Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Different effects of soil bacterial communities affected by biocontrol agent YH-07 on tomato Fusarium wilt inhibition†
Tongtong Tang a,
Xing Suna,
Qin Liu
a,
Xing Suna,
Qin Liu *bc,
Yuanhua Dongbc and
Yuyong Xianga
*bc,
Yuanhua Dongbc and
Yuyong Xianga
aSchool of Biological Science and Food Engineering, Chuzhou University, Chuzhou 239000, China
bInstitute of Soil Science Chinese Academy of Sciences, 71 East Beijing Road, Nanjing 210008, Jiangsu Province, China. E-mail: qliu@issas.ac.cn; Fax: +86 25 86881263; Tel: +86 25 86881388
cUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 21st September 2020
Abstract
Bio-organic fertilizers based on biocontrol microorganisms have been widely applied to suppress soilborne diseases and improve crop yields. Studies on beneficial biocontrol agents have promoted the development of the bio-organic fertilizers in China. Our previous study demonstrated that a biocontrol agent, Erythrobacter sp. YH-07, can inhibit the growth of the plant pathogen Fusarium oxysporum. In the present study, we investigated the effects of this biocontrol agent on tomato wilt and used the illumina-based sequencing approach to characterize the variations in soil bacterial communities in a potted experiment. The aim of our study was to explore the potential correlation among bacterial communities, Fusarium wilt suppression, and soil properties after application of the biocontrol agent YH-07. The results showed that application of Erythrobacter sp. YH-07 effectively controlled outbreaks of tomato Fusarium wilt. The illumina MiSeq sequencing showed that Proteobacteria was the predominant phylum in the soil samples. Bacterial community composition and structure varied under different soil treatments, e.g., the relative abundance of Erythrobacter and Salinimicrobium was significantly increased in the YH treatment, and Acidobacteria were decreased in the YH treatment compared with the CK treatment. Additionally, the correlation results showed that the soil organic matter and available phosphorus and potassium were higher after YH-07 application, and they were positively correlated with bacterial community. The redundancy analysis showed Erythrobacter and Acidobacteria were the dominant genera after YH and CK treatments, respectively, and correlations with tomato Fusarium wilt incidence were negative and positive, respectively.
Introduction
Tomato (Solanum lycopersicum L.) Fusarium wilt caused by Fusarium oxysporum f. sp. lycopersici (Fol) is common worldwide,1 infecting the tomato roots, causing bacterial wilt, and limiting annual tomato yield.2 Organic amendments, crop rotation, and chemical soil fumigation have been suggested for controlling the disease.3,4 However, there is a risk that pathogens may become even more abundant in a short time.5,6 Using antagonistic bacteria to prevent the disease may be the most promising control method, because biocontrol can provide ecosystem protection, food safety, and high efficiency.7–9Changes in soil microbial community composition and the induced effects provide useful information for studying soil quality and health.10 It is particularly important to determine the response of soil bacterial communities to different modifiers because the imbalance in the rhizosphere microbial community is the main cause of soilborne disease.11 The rhizosphere is a battlefield for soilborne pathogens and beneficial microorganisms.12
Liu et al.13 characterized changes in tomato bulk soil microflora in response to different organic additives and their results showed that the continuous application of bio-organic fertilizers (BOF) decreased tomato disease incidence. Microorganisms respond quickly to soil properties, fertilizer management, plant development, and other environmental factors.14 Bonanomi et al.5 suggested that the change in denitrification with fertilizer management was related to the shift in bacterial communities resulting from variation in soil properties, which may cause long-term shifts in the quality and function of the soil. Thus, the interaction between microorganisms, soil, and plants is complex and changeable. Understanding the effects of soil microbial communities on soil properties and the relationship between plant disease and soil microbial communities, that can provide the foundation for soil community manipulation and new opportunities to explore novel strategies to promote plant health in a sustainable way.4 Despite the known key role of bacterial communities in maintaining soil quality and ecosystem sustainability, information on shifts in soil bacterial communities affected by different biocontrol agents is still limited, especially with the emergence of new antagonistic microorganisms.
As described in our previous work, Erythrobacter aureus YH-07 showed antagonistic ability against the pathogen causing tomato wilt disease.15 In this study, we used a deep 16S rRNA pyrosequencing approach to investigate the suppression of tomato bacterial wilt by using YH-07 and investigated the responses of soil microbial communities and physicochemical properties to YH-07 addition. Further, we evaluated whether the disease incidence correlated with composition of the microbial communities and whether disease suppression after YH-07 application correlated with the soil physicochemical properties.
Materials and methods
Biocontrol agent inoculum preparation
Strain YH-07 was cultured in Marine Broth 2216 (MB) at 30 °C for 24 h with agitation in an orbital shaker at 180 rpm, the cells were harvested by centrifugation at 4000g for 20 min, and the pellet was resuspended in sterile filtered double-distilled water. The bacterial suspension concentrations were adjusted to 5 × 107 CFU mL−1.Pathogen inoculum preparation
Fusarium oxysporum lycopersici (Fol) (China General Microbiological Culture Collection Center, Beijing, China) was inoculated in potato dextrose agar (PDA) plates for 4 days at 30 °C. The spores were collected using 5 mL sterile water to flood the PDA plates. The spore concentrations were determined with a hemocytometer and diluted to 5 × 106 CFU mL−1.16Experimental methods
The pot experiments were carried out in the greenhouse of Institute of Soil Science, Chinese Academy of Sciences, Nanjing, Jiangsu Province (32°4′ N, 118°48′ E) during the 2018 growing season. The nursery and pot soils were collected with a shovel at Nanjing Institute of Vegetable Science and had never previously held any plants. At the greenhouse, the soil was homogenized and poured into the sterilized pots (2 kg soil per pot). Tomato seeds (cultivar Hezuo 908) were surface sterilized in H2O2 (5%) for 30 min and washed in sterile water three times (90 s each time), then placed in plates covered with sterile filter papers wet with sterile water (sterile filter paper and sterile water were prepared used high-pressure steam sterilization, 121 °C for 30 min) for germination at 30 °C. The germinated seeds were moved into nursery cups containing 300 g soil, and only one seedling was planted in each nursery cup.16 Plants with three to four true leaves were transplanted to pots and kept in a greenhouse under ambient conditions (28 °C in the day – 14 °C at night; natural light).Two treatments were as follows: (1) in CK treatments, half of the tomato seedlings were irrigated with 20 mL of Fol spore solution, and then 6 hours later these tomato seedlings were irrigated with 50 mL sterile distilled water; (2) in YH treatments, the other half were irrigated with 20 mL of pathogen spore solution, and then 6 hours later these tomato seedlings were irrigated with 50 mL bacterial suspension of strain YH-07. Each treatment contained three replications, each replication consisted of 15 pots, and all the pots were placed with a randomization.
Soil sample collection
Soil sampling for this study was performed at 30, 45, and 60 days after transplantation, according to the method modified from Liu et al.13 Briefly, 5 random pots from each replicate were selected for collecting the soil sample. Soil samples at depths of 0–10 cm were collected using a soil auger (1.5 cm in diameter). The 5 cores from each replicate were thoroughly homogenized to form one soil sample, resulting in 3 samples per treatment. All 18 soil samples were placed into separate sterile self-sealing plastic bags and transported to the laboratory on ice. These soil samples were passed through a 2 mm sieve to remove visible root fragments and stones. Each homogenized soil sample was divided into two random parts. One portion of each sample was placed on trays and air-dried at room temperature for 7 days, then used for soil physicochemical analysis, while the remaining soil was stored at −80 °C for DNA extraction and subsequent analysis.Soil physicochemical analysis
The soil chemical properties were determined according to the method of Bao.17 Soil pH was determined with a glass electrode using a soil-to-water ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 (w/v). The K2Cr2O7 oxidation–reduction titration method was used for soil organic matter (SOM) estimation, and total nitrogen (TN) was determined by the Kjeldahl method. The soil total potassium (TK) was estimated by digesting the samples with concentrated hydrofluoric acid. Available K (AK) in soil was extracted with ammonium acetate and determined with flame photometry. Total phosphorus (TP) was determined by HClO4–H2SO4 digestion, and available phosphorus (AP) in soil was extracted with NaHCO3 and quantified using the molybdenum blue method.
2.5 (w/v). The K2Cr2O7 oxidation–reduction titration method was used for soil organic matter (SOM) estimation, and total nitrogen (TN) was determined by the Kjeldahl method. The soil total potassium (TK) was estimated by digesting the samples with concentrated hydrofluoric acid. Available K (AK) in soil was extracted with ammonium acetate and determined with flame photometry. Total phosphorus (TP) was determined by HClO4–H2SO4 digestion, and available phosphorus (AP) in soil was extracted with NaHCO3 and quantified using the molybdenum blue method.
Assay of tomato disease incidence and biomass
Thirty days after the tomato plantlets were transplanted into the pot, typical Fusarium wilt symptoms appeared, including foliage chlorosis and necrosis.18 Fusarium wilt was recorded at the 30, 45, and 60 days after transplantation. In each replicate, disease incidence was expressed as the percentage of diseased plants among the total number of plants.For plant biomass, whole plants were pulled from the pots after the soil samples were collected at the end of the growing season (60 days after transplantation). The whole plants were washed in sterilized water, and water was removed from the surface of the plant with sterilized filter paper before their fresh weights were recorded. Sterile water and sterile filter paper were prepared using high-pressure steam sterilization, 121 °C for 30 min. Then, each plant was dried in an oven at 85 °C to a constant weight to determine the dry weights.
DNA extraction, PCR amplification, and illumina sequencing
Total genomic DNA was extracted from 0.5 g soil using the E.Z.N.A. Soil DNA Kit (Omega, Carlsbad, CA, USA), according to the manufacturer's instructions.19 The primer pair20 515F (5′-GTGCCAGCMGCCGCG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) was used to amplify the V4 regions of the bacterial 16S ribosomal RNA gene. PCR was performed under the following conditions: 94 °C for 3 min, followed by 35 cycles of 94 °C for 45 s, 50 °C for 60 s, and 72 °C for 90 s, with a final extension at 72 °C for 5 min. The resulting PCR products were separated by 2.0% (w/v) agarose gels and purified by a PCR Purification Kit (Axygen Bio, USA). Then, the purified amplicons were pooled in equimolar concentrations and paired-end sequencing was then performed on the Illumina MiSeq PE300 platform. Split sequences for each sample were merged using FLASH.21 After removing the adaptors and primer sequences, raw bacterial sequences were assembled for each sample according to the unique barcode. The reads were filtered by a QIIME 2.0 quality filter.22 The sequences retained for each sample, referred to as clean paired sequences, were analyzed following the UPARSE pipeline23 to identify operational taxonomic units (OTUs) by making an OTU table. Briefly, sequences with a quality score lower than 0.5 or a length shorter than 200 nt were discarded. Then the retained reads for each sample were assigned to OTUs with a threshold of 97% identity level.24 Finally, the bacterial representative sequences for each OTU were identified for assigning the taxonomic data using the ribosomal database project (RDP) classifier.25All sequences were deposited in the NCBI Sequence Read Archive database (www.ncbi.nlm.nih.gov/Traces/sra) with the accession number SRP067366.
Bioinformatics analyses
The richness index of the Chao estimator (Chao 1)26 and abundance-based coverage estimator (ACE)27 were calculated to estimate the number of OTUs that were observed in the sampling assemblage. The alpha diversity was estimated using Shannon diversity index.28 Good's nonparametric coverage estimator was used to estimate the percentage of the total species that were sequenced in each sample.26 To explore variation in bacterial community structures across treatments, nonmetric multidimensional scaling (NMDS) analysis based on Bray–Curtis distance was performed. Pearson's correlation coefficient was used to evaluate the correlation between selected genera (relative abundance >1%) and tomato disease incidence. ANOSIM analysis was used to verify whether the differences between treatments were significantly greater than the differences within treatments to determine whether the grouping was meaningful. To examine the relationship among the soil bacterial genera, samples, and environmental variables, we performed redundancy analysis (RDA) using CANOCO 5.0.29 For the Mantel tests, Bray–Curtis distance was used to calculate the correlation between the bacterial genera and soil characteristics.Statistical analyses
Soil physicochemical characteristics, alpha diversity indices, bacterial total abundances, and the taxa (phyla and genus) among treatments were analyzed using one-way analysis of variance (ANOVA) at the end of each bioassay, followed by Duncan's multiple range tests. For all parameters, data were calculated using SPSS Statistics software version 21.0 (SPSS Inc., USA). Differences at P < 0.05 were regarded as statistically significant.Results
Disease incidence and tomato biomass
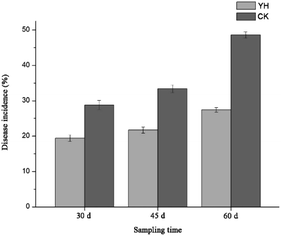
The incidence of tomato wilt disease in the YH treatment at 30, 45, and 60 days was 19.4%, 21.7%, and 27.4%, respectively, which was significantly lower than that in the CK treatment, in which disease incidence was 28.8%, 33.4%, and 48.6%, respectively, at the same time periods (Fig. 1). These results indicated that the biocontrol agent YH-07 more effectively controlled the outbreak of wilt disease in tomato plants.Compared with the CK treatment, YH treatment increased shoot height (17.5%), root length (40.0%), shoot dry weight (19.2%), and root dry weight (16.4%) significantly (Table 1). In general, the biocontrol bacterium YH-07 suppressed disease incidence and increased tomato biomass compared to that with no inoculated treatment.
| Variable | YH | CK |
|---|---|---|
| a Data shown are mean ± one standard error (n = 15) and within each line, treatments that do not share a letter are significantly different (ANOVA; P < 0.05; Duncan's test). | ||
| Shoot height (cm) | 55.3 ± 1.16a | 47.1 ± 1.67b |
| Root length (cm) | 12.3 ± 0.62a | 8.79 ± 0.27b |
| Shoot dry weight (g) | 143 ± 1.58a | 116 ± 2.36b |
| Root dry weight (g) | 80.4 ± 0.32a | 62.1 ± 1.04b |
Soil physicochemical characteristics
The application of antagonistic bacteria significantly affected the soil physicochemical characteristics (Table 2). Compared with the CK treatment, the YH treatment had a significantly (P < 0.05) higher pH, SOM, total K, and available P and K contents. The highest total N and P were found in the CK treatment. Soil physicochemical characteristics were also significantly altered at different sampling dates. Briefly, SOM, total N, and total K were decreased with soil depth, and soil pH and available K were significantly increased in the YH treatment as the growth period increased, while pH, SOM, total N, K, available P and K contents in the CK treatment tended to decrease as the growth period increased (Table 2).| pH | SOM | TN | TP | AP | TK | AK | |
|---|---|---|---|---|---|---|---|
| a Data shown are mean ± one standard error (n = 3) and within each column, treatments that do not share a letter are significantly different (ANOVA; P < 0.05; Duncan's test). | |||||||
| YH30 | 6.88 ± 0.02b | 17.0 ± 0.07b | 2.14 ± 0.19a | 0.57 ± 0.01c | 19.7 ± 0.43c | 10.7 ± 0.22a | 178 ± 2.79b |
| YH45 | 6.83 ± 0.06bc | 18.9 ± 0.15a | 2.12 ± 0.12a | 0.44 ± 0.02d | 27.2 ± 1.90a | 10.5 ± 0.20a | 227 ± 5.08a |
| YH60 | 7.33 ± 0.12a | 18.9 ± 0.14a | 1.92 ± 0.07ab | 0.42 ± 0.01d | 22.1 ± 1.75b | 10.5 ± 0.02a | 222 ± 3.16a |
| CK30 | 6.87 ± 0.01bc | 16.7 ± 0.41b | 2.06 ± 0.06ab | 0.59 ± 0.02b | 18.8 ± 0.38cd | 9.89 ± 0.09b | 169 ± 0.99c |
| CK45 | 6.80 ± 0.07bc | 16.3 ± 0.26c | 1.92 ± 0.03ab | 0.63 ± 0.02a | 18.3 ± 0.37cd | 9.49 ± 0.40bc | 160 ± 4.22d |
| CK60 | 6.72 ± 0.06c | 16.0 ± 0.20c | 1.87 ± 0.12b | 0.64 ± 0.01a | 17.2 ± 0.88d | 9.21 ± 0.27c | 149 ± 0.53e |
Sequencing data and bacterial α-diversity
After filtering the reads based on basal quality and removing the singleton OTUs, we obtained a total of 866![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 427 16S rRNA sequences (342
427 16S rRNA sequences (342![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970
970![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 051 bp bases) with an average length of 395.84 bp across the 18 soil samples. Based on 97% species similarity, in total 3148 bacterial OTUs were found. To correct sampling effects, we used a randomly selected subset of 30
051 bp bases) with an average length of 395.84 bp across the 18 soil samples. Based on 97% species similarity, in total 3148 bacterial OTUs were found. To correct sampling effects, we used a randomly selected subset of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 758 sequences per sample to analyze the richness and diversity of bacterial communities. As shown in Table 3, biocontrol agent application had significant effects on the richness and diversity index. The YH treatments showed a significant decrease in ACE, Chao1, and Shannon diversity index compared to CK treatments (Table 3).
758 sequences per sample to analyze the richness and diversity of bacterial communities. As shown in Table 3, biocontrol agent application had significant effects on the richness and diversity index. The YH treatments showed a significant decrease in ACE, Chao1, and Shannon diversity index compared to CK treatments (Table 3).
| Ace | Chao1 | Shannon | Simpson | Coverage | |
|---|---|---|---|---|---|
| a Data shown are mean ± one standard error (n = 3) and within each column, treatments that do not share a letter are significantly different (ANOVA; P < 0.05; Duncan's test). | |||||
| YH30 | 2377 ± 287c | 2377 ± 287a | 5.04 ± 0.91b | 0.07 ± 0.06c | 0.98 ± 0.00a |
| YH45 | 2248 ± 244c | 1815 ± 281c | 2.53 ± 0.55d | 0.36 ± 0.05a | 0.98 ± 0.00a |
| YH60 | 2079 ± 323c | 1913 ± 180b | 3.50 ± 0.39c | 0.19 ± 0.06b | 0.98 ± 0.00a |
| CK30 | 2680 ± 21.0a | 2663 ± 42.3a | 6.27 ± 0.06a | 0.01 ± 0.00c | 0.98 ± 0.00a |
| CK45 | 2603 ± 104b | 2615 ± 123a | 6.01 ± 0.25a | 0.01 ± 0.00c | 0.98 ± 0.00a |
| CK60 | 2603 ± 111b | 2594 ± 94.3a | 5.44 ± 0.16a | 0.04 ± 0.01c | 0.98 ± 0.00a |
Planting time also had significant effects on the diversity index; CK45 treatment showed a decrease in the Shannon index compared to CK60, while the YH45 treatment showed increase in the Shannon index compared to YH60. The same trend existed in the Chao1 index. Two treatments showed the same Good's query coverage with no significant differences (Table 3).
Bacterial community composition
Bacterial sequences were classified into a total of 37 different phyla across all samples. Overall, the predominant bacterial components of samples from different treatments were similar, and the top 10 species in abundance were consistent (Fig. S1†). Specifically, the two most dominant phyla were Proteobacteria [relative abundance 59.8% and 39.5% in YH and CK, respectively (the same below)] and Bacteroidetes (16.8% and 16.4%), were found in all treatments. Moreover, Actinobacteria (0.07% and 0.07%), Chloroflexi (0.04% and 0.09%), Cyanobacteria (0.03% and 0.08%), Acidobacteria (0.02% and 0.06%), Planctomycetes (0.02% and 0.05%), Gemmatimonadetes (0.02% and 0.03%) and Firmicutes (0.02% and 0.02%) were found in all samples at a relative abundance higher than 1% but lower than 10%.Different treatments had a significant effect on the bacterial taxa distribution, and the relative abundance of Proteobacteria was increased in YH treatments (31%) compared to CK treatments (21%). Bacteroidetes was decreased compared to CK treatments (32%). A similar decreasing trend was found in the relative abundance of Acidobacteria and Chloroflexi (Fig. S1†).
The most abundant classified genera (>1%) for different samples are shown in ESI (Fig. S2†). Among these genera, only Erythrobacter, Ralstonia, Salinimicrobium, Anaerolineaceae, Cytophageceae, Flavisolibacter and Cyanobacteria were represented in all treatments. Moreover, in comparison with CK treatment, higher relative abundances of Erythrobacter and Salinimicrobium and lower relative abundances of Ralstonia, Anaerolineaceae, Flavisolibacter, and Acidobacteria were observed.
At the genera level, clear positive correlations between disease incidence and the relative abundances of Ralastonia (P < 0.05), Anaerolineaceae (P < 0.01), Cyanobacteria (P < 0.05), Acidobacteria (P < 0.01), Comamonadaceae (P < 0.05), and Chryseolinea (P < 0.05) were observed. In contrast, negative correlations for the genera Erythrobacter (P < 0.05) and Salinimicrobium (P < 0.05) were found (Table 4).
| Genus | Disease incidence |
|---|---|
| a Statistical significance at * P < 0.05 and ** P < 0.01. | |
| Erythrobacter | −0.48* |
| Ralstonia | 0.54* |
| Salinimicrobium | −0.46* |
| Anaerolineaceae | 0.59** |
| Cytophagaceae | 0.40 |
| Flavisolibacter | 0.20 |
| Cyanobacteria | 0.53* |
| Acidobacteria | 0.65** |
| Comamonadaceae | 0.46* |
| Xanthomonadaceae | 0.22 |
| Gemmatimonas | 0.40 |
| Chryseolinea | 0.51* |
| Truepera | 0.32 |
Bacterial community structure
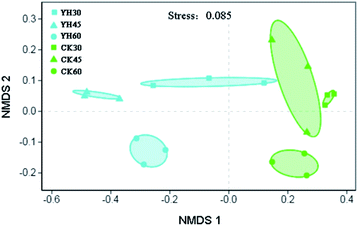
Nonmetric multidimensional scaling (NMDS) analysis based on the Bray–Curtis distance metric showed that the bacterial communities from the same treatment were more similar to each other than those from different treatments, as observed for the six highly supported clusters that were composed of samples from different treatment soils (Fig. 2).Bacterial communities from soil samples that were treated with YH-07 were clustered together, while soil samples from the CK treatment were clustered together based on Bray–Curtis algorithm (Fig. 2). At the same time, ANOSIM results (r = 0.88, P = 0.001) (Table S1†) were basically consistent with the results of NMDS, indicating that the application of biocontrol bacteria YH-07 had a significant impact on bacterial community composition. Bacterial community composition at the different sampling times was differentiated based on the Bray–Curtis algorithm with the same alteration trends for YH and CK treatments (Fig. 2). However, ANOSIM results show no significant difference (Table S1†).
Effects of soil physicochemical characteristics on microbial communities
Mantel tests based on the soil properties and the abundances of the microbial genera data revealed that soil chemical properties were significantly correlated with bacterial communities by selected Bray–Curtis distances (r = 0.80, P < 0.001). Soil available K had the highest correlation with the abundances of the microbial genera (r = 0.79, P < 0.001), while SOM and total N and P showed no correlation (Table 5).| Variable | r | P |
|---|---|---|
| a The bold font numbers indicate a significant difference. | ||
| pH | 0.19 | 0.039 |
| SOM | 0.72 | 0.001 |
| TN | 0.11 | 0.200 |
| TP | −0.01 | 0.876 |
| AP | 0.70 | 0.001 |
| TK | 0.41 | 0.005 |
| AK | 0.79 | 0.001 |
The redundancy analysis (RDA) performed to examine the relationship between the analyzed genera and soil chemical properties, the result showed that the first two components could explain 80.59% of the total variation (Fig. 3). The first component (RDA1), which explained 76.74% of the variance, differentiated the YH and CK treatments from each other, and the second component (RDA2), which differentiated the samples according to different sampling times, explained 3.85% of the variation. As shown in Fig. 3, the dominant microbial genera in the YH treatments were Erythrobacter and Salinimicrobium and related to AP, TK, AK, and SOM. However, the dominant microbial communities in the CK treatment were Ralstinia and Acidobacteria. Moreover, the YH60 samples were clustered around the soil pH axis (Fig. 3).
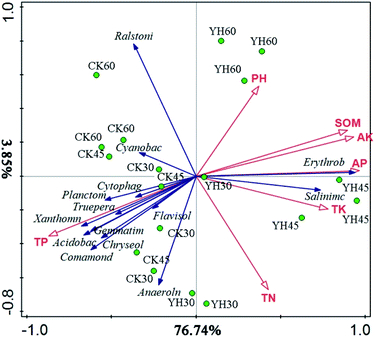 | ||
| Fig. 3 Redundancy analysis of the relationship between the analysed genera, samples and environment variables. | ||
Discussion
In our previous study, the potential mechanism of the biocontrol agent YH-07 for controlling tomato wilt was shown by genome sequencing methods. In this present study, application of biocontrol strain YH-07 significantly suppressed plant disease and improved tomato biomass compared to the uninoculated treatment. Previous studies reporting that the application of biocontrol microorganisms in the soil could control soilborne diseases and improve crop yields in many kinds of plants, such as cucumber,30 tomato,31 and tobacco.32 The results of this paper were consistent with the above reports.Soil bacterial diversity is critical to the function and long-term sustainability of the soil ecosystem.33 In addition, rich soil biodiversity can stabilize the soil ecosystem and enhance the function and biological activity of the soil.34 In the present study, analysis using Chao 1 and ACE showed that the OTU numbers for YH treatment were not significantly higher than for the CK treatment. Moreover, the diversity for YH treatment, which was estimated by the Shannon index, was also not the highest, but the Shannon index of YH45 was significantly higher than that of YH60; at the same time, the Shannon index of CK45 was lower than that of CK60. All the results indicated that short-term application of a biocontrol agent did not significantly affect the richness and diversity of bacterial communities, similar to a previous study reporting that the soil bacterial diversity as a result of bio-organic fertilizer (BIO) treatment was not the highest compared to other treatments, as determined by the Shannon index.35
The present study also demonstrated that biocontrol agent application significantly influenced bacterial community composition and structure. In our study, the results of NMDS analysis showed that the bacterial communities in the soils with different treatments were well differentiated from each other, mainly because of the ability of root exudates to influence the composition of the soil bacterial communities.36
YH-07 application increased the relative abundance of the phylum Proteobacteria as compared with CK treatment. Most Proteobacteria can predominate in a variety of environments,37,38 and many are responsible for symbiotic nitrogen fixation in legumes.39 Deeper taxonomic analyses revealed that the genus Erythrobacter within Proteobacteria had the highest relative abundance in YH treatment. Functional strain YH-07 belongs to genus Erythrobacter and can be detected in the soil, suggesting that YH-07 could survive and colonize the tomato rhizosphere soil. In accordance with our result, Liu et al.40 reported that Bacillus amyloliquefaciens SQR9 could effectively colonize in the soil and suppress cucumber plant disease. Previous study results suggested that the phylum Chloroflexi contains aerobic and anaerobic thermophilic bacteria, filamentous hypoxic phototrophs, and anaerobic organohalide respirators.41,42 In our study, the relative abundance of Chloroflexi was lower in the YH treatment, which coincides with the previous reports, and the relative abundance of this phylum was higher in the chemical fertilizer (CF) treatment than in the CK treatment.43
For the Bacteroidetes phylum, it was reported that increased species abundance of Bacteroides was able to effectively reduce crop morbidity in soil that inhibited wheat total erosion and banana wilt disease.44,45 In this study, the relative abundance of Salinimicrobium (belonging to the Bacteroidetes phylum) was negatively correlated with the incidence of tomato Fusarium wilt and positively correlated with soil pH.
Microorganisms of the genus Salinimicrobium are mostly found in environments such as drought areas, oceans, and salt-rich mediums, and most of them are halophiles. They can grow under NaCl concentrations of 0.5–15% (w/v), which makes them a special group, able to adapt to different salt concentrations. This kind of microorganism can enrich the Na+ ions in the soil because of its salinity. When the saturation of Na+ ions adsorbed on the soil colloid increases to a certain extent, it will cause the hydrolysis of exchangeable cations and produce NaOH in the soil solution, which will increase the soil pH. Therefore, it has been speculated that Salinimicrobium could increase soil pH and create an unfavorable soil environment for pathogens, thus reducing the occurrence of tomato disease.
Some microorganisms have direct or indirect negative effects on plants. For example, Xanthomonas microorganisms can cause bacterial spots and blight on leaves, stems, and fruits of many plants.46 The abundance of Ralstonia in the rhizosphere of plants was relatively high, which would increase the risk of disease of plants.31,47 In this study, it was found that the relative abundance of both Ralstonia and Xanthomonas were significantly positively correlated with the incidence of tomato Fusarium wilt, consistent with previous reports.
In addition, Rosenzweig et al.48 found that the decreasing relative abundance of Acidobacteria could effectively inhibit potato wilt, which means Acidobacteria was positively correlated with the incidence of wilt disease. Similar trends were found in this study: the relative abundance of Acidobacteria in YH treatment was significantly lower than that in the control group.
Previous study results have suggested that the relative abundance of Gemmatimonas was negatively correlated with the plant disease incidence.49 However, in the present study, these taxa did not exhibit a correlation with disease incidence, which suggested that disease incidence depends on other characteristics of the soil.
Soil physicochemical characteristics may also be an important indicator of disease suppression. It is widely known that soil microbial communities have key roles in the soil nutrient cycle, not only affecting soil quality and ecosystem sustainability50 but also influencing the physical and chemical properties of soil.16 In this study, YH treatment had a significantly higher SOM and available K and P contents compared to the CK treatment. Wang et al.43 reported that long-term application of biocontrol fertilizer can effectively accumulate soil nutrients and improve and maintain crop yield. Liu et al.40 found that Fusarium abundance was negatively correlated with SOM in a potato monoculture system.
The resisting salt harm by accumulating potassium is one of the salt tolerance mechanisms of bacteria. Strain YH-07 is a salt-tolerant bacterium, and the genome of strain YH-07 contains a Na+/H+ reverse transporter, which maintains a low concentration of salt in cells by catalyzing the output of univalent cations such as Na+ and K+.51 This salt tolerance mechanism may be the reason strain YH-07 affects the change in K content. The positive correlation between AP content in soil and the biocontrol bacterium YH-07 is obvious, which is consistent with the phosphorylation mechanism of the strain found above, indicating that the biocontrol bacterium YH-07 may be able to mineralize phosphorus in soil, and to some extent, increase the availability of P. Research by Shen et al.52 also indicated that higher soil available P was associated with lower banana Fusarium wilt disease incidence in naturally suppressive soil.
These findings of this study indicated the role of biocontrol agent YH-07 in improving soil nutrients and enhancing the plant's capacity to resist disease. Although the mean difference of soil pH between the three YH and CK samplings is only 0.24, the decrease in pH may significantly affect soil microbial characteristics, nutrient availability, root system, and plant growth.53
To sum up, the change of a single factor cannot accurately reflect the effect of biocontrol agent on disease prevention and control, because the final effect is often the result of the synergistic effects of all factors. Suppression of disease is probably affected by the induced feedback loops among soil properties and soil microbial communities after the application of biocontrol agent YH-07.
Conclusions
In this study, application of Erythrobacter YH-07 in a tomato pot experiment effectively controlled outbreaks of tomato Fusarium wilt and increased the tomato plant biomass compared to CK treatment. Soil microbial composition in this system was significantly affected by the application of biocontrol strain YH-07, and the altered soil microbial communities played a crucial role in suppressing tomato wilt. Specifically, the abundance of Erythrobacter and Salinimicrobium was negatively correlated with disease incidence. Overall, the decrease in disease incidence and increase in tomato plant biomass after YH-07 application might be due to the increase in soil nutrient contents, which modified the bacterial communities by enriching Erythrobacter and Salinimicrobium. The altered microbial community may in turn contribute to greater disease suppression and ensure stable, healthy tomato growth in this system. This study may suggest new approaches for the development of biocontrol strategies. Future studies using long-term experiments would be helpful for clarifying the relationships between soil characteristics, microbiota, and Fusarium wilt.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the anonymous reviewers for their insightful comments and advice on this manuscript. This research was supported by the National Key Research and Development Program of China (2017YFD0200604) and the National Natural Science Foundation of China (no. 31400464).References
- N. Singh and Z. A. Siddiqui, Effects of Bacillus subtilis, Pseudomonas fluorescens and Aspergillus awamori on the wilt-leaf spot disease complex of tomato, Phytoparasitica, 2014, 43, 61–75 CrossRef.
- Q. Y. Xue, J. Q. Li, Y. Zheng, X. Y. Ding and J. H. Guo, Screening tomato-associated bacteria for biological control of grey mold on tomato, Biocontrol Sci. Technol., 2013, 23, 245–259 CrossRef.
- C. F. Ajilogba and O. O. Babalola, Integrated management strategies for tomato Fusarium wilt, Biocontrol Sci., 2013, 18, 117–127 CrossRef.
- L. Wen, S. Y. L. Marzano, L. Ortiz-Ribbing, J. Gruver, G. Hartman and D. M. Eastburn, Suppression of soilborne diseases of soybean with cover crops, Plant Dis., 2017, 101, 1918–1928 CrossRef CAS.
- G. Bonanomi, V. Antignani, M. Capodilupo and F. Scala, Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases, Soil Biol. Biochem., 2010, 42, 136–144 CrossRef CAS.
- M. G. Bakker, J. M. Chaparro, D. K. Manter and J. M. Vivanco, Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays, Plant Soil, 2015, 392, 115–126 CrossRef CAS.
- L. Fu, Y. Ruan, C. Tao, R. Li and Q. Shen, Continous application of bioorganic fertilizer induced resilient culturable bacteria community associated with banana Fusarium wilt suppression, Sci. Rep., 2016, 6, 27731 CrossRef CAS.
- W. Xiong, S. Guo, A. Jousset, Q. Zhao, H. Wu, R. Li, G. A. Kowalchuk and Q. Shen, Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome, Soil Biol. Biochem., 2017, 114, 238–247 CrossRef CAS.
- N. Ling, K. Deng, Y. Song, Y. Wu, J. Zhao, W. Raza, Q. Huang and Q. Shen, Variation of rhizosphere bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer, Microbiol. Res., 2014, 169, 570–578 CrossRef.
- J. Habig and C. Swanepoel, Effects of conservation agriculture and fertilization on soil microbial diversity and activity, Environments, 2015, 2, 358–384 CrossRef.
- M. Ahemad and M. Kibret, Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective, J. King Saud Univ., Sci., 2014, 26, 1–20 CrossRef.
- Y. Pii, T. Mimmo, N. Tomasi, R. Terzano, S. Cesco and C. Crecchio, Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review, Biol. Fertil. Soils, 2015, 15, 403–415 CrossRef.
- H. Liu, W. Xiong, R. Zhang, X. Hang, D. Wang, R. Li and Q. Shen, Continuous application of different organic additives can suppress tomato disease by inducing the healthy rhizospheric microbiota through alterations to the bulk soil microflora, Plant Soil, 2018, 423, 229–240 CrossRef CAS.
- S. Zhao, D. Liu, N. Ling, F. Chen, W. Fang and Q. Shen, Bio-organic fertilizer application significantly reduces the Fusarium oxysporum population and alters the composition of fungi communities of watermelon Fusarium wilt rhizosphere soil, Biol. Fertil. Soils, 2014, 50, 765–774 CrossRef.
- T. Tang, X. Sun, Y. Dong and Q. Liu, Erythrobacter aureus sp. nov., a plant growth-promoting bacterium isolated from sediment in the Yellow Sea, China, 3 Biotech, 2019, 9, 430 CrossRef.
- Q. Zhao, C. Dong, X. Yang, X. Mei, W. Ran, Q. Shen and Y. Xu, Biocontrol of Fusarium wilt disease for Cucumis melo melon using bio-organic fertilizer, Appl. Soil Ecol., 2011, 47, 67–75 CrossRef.
- S. Bao, Soil and agro-chemical analytical methods, China Agriculture Press, Beijing, 2010 Search PubMed.
- M. Faheem, W. Raza, W. Zhong, Z. Nan, Q. Shen and Y. Xu, Evaluation of the biocontrol potential of Streptomyces goshikiensis YCXU against Fusarium oxysporum f. sp. Niveum, Biol. Control, 2015, 81, 101–110 CrossRef.
- P. D. Schloss, S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn and C. F. Weber, Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol., 2009, 75, 7537–7541 CrossRef CAS.
- M. J. Claesson, O. O'Sullivan, Q. Wang, J. Nikkilä, J. R. Marchesi, H. Smidt, W. M. de Vos, R. P. Ross and P. W. O'Toole, Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine, PLoS One, 2009, 4, e0006669 CrossRef.
- J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, C. A. Lozupone, P. J. Turnbaugh, N. Fierer and R. Knight, Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4516–4522 CrossRef CAS.
- J. Sun, Q. Zhang, J. Zhou and Q. Wei, Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site, PLoS One, 2014, 9, e111744 CrossRef.
- R. C. Edgar, UPARSE: Highly accurate OTU sequences from microbial amplicon reads, Nat. Methods, 2013, 10, 996–998 CrossRef CAS.
- E. Pruesse, C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies and F. O. Glöckner, SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB, Nucleic Acids Res., 2007, 35, 7188–7196 CrossRef CAS.
- J. R. Cole, Q. Wang, J. A. Fish, B. Chai, D. M. McGarrell, Y. Sun, C. T. Brown, A. Porras-Alfaro, C. R. Kuske and J. M. Tiedje, Ribosomal Database Project: Data and tools for high throughput rRNA analysis, Nucleic Acids Res., 2014, 42, D633–D642 CrossRef CAS.
- J. Bunge and K. Barger, Parametric models for estimating the number of classes, Biom. J., 2008, 50, 971–982 CrossRef.
- C. Wang, J. Zhou, J. Liu, K. Jiang, H. Xiao and D. Du, Responses of the soil fungal communities to the co-invasion of two invasive species with different cover classes, Plant Biol., 2017, 20, 151–159 CrossRef.
- H. G. Washington, Diversity, biotic and similarity indices. A review with special relevance to aquatic ecosystems, Water Res., 1984, 18, 653–694 CrossRef.
- C. Morris, Analysis of Ecological Data Using Canoco 5, Proceedings of the Annual Congresses of the Grassland Society of Southern Africa, 2nd edn, 2015, vol. 32, pp. 289–290 Search PubMed.
- X. Huang, L. Chen, W. Ran, Q. Shen and X. Yang, Trichoderma harzianum strain SQR-T37 and its bio-organic fertilizer could control Rhizoctonia solani damping-off disease in cucumber seedlings mainly by the mycoparasitism, Appl. Microbiol. Biotechnol., 2011, 91, 741–755 CrossRef CAS.
- Z. Wei, X. Yang, S. Yin, Q. Shen, W. Ran and Y. Xu, Efficacy of Bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field, Appl. Soil Ecol., 2011, 48, 152–159 CrossRef.
- Y. Liu, J. Shi, Y. Feng, X. Yang, X. Li and Q. Shen, Tobacco bacterial wilt can be biologically controlled by the application of antagonistic strains in combination with organic fertilizer, Biol. Fertil. Soils, 2013, 49, 447–464 CrossRef.
- A. C. Kennedy and K. L. Smith, Soil microbial diversity and the sustainability of agricultural soils, Plant Soil, 1995, 170, 75–86 CrossRef CAS.
- G. Chaer, M. Fernandes, D. Myrold and P. Bottomley, Comparative resistance and resilience of soil microbial communities and enzyme activities in adjacent native forest and agricultural soils, Microb. Ecol., 2009, 58, 414–424 CrossRef CAS.
- C. Xue, Y. Ruan, J. Zhang, Q. Shen, D. Wang, Z. Shen and R. Li, Deep 16S rRNA pyrosequencing reveals a bacterial community associated with banana Fusarium wilt disease suppression induced by bio-organic fertilizer application, PLoS One, 2014, 9, e98420 CrossRef.
- J. M. Chaparro, D. V. Badri and J. M. Vivanco, Rhizosphere microbiome assemblage is affected by plant development, ISME J., 2014, 8, 790 CrossRef CAS.
- J. D. Neufeld and W. W. Mohn, Unexpectedly high bacterial diversity in arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags, Appl. Environ. Microbiol., 2005, 71, 5710–5718 CrossRef CAS.
- P. Curtis, C. H. Nakatsu and A. Konopka, Aciduric Proteobacteria isolated from pH 2.9 soil, Arch. Microbiol., 2002, 178, 65–70 CrossRef CAS.
- J. Raymond, J. L. Siefert, C. R. Staples and R. E. Blankenship, The natural history of nitrogen fixation, Mol. Biol. Evol., 2004, 21, 541–554 CrossRef CAS.
- Y. Liu, N. Zhang, M. Qiu, H. Feng, J. M. Vivanco, Q. Shen and R. Zhang, Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection, FEMS Microbiol. Lett., 2014, 353, 49–56 CrossRef CAS.
- S. Hanada, A. Hiraishi, K. Shimada and K. Matsuura, Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement, Int. J. Syst. Bacteriol., 1995, 45, 676–681 CrossRef CAS.
- P. Hugenholtz and E. Stackebrandt, Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description), Int. J. Syst. Evol. Microbiol., 2004, 54, 2049–2051 CrossRef.
- L. Wang, J. Li, F. Yang, E. Yaoyao, W. Raza, Q. Huang and Q. Shen, Application of bioorganic fertilizer significantly increased apple yields and shaped bacterial community structure in orchard soil, Microb. Ecol., 2017, 73, 404–416 CrossRef CAS.
- H. Sanguin, L. Kroneisen, K. Gazengel, M. Kyselková, B. Remenant, C. Prigent-Combaret, G. L. Grundmann, A. Sarniguet and Y. Moënne-Loccoz, Development of a 16S rRNA microarray approach for the monitoring of rhizosphere Pseudomonas populations associated with the decline of take-all disease of wheat, Soil Biol. Biochem., 2008, 40, 1028–1039 CrossRef CAS.
- Z. Shen, B. Wang, N. Lv, Y. Sun, X. Jiang, R. Li, Y. Ruan and Q. Shen, Effect of the combination of bio-organic fertiliser with Bacillus amyloliquefaciens NJN-6 on the control of banana Fusarium wilt disease, crop production and banana rhizosphere culturable microflora, Biocontrol Sci. Technol., 2015, 25, 716–731 CrossRef.
- J. Boch and U. Bonas, Xanthomonas AvrBs3 family-type III effectors: discovery and function, Annu. Rev. Phytopathol., 2010, 48, 419–436 CrossRef CAS.
- V. Shanmugam, P. Chugh and P. Sharma, Cold-tolerant Trichoderma species for the management of Fusarium wilt of tomato plants, Ann. Microbiol., 2015, 65, 543–551 CrossRef.
- N. Rosenzweig, J. M. Tiedje, J. F. Quensen III, Q. Meng and J. J. Hao, Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses, Plant Dis., 2012, 96, 718–725 CrossRef.
- C. L. Rivard, S. O'Connell, M. M. Peet, R. M. Welker and F. J. Louws, Grafting tomato to manage bacterial wilt caused by ralstonia solanacearum in the southeastern United States, Plant Dis., 2012, 96, 973–978 CrossRef CAS.
- S. Zhong, Q. Shen, Y. Ruan, Z. Shen, R. Li, B. Wang, Y. Wang and X. Mei, Induced soil microbial suppression of banana Fusarium wilt disease using compost and biofertilizers to improve yield and quality, Eur. J. Soil Biol., 2013, 57, 1–8 Search PubMed.
- C. Hunte, E. Screpanti, M. Venturi, A. Rimon, E. Padan and H. Michel, Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH, Nature, 2005, 435, 1197–1202 CrossRef CAS.
- Z. Shen, Y. Ruan, B. Wang, S. Zhong, L. Su, R. Li and Q. Shen, Effect of biofertilizer for suppressing Fusarium wilt disease of banana as well as enhancing microbial and chemical properties of soil under greenhouse trial, Applied Soil Ecology, 2015, 93, 111–119 CrossRef.
- A. D. van Diepeningen, O. J. de Vos, G. W. Korthals and A. H. C. van Bruggen, Effects of organic versus conventional management on chemical and biological parameters in agricultural soils, Applied Soil Ecology, 2006, 31, 120–135 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05452f |
| This journal is © The Royal Society of Chemistry 2020 |