, Northwest University, No. 229 Taibai Beilu, Xi'an, 710069, China. E-mail: zhengli@nwu.edu.cn
bDepartment of Endocrinology, Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an, 710004, China
cDepartment of Pharmaceutical Analysis, School of Pharmaceutical Sciences, Soochow University, Suzhou, Jiangsu, China. E-mail: Yangs2020@suda.edu.cn
First published on 1st November 2020
Abstract
Glycosylation is involved in several biological processes, and its alterations can reflect the process of certain diseases. Type 2 diabetes mellitus (T2DM) has attained the status of a global pandemic; however, the difference in salivary protein glycosylation between healthy subjects and patients with T2DM has not been fully understood. In the present study, salivary specimens from patients with T2DM (n = 72) and healthy volunteers (HVs, n = 80) were enrolled and divided into discovery and validation cohorts. A method combining the lectin microarray and lectin blotting was employed to investigate and confirm the altered glycopatterns in salivary glycoproteins. Then, lectin-mediated affinity capture of glycoproteins and MALDI-TOF/TOF-MS were performed to obtain the precise structural information of the altered glycans. As a result, the glycopatterns recognized by 5 lectins (LEL, VVA, Jacalin, RCA120 and DSA) showed significant alteration in the saliva of T2DM patients. Notably, the glycopattern of Galβ-1,4GlcNAc (LacNAc) recognized by LEL exhibited a significant increase in T2DM patients compared to HVs in both discovery and validation cohorts. The MALDI-TOF/TOF-MS results indicated that there were 10 and 7 LacNAc-containing N/O-glycans (e.g. m/z 1647.586, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 688.613 and 1562.470) that were identified only in T2DM patients. Besides, the relative abundance of 3 LacNAc-containing N-glycans and 10 LacNAc-containing O-glycans showed an increase in the glycopattern in T2DM patients. These results indicated that the glycopattern of LacNAc is increased in salivary glycoproteins from T2DM patients, and an increase in LacNAc-containing N/O-glycans may contribute to this alteration. Our findings provide useful information to understand the complex physiological changes in the T2DM patients.
688.613 and 1562.470) that were identified only in T2DM patients. Besides, the relative abundance of 3 LacNAc-containing N-glycans and 10 LacNAc-containing O-glycans showed an increase in the glycopattern in T2DM patients. These results indicated that the glycopattern of LacNAc is increased in salivary glycoproteins from T2DM patients, and an increase in LacNAc-containing N/O-glycans may contribute to this alteration. Our findings provide useful information to understand the complex physiological changes in the T2DM patients.
1. Introduction
The global prevalence of T2DM is rapidly increasing, and the number of people with T2DM worldwide has more than doubled over the past three decades.1–3 Epidemiological studies indicate that chronic hyperglycemia, insulin resistance, and a relative insulin secretion defect are the characteristics of T2DM. Furthermore, overweight, obesity,4–6 genetic susceptibility,7–9 lifestyle and environmental risk factors10,11 are important risk factors for T2DM. Although it is known that the pathogenesis of T2DM can be described simplistically as problems with insulin resistance and compensatory hyperinsulinemia, the mechanism of T2DM also remains to be determined.As one of the most complex and important body fluids, saliva is secreted by three pairs of major glands and other minor submucosal salivary glands. The proteins in saliva are derived via two ways: secreted from salivary glands or from blood via passive diffusion, active transport.12,13 Recent studies have elucidated that saliva can be seen in many cases as a reflection of the physiological function of the body and can be used for the non-invasive detection of diseases such as Sjogren's syndrome, cystic fibrosis and cancer.14–17 Our previous studies have demonstrated that the glycosylation of salivary glycoproteins showed alterations between healthy volunteers and cancer patients. Furthermore, the diagnostic models constructed based on these alterations showed a good diagnostic performance in breast and gastric cancer detection.18,19
Glycosylation is one of the most common and complex forms of post-translational modification. It is known that alterations of glycosylation are common features of certain diseases such as cancer, Alzheimer's disease and inflammatory diseases.20–22 In T2DM, alterations of protein glycosylation are also associated with T2DM progression. The elevated concentration of free fatty acids by the administration of a high-fat diet caused a deficit of GnT-4a glycosyltransferase expression in pancreatic beta cells that produced signs of metabolic diseases including hyperglycemia, impaired glucose tolerance and diminished insulin action in muscle and adipose tissues, and this pathogenic process was active in human islet cells obtained from donors with T2DM.23 Besides, O-GlcNAc-modified CaMKII is increased in the heart and brain of diabetic humans and rats, and an increased glucose concentration significantly enhances CaMKII-dependent activation of spontaneous sarcoplasmic reticulum Ca2+ release events that can contribute to cardiac mechanical dysfunction and arrhythmias in cardiomyocytes.24 In serum, the N-glycan composition of T2DM patients are different compared to healthy control subjects, including the decrease of α(1,6)-linked arm monogalactosylated, core-fucosylated diantennary N-glycans in T2DM compared with healthy subjects, which suggested that the serum N-glycan levels could therefore provide a non-invasive alternative marker for T2DM.25
Here, an integrated glycomics strategy was employed to investigate and verify the altered glycopatterns in salivary protein from HVs and patients with T2DM. Subsequently, the glycoproteins were isolated by lectin-mediated affinity capture, and glycans were released and characterized by MALDI-TOF/TOF-MS to acquire precise structural information of the altered glycans. This study aims to clarify the altered salivary glycosylation associated with T2DM and identify the precise alterations of salivary protein glycopatterns, which may help us to understand the complex physiological changes in T2DM.
2. Experimental
2.1 Ethics statements
The collection of human salivary specimens was carried out in accordance with the approved guidelines, approved by the Human Ethics Committee of all participating units (Northwest University, The Second Affiliated Hospital of Xi'an Jiaotong University, Shannxi). Written informed consent was received from participants. This study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki.2.2 Study population and whole saliva collection
T2DM was diagnosed according to the diagnostic criteria by the American Diabetes Association.26 To investigate the difference in glycosylation, saliva samples from HVs and patients with T2DM were collected. HVs who underwent a medical examination and were confirmed to have neither diabetes nor systemic diseases were enrolled as control subjects. In total, 72 patients with T2DM and 80 HVs were enrolled in this study between October 2017 and March 2018. The above HVs and patients were divided into discovery cohort and validation cohort. A summary of the patient and HV clinical characteristics and the detailed information of the study cohorts are provided in Table 1.| Discovery cohort | Validation cohort | |||
|---|---|---|---|---|
| HVs | T2DM patients | HVs | T2DM patients | |
| a Mean ± SD. | ||||
| Subject number | 40 | 35 | 40 | 37 |
Sex (male![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) female) female) |
25/15 | 20/15 | 22/18 | 23/14 |
| Agea (year) | 56.92 ± 7.43 | 58.17 ± 12.35 | 57.28 ± 6.33 | 54.83 ± 11.17 |
| BMI (kg m−2) | 21.45 ± 4.32 | 25.41 ± 2.66 | 21.74 ± 4.68 | 26.43 ± 3.74 |
| Duration of diabetes (years) | — | 14.29 ± 6.41 | — | 12.59 ± 8.10 |
| HbA1c (%) | — | 7.19 ± 1.12 | — | 7.77 ± 1.58 |
The collection protocol has been described in previous reports.18,27,28 Briefly, unstimulated saliva was collected between 9 a.m. and 11 a.m. at least 3 h after the last intake of food. The mouth was rinsed with sterile physiological saline immediately before the collection. Whole saliva was collected and placed on ice immediately and then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 15 min to remove insoluble precipitate. The supernatant was transferred to a new tube, and 10 μL of protease cocktail inhibitor (1
000 × g at 4 °C for 15 min to remove insoluble precipitate. The supernatant was transferred to a new tube, and 10 μL of protease cocktail inhibitor (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (v/v), Sigma-Aldrich) was added into the supernatant. The protein concentration of each salivary specimen was determined by a BCA assay (Beyotime Biotechnology, China). After that, the processed salivary specimens were stored at −80 °C until use.
100 (v/v), Sigma-Aldrich) was added into the supernatant. The protein concentration of each salivary specimen was determined by a BCA assay (Beyotime Biotechnology, China). After that, the processed salivary specimens were stored at −80 °C until use.
2.3 Lectin microarrays and data analysis
The manufacture of lectin microarray and data acquisition were performed as described previously.28–31 First, salivary protein was labelled with Cy3 dye (GE Healthcare); after purification, 4 μg of Cy3 labelled salivary protein was mixed with 120 μL of incubation buffer and applied to the lectin microarray and incubated at 37 °C for 3 h. Subsequently, microarrays were washed and centrifuged to dry. The fluorescence intensity of each spot was extracted using the Gene Pix software (version 6.0; Axon Instruments Inc., Sunnyvale, CA). To eliminate the influence of non-specific adsorption, the signal values less than average background + standard deviations (SD) were excluded for further analysis, and a global normalization method was utilized to eliminate fluoresce bias. The processed data of the parallel data sets were compared with each other based on fold-changes according to the following criteria: fold changes ≥2 or <0.5 and p < 0.05 in the pairs indicated up- or down-regulation of certain kind of glycopatterns respectively. Differences in each lectin between two groups were tested using the Mann–Whitney test using the GraphPad Prism software (Version 7.0, GraphPad Software, Inc., San Diego, CA). Then, the normalized data were further analyzed by unsupervised average hierarchical cluster analysis (HCA) using Expander 6.0 (http://acgt.cs.tau.ac.il/expander/) and principal component analysis (PCA) using the Multi-Variate Statistical Package (UK).2.4 Lectin blotting analysis
The lectin blotting was performed as described previously.18,28,30 Briefly, 40 μg of pooled salivary protein from HVs and patients with T2DM was separated by using 10% polyacrylamide resolving gel and 3% stacking gel, respectively. Then, the proteins in the gel were transferred onto a PVDF membrane (0.22 μm Millipore, Bedford, MA, USA) and blocked by 1× carbo-free blocking solution (Vector Labs, Burlingame, CA) for 1 h. Then, the membranes were incubated with Cy5-labeled Lycopersicon esculentum Lectin (LEL) (2 μg mL−1 in Carbo-Free Blocking Solution) with gentle shaking at 4 °C overnight in the darkness. After washing three times with TBST buffer, the image was acquired using a STORM fluorImager (Molecular Dynamics, Sunnyvale, CA, USA). The gray value of the protein bands was measured using the ImageJ software (NIH).2.5 Selective isolation of glycoprotein fractions from saliva by LEL-magnetic particle conjugates
The LacNAc-containing salivary glycoproteins were isolated from HVs and patients with T2DM using LEL-magnetic particle conjugates as described previously.31,32 Briefly, 2 mg of pooled salivary proteins from HVs and patients with T2DM was diluted to 600 μL with the binding buffer supplemented with 6 μL proteinase inhibitor cocktail (Sigma-Aldrich), respectively. Subsequently, the conjugates were incubated with diluted salivary proteins and incubated for 3 h at room temperature under gentle shaking. Afterwards, the conjugates were washed three times with a washing buffer to remove unbound proteins. The glycoproteins bound to the conjugates were eluted with an eluting buffer (8 M urea, 40 mM NH4HCO3) at room temperature for 1 h with shaking. The concentration of isolated proteins was determined by the BCA assay (Beyotime).2.6 Sialic acid ethyl esterification
The sialic acid linkage specificity of the derivatization reaction was performed as described previously with minor modifications.33 Briefly, 400 μg of isolated proteins was concentrated and desalted by size-exclusion spin 10 kDa ultrafiltration (Amicon Ultra-0.5 mL 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MW cut off, Millipore). Subsequently, glycoproteins were collected and lyophilized by CentriVap (Labconco, America). After that, the sialic acids of glycoproteins were derivatized at 60 °C for 1 h using 500 μL of dimethylamidation reagent (250 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, ≥99.0%, Sigma-Aldrich), 500 mM 1-hydroxybenzotriazole hydrate (HOBt, J&K Scientific Ltd) and 250 mM dimethylamine solution (purum, 33% in absolute ethanol, Sigma-Aldrich) in dimethyl sulfoxide (DMSO, 99%, Sigma-Aldrich)). Then, 200 μL of ammonia (Sigma-Aldrich) (1
000 MW cut off, Millipore). Subsequently, glycoproteins were collected and lyophilized by CentriVap (Labconco, America). After that, the sialic acids of glycoproteins were derivatized at 60 °C for 1 h using 500 μL of dimethylamidation reagent (250 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, ≥99.0%, Sigma-Aldrich), 500 mM 1-hydroxybenzotriazole hydrate (HOBt, J&K Scientific Ltd) and 250 mM dimethylamine solution (purum, 33% in absolute ethanol, Sigma-Aldrich) in dimethyl sulfoxide (DMSO, 99%, Sigma-Aldrich)). Then, 200 μL of ammonia (Sigma-Aldrich) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 (v/v) derivatization solution–ammonia) was added and the samples were incubated at 60 °C for further 2 h. Following derivatization, the glycoproteins were desalted by 10 kDa Amicon Ultra unit (Millipore).
0.4 (v/v) derivatization solution–ammonia) was added and the samples were incubated at 60 °C for further 2 h. Following derivatization, the glycoproteins were desalted by 10 kDa Amicon Ultra unit (Millipore).
2.7 PNGase F release of N-linked glycans
N-Glycans were released by PNGase F glycosidase (New England Biolabs, Beverly, MA) according to previous protocols.32,34 Briefly, the derivatized glycoproteins were denatured with 8 M urea (Sigma-Aldrich), 10 mM DTT (dithiothreitol, Sigma-Aldrich) and 10 mM IAM (iodoacetamide, Sigma-Aldrich), and exchanged buffer into 40 mM NH4HCO3 by 10 kDa centrifugal ultrafiltration. After that, 2 μL of PNGase F (NEB) was added and incubated with shaking at 37 °C overnight to release the N-linked glycans. The reaction was terminated by incubating the mixture at 80 °C for 5 min. By centrifuging at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min, the mixture of N-linked glycans was collected. This eluting step was replicated twice with 40 mM NH4HCO3, and N-glycans were then evaporated to dryness using a vacuum centrifuge.
000 × g for 10 min, the mixture of N-linked glycans was collected. This eluting step was replicated twice with 40 mM NH4HCO3, and N-glycans were then evaporated to dryness using a vacuum centrifuge.
2.8 NaClO release of O-linked glycans
The release of O-linked glycans was carried out as described previously with minor modifications.35,36 The derivatized glycoproteins were concentrated using a new 10 kDa MWCO ultrafiltration unit and rinsed 6 times with ultrapure water, and the volume was adjusted to 200 μL with ultrapure water. Then, 100 μL of 6% NaClO (J&K) was added under gentle shaking for 30 min. After that, 15 μL of 10% formic acid (J&K) was added slowly to the reaction mixture and cooled on ice for 10 min. The mixture was centrifuged and rinsed six times with ultrapure water. The volume of the mixture was adjusted to 500 μL with H2O, and the pH was adjusted to 7.6. Then, 6.64 μL of 6% NaClO was added into the mixture and reacted at room temperature for 24 h to release O-glycans. Subsequently, 8 μL of 10% formic acid was added slowly to the mixture and cooled on ice for 10 min to terminate the reaction. The O-linked glycan-acids were collected after centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min. This eluting step was repeated twice with ultrapure water, and O-glycans were evaporated to dryness.
000 × g for 10 min. This eluting step was repeated twice with ultrapure water, and O-glycans were evaporated to dryness.
2.9 Purification of released N-/O-linked glycans
The released glycans were purified using HyperSep Hypercarb SPE cartridges (25 mg, 1 mL; Thermo Scientific) according to the manufacturer's recommendation. Briefly, the cartridge was washed three times with 3 mL of water, and then equilibrated with 3 mL of 50% acetonitrile (ACN, Sigma-Aldrich)/0.1% trifluoroacetic acid (TFA, Sigma-Aldrich) and 3 mL of 5% ACN/0.1% TFA. The glycans were re-dissolved in 500 μL of 0.1% TFA and loaded onto SPE cartridges for three times. Then, they were washed with 3 mL of HPLC-grade water and 3 mL of 5% ACN/0.1% TFA. The glycans were eluted with 0.5 mL of elution solution (50% (v/v) ACN with 0.1% (v/v) TFA). The purified glycans were collected and lyophilized.2.10 Characterization of N-glycans by MALDI-TOF/TOF-MS
The purified N-linked glycans and O-glycan acids were characterized by matrix-assisted laser desorption ionization time-of-flight/time-of-flight mass spectroscopy (MALDI-TOF/TOF-MS, UltrafleXtreme, Bruker Daltonics; Bremen, Germany) as described previously.32,34,36 Briefly, lyophilized N-glycans or O-glycan-acids were resuspended and spotted onto an MTP AnchorChip sample target. Then, 2 μL of 20 mg mL−1 2,5-dihydroxybenzoic acid (DHB) in 50% methanol solution with 1 mM NaCl was spotted to recrystallize the glycans. Ionization was performed by irradiation of a nitrogen laser (337 nm) operating at 1 kHz, and mass calibration was performed using peptide calibration standards (250 calibration points; Bruker). Measurements were taken in positive and reflectron modes, and the intense ions from MS spectra were subsequently selected and subjected to MS/MS. Representative MS spectra of N-glycans or O-glycan acids with signal-to-noise ratios > 3 were chosen and annotated using the GlycoWorkbench software.32,37–39 In addition, the relative intensities (RIs) of each peak were calculated using the GlycoWorkbench software. It is known that O-glycans are linked to serine or threonine residues, and as a result, glycolic acid or lactic acid was derived at the reducing end of the released O-glycans according to the amino acid residue. The O-glycan acids were calibrated with an increase of 58 Da or 72 Da in molecular mass over the corresponding free reducing glycans based on which amino acids (serine or threonine) are linked to the glycan. The glycolic acid and lactic acid are represented as A and B respectively.3. Results
3.1 Comparison of salivary glycopatterns between HVs and patients with T2DM
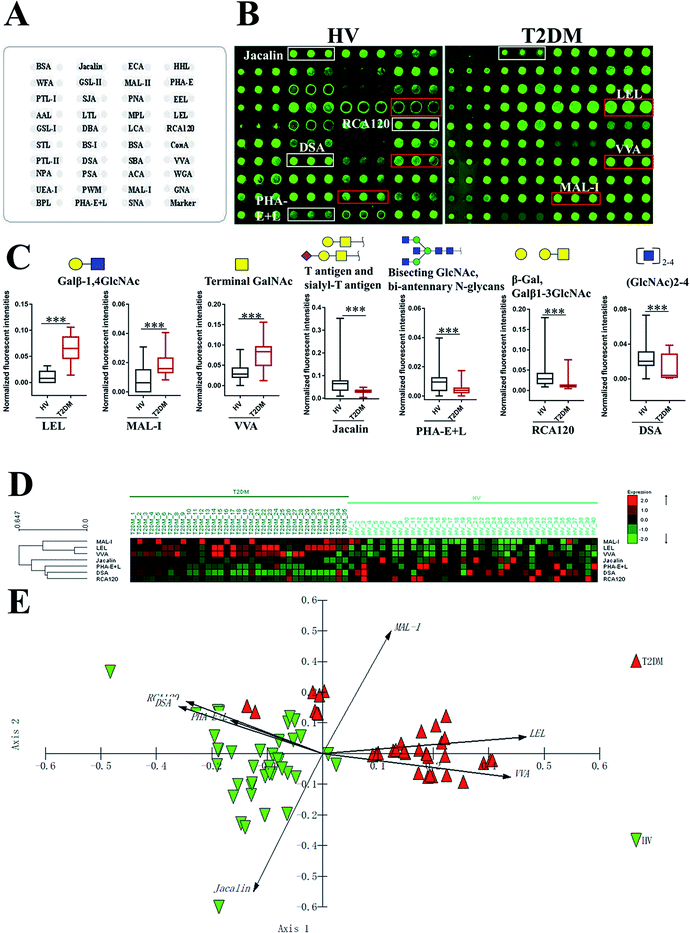
To investigate the alteration of salivary glycopatterns associated with T2DM, 75 salivary samples of HVs (n = 40) and T2DM patients (n = 35) in the discovery cohort were subjected to lectin microarray analysis. The layouts of the lectin microarray and glycopatterns of Cy3-labelled salivary proteins from HVs and patients with T2DM bound to the lectin microarrays are shown in Fig. 1A and B. The normalized fluorescent intensities (NFIs) for each lectin are summarized in ESI Table S1.† The ratio analysis revealed that the patients with T2DM showed higher levels of Galβ-1,4GlcNAc (LacNAc) and poly-LacNAc (bound by lectins: LEL, MAL-I), and terminal GalNAc (VVA), and lower levels of T antigen and sialyl-T antigen (Jacalin), bisecting GlcNAc structure (PHA-E), β-Gal, Galβ1-3GlcNAc (RCA120) and oligomers of GlcNAc (DSA) than HVs (Fig. 1C). Hierarchical cluster analysis (HCA) and principal component analysis (PCA) were performed to visualize these differences respectively. The result of HCA, MAL-I, LEL and VVA were classified into one category and Jacalin, PHA-E + L, DSA and RCA120 were classified into another category, which indicate the glycopatterns identified by these lectins were different between HVs and patients with T2DM (Fig. 1D). The results of PCA revealed that, although there was a small overlapping area between patients with T2DM and HVs, most of patients and HVs were separated, which indicated that the differentiation of most of the patients with T2DM from HVs based on altered salivary glycopatterns is possible (Fig. 1E).3.2 Validation of the altered salivary glycopatterns
To validate the altered salivary glycopatterns, the samples in the validation cohort from 37 T2DM patients and 40 HVs were further subjected to lectin microarray analysis. The NFIs of each lectin in validation set are summarized as the mean with 95% confidence interval in Table S1.† Compared to HVs, The NFIs of 6 lectins (e.g. LEL, VVA and STL) were significantly altered in T2DM patients in the validation cohort (data were not shown). Of which, the NFIs of 5 lectins (LEL, VVA, Jacalin, RCA120 and DSA) exhibited similar tendency in both discovery and validation cohorts (Table 2).| Lectin | Specificity | Discovery cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|---|
| HVs (n = 40) | T2DM patients (n = 35) | Fold changeb | HVs (n = 40) | T2DM patients (n = 37) | Fold changeb | ||
| a The NFIs of lectins are represented as mean with 95% confidence interval.b *p < 0.05, **p < 0.01, and ***p < 0.001. | |||||||
| LEL | Galβ-1,4GlcNAc(LacNAc) and poly LacNAc | 0.012 (0.008–0.015) | 0.063 (0.053–0.072) | 5.375*** | 0.012 (0.009–0.015) | 0.057 (0.053–0.062) | 4.845*** |
| VVA | Terminal GalNAc, GalNAcα-Ser/Thr(Tn), GalNAcα1-3Gal | 0.032 (0.026–0.038) | 0.075 (0.063–0.088) | 2.375*** | 0.034 (0.028–0.040) | 0.083 (0.069–0.096) | 2.445*** |
| Jacalin | Galβ1-3GalNAcα-Ser/Thr(T), GlcNAcβ1-3-GalNAcα-Ser/Thr(Core3), sialyl-T(ST) | 0.071 (0.051–0.090) | 0.030 (0.026–0.033) | 0.420*** | 0.066 (0.046–0.086) | 0.033 (0.030–0.037) | 0.491** |
| RCA120 | β-Gal, Galβ1-3GlcNAc (type I) | 0.042 (0.029–0.054) | 0.015 (0.010–0.020) | 0.357*** | 0.043 (0.030–0.056) | 0.016 (0.012–0.019) | 0.362*** |
| DSA | (GlcNAc)2-4 | 0.024 (0.019–0.029) | 0.011 (0.007–0.015) | 0.471*** | 0.022 (0.018–0.026) | 0.008 (0.005–0.011) | 0.281*** |
3.3 Lectin blotting analysis
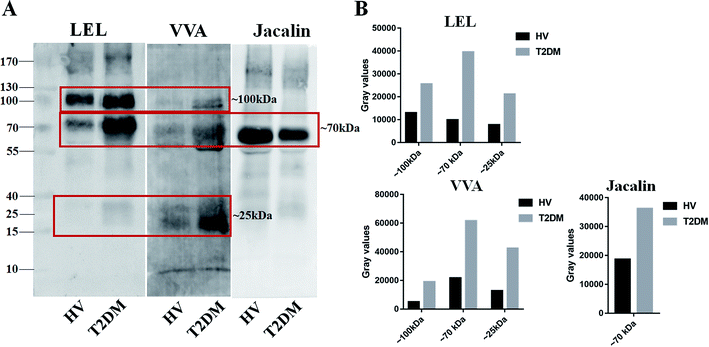
Lectin blotting analyses were performed using three selected lectins (LEL, VVA and Jacalin) to confirm the different abundances of glycopatterns between HVs and T2DM patients. The protein bands showed that the differences between T2DM and HVs were marked with red frames (Fig. 2A). LEL showed a stronger binding to two apparent bands (about 100 kDa and 70 kDa) in pooled salivary samples from T2DM patients than from HVs. In addition, the protein band about 25 kDa showed a staining signal in T2DM samples but has not been observed in HVs. VVA exhibited a stronger binding to three bands (about 100 kDa, 70 kDa and 25 kDa) in T2DM samples than in HV samples. Conversely, Jacalin showed a weaker binding to the protein band (about 70 kDa) in T2DM samples than in HV samples. The signal intensity of the bands for HVs and T2DM patients was measured using Image J (Fig. 2B). Generally, the results of lectin blottings and lectin microarrays exhibited a similar alteration in these lectins.3.4 N-Glycan profiles of the isolated salivary glycoproteins in HVs and patients with T2DM
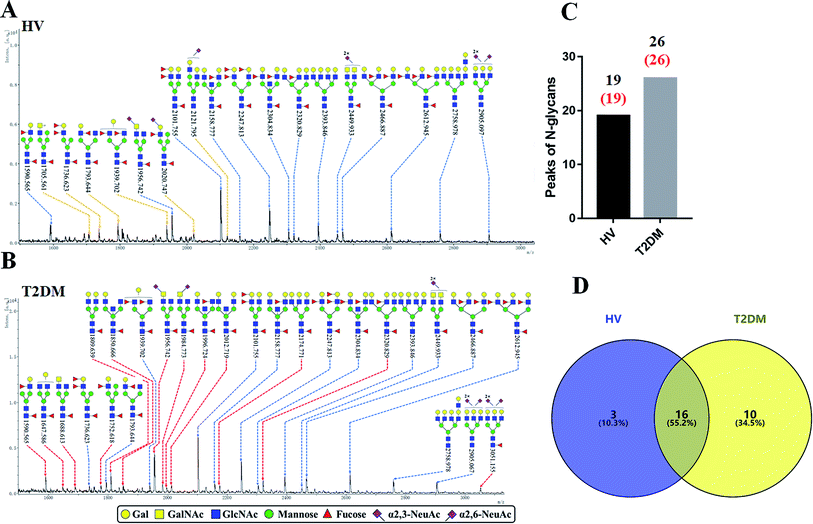
The glycopattern of LacNAc identified by LEL exhibited a significant increase in salivary proteins of patients with T2DM compared to HVs. To acquire detailed structural information of this glycopattern, LacNAc-containing glycoproteins were isolated from pooled salivary protein using the LEL-magnetic particle conjugates. Then, N-glycans were released by PNGase F and were characterized by MALDI-TOF/TOF-MS. MS profiles and proposed structures of the N-glycans are shown in Fig. 3A and B. A total of 19 and 26 N-glycan peaks from HVs and patients with T2DM were identified and annotated with proposed structures, all of which are LacNAc or LacdiNAc-containing glycans (Fig. 3C). The detailed information about these N-glycans is summarized in Table S3.† Of these, three (m/z 1705.561, 2020.747 and 2121.795) and ten (e.g., m/z 1647.586, 1688.613 and 1809.639) N-glycans were only presented in HVs or T2DM patients uniquely, and 16 N-glycans (e.g., m/z 1590.565, 1736.623 and 1793.644) were presented in both HVs and patients with T2DM (Fig. 3D). Among these overlap glycans, the relative intensity (RI) for the glycans at m/z 1590.565, 1956.742 and 2320.829 resulted in a higher signal in T2DM patients (all fold change > 1.5), and the glycans at m/z 1736.623, 1793.644 and 1939.702 were lower in T2DM patients than in HVs (all fold change < 0.67). The detailed information about these altered glycans is summarized in Tables 3 and 4. These results indicated that neo-LacNAc-containing N-glycans correlated with the elevated relative abundance of LacNAc in salivary glycoprotein from T2DM patients. It is known that sialic acid could further modify based on LacNAc or LacdiNAc to form sialylated glycans. The linkage-specific sialic acid derivatization method provides more structural information about sialylation of glycans in salivary glycoproteins from T2DM patients. A total of 5 sialylated N-glycans were identified from HVs and patients with T2DM respectively. Among these, the sialylated N-glycans Hex5HexNAc3(α2,3) NeuAc1dHex2 at m/z 2020.747 and Hex6HexNAc4(α2,6)NeuAc1 at m/z 2121.795 were presented in HVs uniquely, and two sialylated N-glycans Hex3HexNAc5(α2,6)NeuAc1dHex1 at m/z 1984.733 and Hex6HexNAc5(α2,3)NeuAc2(α2,6)NeuAc1dHex1 at m/z 3051.155 were only observed in patients with T2DM. To provide insights into the substitution and branching pattern of the monosaccharide constituents, the glycan peaks observed in the MS spectrum were assigned to tandem MS analysis. The MS/MS spectra of the precursor ions m/z: 1793.644, 1809.639, 2101.755, 2320.829 and 2612.945 are illustrated in Fig. S1.†| No | Calculated m/z | Ion type | Predicted components | Proposed N-glycan structureb | Relative intensities of N-glycans | Fold changec (T2DM patients/HVs) | |
|---|---|---|---|---|---|---|---|
| HVs (%) | T2DM patients (%) | ||||||
a  .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software.c —, the denominator of the fold-change is zero. .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software.c —, the denominator of the fold-change is zero. |
|||||||
| 1 | 1647.586 | Na | Hex4HexNAc4dHex1 | 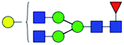 |
ND | 14.490 | — |
| 2 | 1688.613 | Na | Hex3HexNAc5dHex1 | 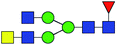 |
ND | 14.926 | — |
| 3 | 1752.618 | Na | Hex5HexNAc3dHex2 | 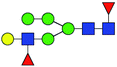 |
ND | 9.530 | — |
| 4 | 1809.639 | Na | Hex5HexNAc4dHex1 | 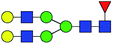 |
ND | 31.281 | — |
| 5 | 1850.666 | Na | Hex4HexNAc5dHex1 | 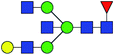 |
ND | 10.832 | — |
| 6 | 1984.773 | H | Hex3HexNAc5(α2,6)NeuAc1dHex1 | 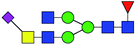 |
ND | 11.422 | — |
| 7 | 1996.724 | Na | Hex4HexNAc5dHex2 | 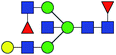 |
ND | 8.431 | — |
| 8 | 2012.719 | Na | Hex5HexNAc5dHex1 | 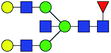 |
ND | 13.422 | — |
| 9 | 2174.771 | Na | Hex6HexNAc5dHex1 | 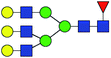 |
ND | 12.953 | — |
| 10 | 3051.155 | H | Hex6HexNAc5(α2,3)NeuAc2(α2,6)NeuAc1dHex1 | 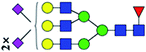 |
ND | 6.360 | — |
| 11 | 1590.565 | Na | Hex4HexNAc3dHex2 | 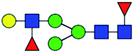 |
15.567 | 24.536 | 1.576 |
| 12 | 1956.742 | H | Hex3HexNAc5(α2,3)NeuAc1dHex1 | 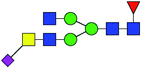 |
42.020 | 69.623 | 1.657 |
| 13 | 2320.829 | Na | Hex5HexNAc5dHex3 | 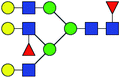 |
11.847 | 19.707 | 1.663 |
| No | Calculated m/z | Ion type | Predicted components | Proposed N-glycan structureb | Relative intensities of N-glycans | Fold change (T2DM patients/HVs) | |
|---|---|---|---|---|---|---|---|
| HVs (%) | T2DM patients (%) | ||||||
a  .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software. .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software. |
|||||||
| 1 | 1705.561 | H | Hex4HexNAc4S1dHex1 | 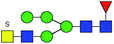 |
3.165 | ND | 0 |
| 2 | 2020.747 | H | Hex5HexNAc3(α2,3)NeuAc1dHex2 | 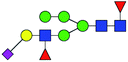 |
2.259 | ND | 0 |
| 3 | 2121.795 | H | Hex6HexNAc4(α2,6)NeuAc1 | 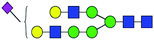 |
2.350 | ND | 0 |
| 4 | 1736.623 | Na | Hex4HexNAc3dHex3 | 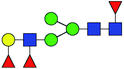 |
23.095 | 11.651 | 0.504 |
| 5 | 1793.644 | Na | Hex4HexNAc4dHex2 | 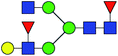 |
30.949 | 14.696 | 0.475 |
| 6 | 1939.702 | Na | Hex4HexNAc4dHex3 | 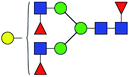 |
30.112 | 9.331 | 0.310 |
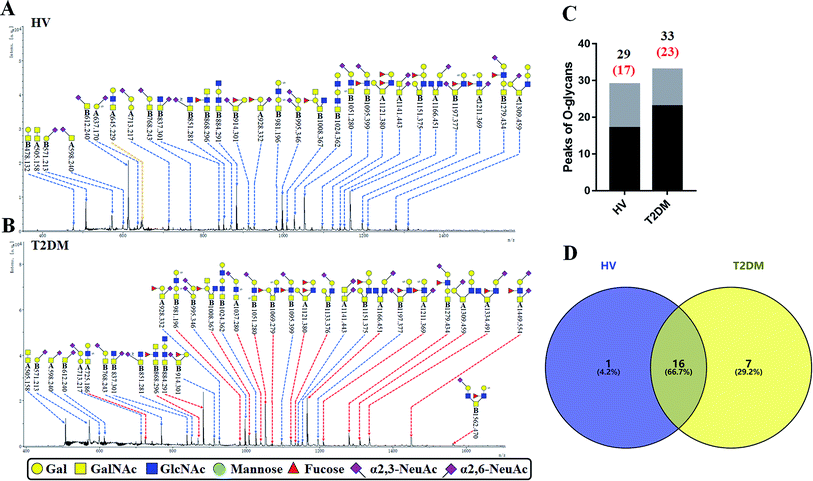
3.5 O-Glycan profiles of the isolated salivary glycoproteins in HVs and patients with T2DM
The O-glycan-acids were released from the isolated glycoproteins by oxidation with NaClO and characterized by MALDI-TOF/TOF-MS. MS profiles, and the proposed structures of O-glycans are shown in Fig. 4A and B. A total of 29 and 33 O-glycan peaks from HVs and T2DM patients were identified and annotated with proposed structures (Fig. 4C and Table S4†). Among these, 17 and 23 LacNAc or LacdiNAc modified O-glycans that were identified in HVs and patients with T2DM respectively (Fig. 4C). In addition, 16 LacNAc or LacdiNAc modified O-glycans (e.g. m/z 837.301, 851.281 and 868.296) that annotated in both HVs and patients with T2DM. Of these, the RI for 10 LacNAc-containing O-glycans (e.g. m/z at 868.296, 884.291 and 981.196) were increased in T2DM patients compared to HVs. Besides, the LacNAc-containing O-glycan at m/z 645.229 was presented only in HVs, and 7 LacNAc-containing O-glycans (e.g. m/z 725.186, 1037.280 and 1069.279) existed only in patients with T2DM (Fig. 4D). The information of these altered glycans is summarized in Tables 5 and 6. Our results revealed that 8 out of 17 O-glycans which showed an increase uniquely in T2DM patients were further modified with sialic acids based on LacNAc. Among these, five glycans are α-2,3-sialylated and three glycans are α-2,6-sialylated O-glycans. It suggested that the increase of LacNAc may further provide more substrate for the modification of sialic acid.| No | Calculated m/z | Ion type | Predicted components | Proposed N-glycan structureb | Relative intensities of O-glycans | Fold-changec (T2DM patients/HVs) | |
|---|---|---|---|---|---|---|---|
| HVsc (%) | T2DM patients (%) | ||||||
a  .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software. In O-glycans, the reducing end A or B was residue of –CH2COOH from serine or residue of –CH(CH3)COOH from threonine.c —, the denominator of the fold-change is zero. .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software. In O-glycans, the reducing end A or B was residue of –CH2COOH from serine or residue of –CH(CH3)COOH from threonine.c —, the denominator of the fold-change is zero. |
|||||||
| 1 | 725.186 | H | Hex1HexNAc2S1 |  |
ND | 0.931 | — |
| 2 | 1037.280 | Na | Hex1HexNAc2(α2,3)NeuAc1S1 | 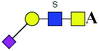 |
ND | 0.510 | — |
| 3 | 1069.279 | Na | Hex2HexNAc2S1dHex1 | 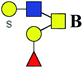 |
ND | 0.644 | — |
| 4 | 1133.376 | Na | Hex2HexNAc2(α2,3)NeuAc1 | 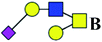 |
ND | 0.734 | — |
| 5 | 1334.491 | Na | Hex1HexNAc3(α2,6)NeuAc1dHex1 | 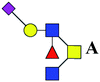 |
ND | 2.329 | — |
| 6 | 1449.554 | Na | Hex1HexNAc2(α2,6)NeuAc2dHex1 | 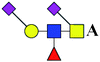 |
ND | 2.059 | — |
| 7 | 1562.470 | Na | Hex2HexNAc3(α2,3)NeuAc1S1dHex1 | 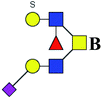 |
ND | 0.612 | — |
| 8 | 868.296 | Na | HexNAc3dHex1 | 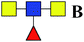 |
6.422 | 13.063 | 2.034 |
| 9 | 884.291 | Na | Hex1HexNAc3 |  |
64.770 | 100.000 | 1.544 |
| 10 | 981.196 | H | Hex2HexNAc2S2 |  |
5.656 | 8.517 | 1.506 |
| 11 | 1008.367 | H | Hex1HexNAc3dHex1 | 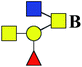 |
11.611 | 19.463 | 1.676 |
| 12 | 1051.28 | Na | Hex1HexNAc2(α2,3)NeuAc1S1 | 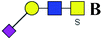 |
50.187 | 78.874 | 1.572 |
| 13 | 1121.38 | Na | Hex2HexNAc2dHex2 | 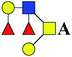 |
5.500 | 11.368 | 2.067 |
| 14 | 1166.451 | H | Hex1HexNAc3(α2,6)NeuAc1 | 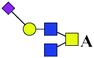 |
57.772 | 91.382 | 1.582 |
| 15 | 1211.369 | Na | Hex1HexNAc2(α2,6)NeuAc1S1dHex1 | 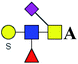 |
5.500 | 9.609 | 1.747 |
| 16 | 1279.434 | Na | Hex2HexNAc2(α2,3)NeuAc1dHex1 | 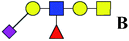 |
8.642 | 24.486 | 2.833 |
| 17 | 1309.459 | Na | Hex3HexNAc2(α2,3)NeuAc1 | 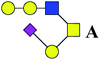 |
6.621 | 11.573 | 1.748 |
| No | Calculated m/z | Ion type | Predicted components | Proposed N-glycan structureb | Relative intensities of O-glycans | Fold-change (T2DM patients/HVs) | |
|---|---|---|---|---|---|---|---|
| HVs (%) | T2DM patients (%) | ||||||
a  .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software. In O-glycans, the reducing end A or B was residue of –CH2COOH from serine or residue of –CH(CH3)COOH from threonine. .b Proposed glycan structure, monosaccharides are represented according to MS-tools from the GlycoWorkbench software. In O-glycans, the reducing end A or B was residue of –CH2COOH from serine or residue of –CH(CH3)COOH from threonine. |
|||||||
| 1 | 645.229 | H | Hex1HexNAc2 |  |
1.779 | ND | 0 |
4. Discussion
The moieties of glycans on the cell surface are involved in many biological processes such as cellular recognition, migration and endocytosis.40–42 One of the most fascinating aspects of glycosylation is heterogeneity. The structural heterogeneity of glycans has been observed in various diseases, which is known as aberrant glycosylation associated with diseases. Previous studies had indicated that aberrant glycosylation of proteins could reflect the process of diseases. Many of these altered glycosylation patterns found in cancer have been termed ‘oncofetal’ as they resemble patterns often seen in early development. During the tumor development, glycan composition can change in parallel with changes in cellular metabolism.43,44 Recent studies have demonstrated that the glycosylation of certain proteins is changed as diabetes mellitus occurred. The glycan moieties of glycodelin-A, but not the protein core showed difference between patients with gestational diabetes mellitus and healthy controls.45 The O-linked β-N-acetylglucosamine (O-GlcNAc) modification of CaMKII (Ca2+/calmodulin-dependent protein kinase II) is increased in the heart and brain from diabetic rats and humans compared to controls. This alteration of CaMKII may contribute to cardiac mechanical dysfunction and arrhythmias.46 Furthermore, it also reported that the elevated level of aortic O-GlcNAcylation was associated with impaired aortic compliance in diabetic mice induced by low-dose streptozotocin.47 These findings indicated that the altered glycosylation contributes to diabetic complications.Saliva is an ideal biological fluid that is befitting for novel approaches to clinical diagnosis and monitoring of patients with both oral and systemic diseases.48 Saliva contains multiple biomarkers (including antibodies and neoplasma markers) that make it useful for early detection of diseases.49 The studies revealed that diabetes influences the composition of saliva (salivary glucose, amylase and immunoglobulin A) and that saliva can be used as a non-invasive biomarker for monitoring the process of diabetes mellitus.50 In the present study, the integrated glycomics methods are used to investigate the difference in glycosylation of salivary glycoproteins between HVs and patients with T2DM. We found that the glycopatterns of Galβ-1,4GlcNAc(LacNAc) and terminal GalNAc, which were recognized by LEL and VVA, showed a significant increase in patients with T2DM. In contrast, Galβ1-3GalNAcα-Ser/Thr(T), β-Gal, Galβ1-3GlcNAc and oligomers of GlcNAc, which were recognized by Jacalin, RCA120 and DSA showed a significant decrease in patients with T2DM compared to HVs. These findings indicated that the glycan profiles of salivary glycoproteins from HVs and patients with T2DM were different. Compared with healthy subjects, the N-glycan composition of sera from T2DM patients was altered, particularly the α-1,6-linked arm monogalactosylated, core-fucosylated diantennary N-glycans were significantly reduced in T2DM patients.25 The studies by Mise et al. indicated that the glycopatterns including LacNAc in urinary protein were correlated with a decrease in the estimated glomerular filtration rate (eGFR); as a result, urinary glycan profile may be useful for predicting renal prognosis in patients with type 2 diabetes.51
It is established that the change in glycosylation is attributed to the altered expression of glycoproteins, as well as the composition and structure of glycans.20,40 In this study, we demonstrated that the relative abundance of LacNAc recognized by LEL showed a significant increase in patients with T2DM compared to HVs in both discovery and validation cohorts. Furthermore, the result of lectin blotting revealed that two protein bands exhibited stronger binding signals in T2DM patients than in HVs. The glycoprotein bands at 100 kDa and 70 kDa showed increased staining signals in samples from T2DM patients. In addition, the results of MS showed that most of the glycan peaks identified in HVs were also observed in patients with T2DM. However, 10 LacNAc-containing N-glycans and 7 LacNAc-containing O-glycans were annotated in T2DM patients uniquely. These findings suggested that these neo-LacNAc-modified glycans may also be correlated with the increase in LacNAc in salivary glycoproteins of patients with T2DM. Several studies have reported that the high branching, triantennary, tri-galactosylated and antennary fucosylated N-glycan structures were increased in plasma from patients with T2DM compared to controls. These findings suggested that the complexity of plasma N-glycome is increased during the development of T2DM.52 Moreover, the high level of galactosylated N-glycans in plasma protein showed correlation with a higher risk of developing type 2 diabetes and poorer blood glucose management.53
It indicated that a catalogue of 2290 proteins were identified in whole saliva, of which, approximately 27% are found in plasma.54 IgG is known as a high-abundant protein in serum, and it also diffuses into saliva through gingival crevices or produced locally.55 The level of monogalactosylated N-glycans with bisecting GlcNAc on IgG has been reported to be associated with T2DM, which reflects the process.56 The high-level HbA1c is associated with a higher relative abundance of galactosylated N-glycans on serum IgG of patients with kidney dysfunction.57 Sialic acid occurs to the end of glycan in a glycoprotein, which is crucial for a variety of functions, such as cell recognition and metastasis. It is reported that N-glycans with sialic acids increased in the vitreous samples of the proliferative diabetic retinopathy group compared to the control group.58 In the terminal of glycan, LacNAc can serve as a scaffold to be modified with sialic acid. Our results indicated that 8 out of 17 O-glycans, which showed an increase or uniqueness in T2DM patients, were further modified with sialic acids based on LacNAc. Of these glycans, five are α-2,3-sialylated and three are α-2,6-sialylated O-glycans. Several IgG glycans were firmly associated with type 2 diabetes; moreover, the galactosylation and sialylation of the IgG glycans were associated with kidney dysfunction.59 The studies by Dotz et al. reported that the N-glycosylation features of plasma protein were significantly associated with type 2 diabetes. Moreover, α-2,6-linked sialylation was increased in patients with T2DM, and glycosylation changes were reflective of inflammation.52 Accumulating evidence indicated that an interindividual variation in protein N-glycosylation might be a novel risk factor contributing to diabetes development.60
5. Conclusions
In summary, we have established that the glycopattern of LacNAc significantly increased in salivary glycoproteins from patients with T2DM compared to HVs. Our data suggested that an increase of LacNAc-containing N/O-glycans may contribute to the accumulation of LacNAc in salivary glycoproteins, which are associated with T2DM. Our findings suggested that altered glycopatterns in salivary glycoproteins are sensitive to the presence of diabetes, which could provide a non-invasive alternative indicator for T2DM. Moreover, further studies are needed to investigate the relationship between the altered glycosylation in salivary proteins as well as occurrence and the development of T2DM.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by (1) National Natural Science Foundation of China (Grant No. 81871955); (2) Science and Technology Project of Shaanxi Province (No. 2017SF-194).References
- P. Zimmet, K. G. Alberti and J. Shaw, Nature, 2001, 414, 782–787 CrossRef
.
- G. Danaei, M. M. Finucane, Y. Lu, G. M. Singh, M. J. Cowan, C. J. Paciorek, J. K. Lin, F. Farzadfar, Y. H. Khang, G. A. Stevens, M. Rao, M. K. Ali, L. M. Riley, C. A. Robinson, M. Ezzati and Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Blood Glucose), Lancet, 2011, 378, 31–40 CrossRef
.
- L. Chen, D. J. Magliano and P. Z. Zimmet, Nat. Rev. Endocrinol., 2011, 8, 228–336 CrossRef
.
- V. S. Malik, B. M. Popkin, G. A. Bray, J. P. Després and F. B. Hu, Circulation, 2010, 121, 1356–1364 CrossRef
.
- F. B. Hu, Diabetes Care, 2011, 34, 1249–1257 CrossRef
.
- L. L. Liu, J. M. Lawrence, C. Davis, A. D. Liese, D. J. Pettitt, C. Pihoker, D. Dabelea, R. Hamman, B. Waitzfelder, H. S. Kahn and SEARCH for Diabetes in Youth Study Group, Pediatr. Diabetes, 2010, 11, 4–11 CrossRef
.
- M. F. Hivert, J. L. Vassy and J. B. Meigs, Nat. Rev. Endocrinol., 2014, 10, 198–205 CrossRef
.
- H. Mao, Q. Li and S. Gao, PLoS One, 2012, 7, e45882 CrossRef
.
- M. Murea, L. Ma and B. I. Freedman, Rev. Diabet. Stud., 2012, 9, 6–22 CrossRef
.
- T. Temelkova-Kurktschiev and T. Stefanov, Exp. Clin. Endocrinol. Diabetes, 2012, 120, 1–6 CrossRef
.
- P. F. Coogan, L. F. White, M. Jerrett, R. D. Brook, J. G. Su, E. Seto, R. Burnett, J. R. Palmer and L. Rosenberg, Circulation, 2012, 125, 767–772 CrossRef CAS
.
- T. Pfaffe, J. Cooper-White, P. Beyerlein, K. Kostner and C. Punyadeera, Clin. Chem., 2011, 57, 675–687 CrossRef
.
- R. Haeckel and P. Hanecke, Ann. Biol. Clin., 1993, 51, 903–910 Search PubMed
.
- S. Katsiougiannis and D. T. Wong, Rheum. Dis. Clin. North Am., 2016, 42, 449–456 CrossRef
.
- M. Castagnola, E. Scarano, G. C. Passali, I. Messana, T. Cabras, F. Iavarone, G. Di Cintio, A. Fiorita, E. De Corso and G. Paludetti, Acta Otorhinolaryngol. Ital., 2017, 37, 94–101 Search PubMed
.
- A. C. Gonçalves, F. A. Marson, R. M. Mendonça, J. D. Ribeiro, A. F. Ribeiro, I. A. Paschoal and C. E. Levy, Diagn. Pathol., 2013, 19, 46 CrossRef
.
- S. Saxena, B. Sankhla, K. S. Sundaragiri and A. Bhargava, Adv. Biomed. Res., 2017, 28, 90 CrossRef
.
- X. Liu, H. Yu, Y. Qiao, J. Yang, J. Shu, J. Zhang, Z. Zhang, J. He and Z. Li, EBioMedicine, 2018, 28, 70–79 CrossRef
.
- J. Shu, H. Yu, X. Li, D. Zhang, X. Liu, H. Du, J. Zhang, Z. Yang, H. Xie and Z. Li, Oncotarget, 2017, 8, 35718–35727 CrossRef
.
- S. R. Stowell, T. Ju and R. D. Cummings, Annu. Rev. Pathol., 2015, 10, 473–510 CrossRef
.
- M. Frenkel-Pinter, M. D. Shmueli, C. Raz, M. Yanku, S. Zilberzwige, E. Gazit and D. Segal, Sci. Adv., 2017, 3, e1601576 CrossRef
.
- J. H. Dewald, F. Colomb, M. Bobowski-Gerard, S. Groux-Degroote and P. Delannoy, Cells, 2016, 5, E43 CrossRef
.
- K. Ohtsubo, M. Z. Chen, J. M. Olefsky and J. D. Marth, Nat. Med., 2011, 17, 1067–1075 CrossRef
.
- J. R. Erickson, L. Pereira, L. Wang, G. Han, A. Ferguson, K. Dao, R. J. Copeland, F. Despa, G. W. Hart, C. M. Ripplinger and D. M. Bers, Nature, 2013, 502, 372–376 CrossRef
.
- R. Testa, V. Vanhooren, A. R. Bonfigli, M. Boemi, F. Olivieri, A. Ceriello, S. Genovese, L. Spazzafumo, V. Borelli, M. G. Bacalini, S. Salvioli, P. Garagnani, S. Dewaele, C. Libert and C. Franceschi, PLoS One, 2015, 10, e0119983 CrossRef
.
- American Diabetes Association, Diabetes Care, 2018, 41, S13–S27 CrossRef
.
- S. Bandhakavi, S. K. Van Riper, P. N. Tawfik, M. D. Stone, T. Haddad, N. L. Rhodus, J. V. Carlis and T. J. Griffin, J. Proteome Res., 2011, 10, 1052–1061 CrossRef
.
- Y. Qin, Y. Zhong, M. Zhu, L. Dang, H. Yu, Z. Chen, W. Chen, X. Wang, H. Zhang and Z. Li, J. Proteome Res., 2013, 12, 2742–2754 CrossRef
.
- H. Yu, M. Zhu, Y. Qin, Y. Zhong, H. Yan, Q. Wang, H. Bian and Z. Li, J. Proteome Res., 2012, 11, 5277–5285 CrossRef
.
- Y. Zhong, Y. Qin, H. Yu, J. Yu, H. Wu, L. Chen, P. Zhang, X. Wang, Z. Jia, Y. Guo, H. Zhang, J. Shan, Y. Wang, H. Xie, X. Li and Z. Li, Sci. Rep., 2015, 5, 8971 CrossRef
.
- G. Yang, T. Cui, Y. Wang, S. Sun, T. Ma, T. Wang, Q. Chen and Z. Li, Proteomics, 2013, 13, 1481–1498 CrossRef
.
- J. Zhang, Y. Zhong, P. Zhang, H. Du, J. Shu, X. Liu, H. Zhang, Y. Guo, Z. Jia, L. Niu, F. Yang and Z. Li, Glycobiology, 2019, 29, 242–259 CrossRef
.
- S. Holst, B. Heijs, N. de Haan, R. J. van Zeijl, I. H. Briaire-de Bruijn, G. W. van Pelt, A. S. Mehta, P. M. Angel, W. E. Mesker, R. A. Tollenaar, R. R. Drake, J. V. Bovée, L. A. McDonnell and M. Wuhrer, Anal. Chem., 2016, 88, 5904–5913 CrossRef
.
- X. Qin, Y. Guo, H. Du, Y. Zhong, J. Zhang, X. Li, H. Yu, Z. Zhang, Z. Jia and Z. Li, Front. Physiol., 2017, 8, 596 CrossRef
.
- X. Song, J. Hong, L. Yi, D. F. Smith and R. D. Cummings, Nat. Methods, 2016, 13, 528–534 CrossRef
.
- J. Shu, H. Yu, H. Du, J. Zhang, K. Zhang, X. Li, H. Xie and Z. Li, Cancer Biomarkers, 2018, 22, 669–681 Search PubMed
.
- R. Ranzinger and W. S. York, Methods Mol. Biol., 2014, 1273, 109–124 CrossRef
.
- D. Damerell, A. Ceroni, K. Maass, R. Ranzinger, A. Dell and S. M. Haslam, Methods Mol. Biol., 2015, 1273, 3 CrossRef
.
- A. Varki, R. D. Cummings, M. Aebi, N. H. Packer, P. H. Seeberger, J. D. Esko, P. Stanley, G. Hart, A. Darvill, T. Kinoshita, J. J. Prestegard, R. L. Schnaar, H. H. Freeze, J. D. Marth, C. R. Bertozzi, M. E. Etzler, M. Frank, J. F. Vliegenthart, T. Lütteke, S. Perez, E. Bolton, P. Rudd, J. Paulson, M. Kanehisa, P. Toukach, K. F. Aoki-Kinoshita, A. Dell, H. Narimatsu, W. York, N. Taniguchi and S. Kornfeld, Glycobiology, 2015, 25, 1323–1324 CrossRef
.
- K. Ohtsubo and J. D. Marth, Cell, 2006, 126, 855–867 CrossRef
.
- H. B. Guo, I. Lee, M. Kamar, S. K. Akiyama and M. Pierce, Cancer Res., 2002, 62, 6837–6845 Search PubMed
.
- E. A. Partridge, C. Le Roy, G. M. Di Guglielmo, J. Pawling, P. Cheung, M. Granovsky, I. R. Nabi, J. L. Wrana and J. W. Dennis, Science, 2004, 306, 120–124 CrossRef
.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Nat. Rev. Nephrol., 2019, 15, 346–366 CrossRef
.
- L. Oliveira-Ferrer, K. Legler and K. Milde-Langosch, Semin. Cancer Biol., 2017, 44, 141–152 CrossRef
.
- C. L. Lee, P. C. Chiu, P. C. Pang, I. K. Chu, K. F. Lee, R. Koistinen, H. Koistinen, M. Seppälä, H. R. Morris, B. Tissot, M. Panico, A. Dell and W. S. Yeung, Diabetes, 2011, 60, 909–917 CrossRef
.
- J. R. Erickson, L. Pereira, L. Wang, G. Han, A. Ferguson, K. Dao, R. J. Copeland, F. Despa, G. W. Hart, C. M. Ripplinger and D. M. Bers, Nature, 2013, 502, 372–376 CrossRef
.
- J. M. Heath, Y. Sun, K. Yuan, W. E. Bradley, S. Litovsky, L. J. Dell'Italia, J. C. Chatham, H. Wu and Y. Chen, Circ. Res., 2014, 114, 1094–1102 CrossRef CAS
.
- D. Malamud, Dent. Clin. North Am., 2011, 55, 159–178 CrossRef
.
- S. Chojnowska, T. Baran, I. Wilińska, P. Sienicka, I. Cabaj-Wiater and M. Knaś, Adv. Med. Sci., 2018, 63, 185–191 CrossRef
.
- S. E. Abd-Elraheem, A. M. El Saeed and H. H. Mansour, Diabetes Metab. Syndr., 2017, 11, S637–S641 CrossRef
.
- K. Mise, M. Imamura, S. Yamaguchi, S. Teshigawara, A. Tone, H. A. Uchida, J. Eguchi, A. Nakatsuka, D. Ogawa, M. Yoshida, M. Yamada, K. Shikata and J. Wada, Diabetes Care, 2018, 41, 1765–1775 CAS
.
- V. Dotz, R. Lemmers, K. R. Reiding, A. L. Hipgrave Ederveen, A. G. Lieverse, M. T. Mulder, E. Sijbrands, M. Wuhrer and M. van Hoek, Biochim. Biophys. Acta, Gen. Subj., 2018, 1862, 2613–2622 CrossRef CAS
.
- T. Keser, I. Gornik, F. Vučković, N. Selak, T. Pavić, E. Lukić, I. Gudelj, H. Gašparović, B. Biočina, T. Tilin, A. Wennerström, S. Männistö, V. Salomaa, A. Havulinna, W. Wang, J. F. Wilson, N. Chaturvedi, M. Perola, H. Campbell, G. Lauc and O. Gornik, Diabetologia, 2017, 60, 2352–2360 CrossRef CAS
.
- J. A. Loo, W. Yan, P. Ramachandran and D. T. Wong, J. Dent. Res., 2010, 89, 1016–1023 CrossRef CAS
.
- P. Brandtzaeg, Ann. N. Y. Acad. Sci., 2007, 1098, 288–311 CrossRef CAS
.
- R. Lemmers, M. Vilaj, D. Urda, F. Agakov, M. Šimurina, L. Klaric, I. Rudan, H. Campbell, C. Hayward, J. F. Wilson, A. G. Lieverse, O. Gornik, E. Sijbrands, G. Lauc and van M. Hoek, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 2240–2249 CrossRef CAS
.
- M. L. Bermingham, M. Colombo, S. J. McGurnaghan, L. A. K. Blackbourn, F. Vučković, M. Pučić Baković, I. Trbojević-Akmačić, G. Lauc, F. Agakov, A. S. Agakova, C. Hayward, L. Klarić, C. N. A. Palmer, J. R. Petrie, J. Chalmers, A. Collier, F. Green, R. S. Lindsay, S. Macrury, J. A. McKnight, A. W. Patrick, S. Thekkepat, O. Gornik, P. M. McKeigue, H. M. Colhoun and SDRN Type 1 Bioresource Investigators, Diabetes Care, 2018, 41, 79–87 CrossRef CAS
.
- S. Inafuku, K. Noda, M. Amano, T. Ohashi, C. Yoshizawa, W. Saito, M. Murata, A. Kanda, S. Nishimura and S. Ishida, Invest. Ophthalmol. Visual Sci., 2015, 56, 5316–5322 CrossRef CAS
.
- C. Barrios, J. Zierer, I. Gudelj, J. Štambuk, I. Ugrina, E. Rodríguez, M. J. Soler, T. Pavić, M. Šimurina, T. Keser, M. Pučić-Baković, M. Mangino, J. Pascual, T. D. Spector, G. Lauc and C. Menni, J. Am. Soc. Nephrol., 2016, 27, 933–941 CrossRef CAS
.
- N. Rudman, O. Gornik and G. Lauc, FEBS Lett., 2019, 593, 1598–1615 CrossRef CAS
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05466f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |