Open Access Article
Open Access ArticleUnderstanding the relevance of protein corona in nanoparticle-based therapeutics and diagnostics
Debolina Chakraborty
a,
K. R. Ethiraj
*a and
Amitava Mukherjee
 *b
*b
aSchool of Advanced Sciences, Vellore Institute of Technology, Vellore, 632014, India. E-mail: ethukr@gmail.com; krethiraj@vit.ac.in
bCentre for Nanobiotechnology, Vellore Institute of Technology, Vellore, 632014, India. E-mail: amit.mookerjea@gmail.com; amitav@vit.ac.in
First published on 21st July 2020
Abstract
Over the past few decades, nanoparticle-based therapeutic and diagnostic systems have gained immense recognition. A relative improvement in the status of the global cancer burden has been successful due to the advent of nanoparticle-based formulations. However, exposure of nanoparticles (NPs) to a real-time biological media alters its native identity due to the formation of the biomolecular corona. Such biological interactions hinder the efficiency of the NPs system. The parameters that govern such intricate interaction are generally overlooked while designing nano drugs and delivery systems (nano-DDS). Fabricating nano-DDS with prolonged circulation time, enhanced drug-loading, and release capacity along with efficient clearance, remain the primary concerns associated with cancer therapeutics. This present review firstly aims to summarize the critical aspects that influence protein coronation on therapeutic nanoparticles designed for anti-cancer therapy. The role of protein corona in modifying the overall pharmacodynamics of the nanoparticle-based DDS has been discussed. Further, the studies and patents that extend the concept of protein corona into diagnostics have been elaborated. An understanding of the pros and cons associated with protein coronation would not only help us gain better insights into the fabrication of effective anti-cancer drug-delivery systems but also improve the shortcomings related to the clinical translation of these nanotherapeutics.
1. Introduction
Extensive research in the field of nanotechnology has led to substantial use of various forms of nanoparticles (NPs) in environmental, agricultural, industrial, and medicinal fields.1–4 NPs-based systems have enabled researchers to solve several critical aspects associated with disease diagnostics and therapeutics.5 The administration of the NPs to the complex biological media exposes its pristine surface to several biomolecules. As suggested by K. A. Dawson, the biomolecules present in the immediate environment inevitably adsorbs onto the NPs and forms a corona.6 The corona formation eventually alters the native property and desired functionality of the NPs.7–9 Thus only a few nanoparticle-based formulations are successfully translated into the clinical phase.To narrow the gap between this bench-to-bedside researches, a proper understanding of the biological interactions with the NPs is obligatory. A simple PubMed search shows that over a while, there has been a tremendous increase in the attempt to understand protein corona. Several research groups have categorically reported various aspects related to the formation of protein corona on different NPs (Fig. 1). Others have emphasized on diverse detection strategies employed to evaluate protein corona either quantitatively or qualitatively.10,11 Elaborate studies conducted by our group has helped in understanding the protein corona formation based on variation in surface functionalization over a dumbbell-, rod-bimetallic-gold nanorods and also selenium NPs.12–15 With the conclusions from our previous study, we further strategically applied the coronated nanostructures as an effective drug delivery agent for cancer chemotherapy.16 Over the past 5 years, several review articles have collectively summarized the principles and parameters that govern corona formation.8,17,18 However, there are very few reports that illustrate the utility of the protein corona in the emerging field of nanotherapeutics and diagnostics. For successful clinical trials, it is necessary to tactfully deal with the aspects of protein corona that determine the efficiency of the final product in vivo.
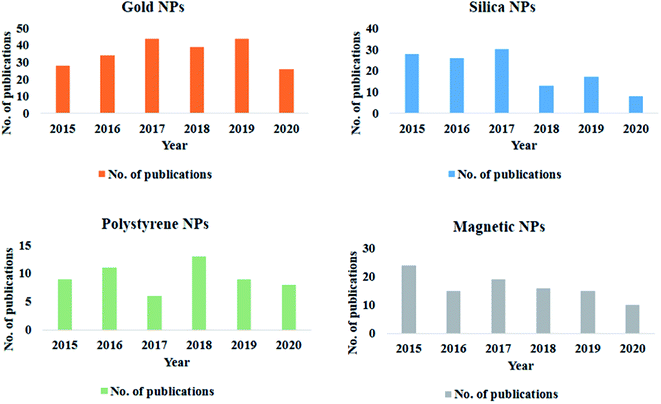 | ||
| Fig. 1 Graphical representation of studies on protein corona formation on different nanoparticles over the past 5 years. | ||
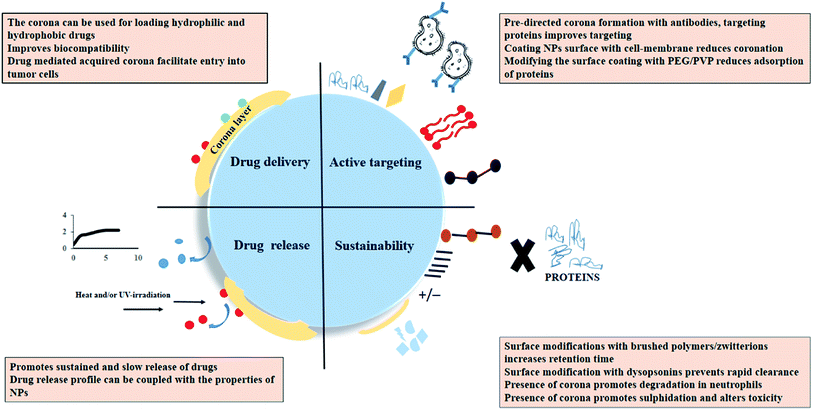
The current review firstly gives a comprehensive overview of the theories of protein corona formation and briefly summarizes the characteristics of the NPs that influence the same. The performance of a therapeutic or diagnostic NP is constantly challenged by the complex nature of an individual's physiological system. The review discusses the specifications of the exposure conditions that directly regulate coronation on different NPs. The analysis of protein corona as a potential drug delivery tool and its impact on pharmacokinetics and pharmacodynamics of NPs in a biological system has been elaborated. Other important takeaways from the review are the possible application of protein coronated NPs as an advanced diagnostic tool. The concept of personalized corona from an individual can give a specific molecular identity to the NPs which in turn can be utilized for early-stage disease screening or even post-treatment when the majority of the disease-specific biomarkers are removed. In this regard, some of the recent patents have also been discussed. The summarized details aim to highlight the critical facets governing nanobio-interactions and the strategies employed to use such parameters in diagnostics and therapeutics.
2. Understanding the theories of protein corona formation
The introduction of NPs in the physiological system leads to the adsorption of biomolecules on the NPs surface. Proteins being the most abundant biomolecule, layers over the surface of the NPs, and form an NP–protein corona complex (Scheme 1).19 Terminologies like hard and soft corona classify the corona layer, in terms of their binding affinities towards the NPs surface.20 The characteristics of the hard and soft corona have been listed in Table 1.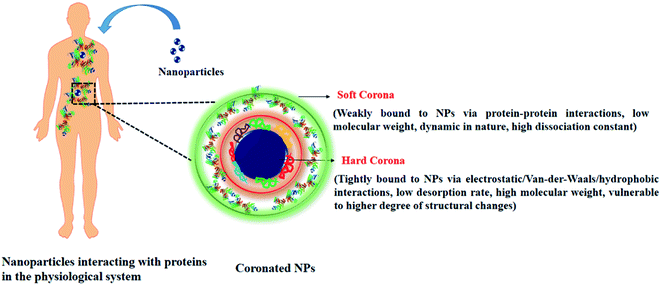 | ||
| Scheme 1 Schematic representation showing nanoparticle–protein interaction and properties of bound corona proteins. | ||
| Parameter | Soft corona | Hard corona |
|---|---|---|
| Type of adherence to the NPs | Loose (indirectly associated with NPs via protein–protein interaction) | Tight (directly associated with the NPs via electrostatic, van-der-Waals, hydrophobic interactions |
| Dissociation constant | High, dynamic in nature | Low, strongly associated with the NPs surface |
| Structural changes | A lower degree of conformational changes observed | A higher degree of conformational changes are observed |
| Mode of detection | Liquid chromatography with tandem mass spectrometry (LC-MS/MS), protein quantification by bicinchoninic acid (BCA)/Bradford assay | Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), LC-MS/MS, transmission electron microscopy (TEM), fluorescence correlation spectroscopy (FCS) |
| Isolation from NPs surface | Sucrose-gradient centrifugation, ultra-centrifugation | Treatment with laemmli buffer followed by heating and centrifugation |
The dynamics of protein corona formation is governed by several forces such as electrostatic, van-der-Waals, and hydrophobic interaction.19,20 Different conclusions regarding the mechanism of protein corona formation have been reported. A simplistic diffusion-based approach was put forward by Leo Vroman that described adsorption kinetics of proteins for several flat and curved NPs surfaces. The Vroman effect states that during NPs incubation in a complex multiprotein system, the high mobility proteins (having low molecular weight) arrive first and adsorb on the NPs surface, in the due course of time the adsorbed proteins are replaced by more stable less mobile proteins that have a higher binding affinity for the NPs.21,22 To put it simply, a critical balance between the kinetics and thermodynamics of proteins determines the corona formation on the NPs surface. In another theory put forward by Hirish et al. it is described that the entire dynamic exchange occurs via the formation of “transient complex”. Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS) and Atomic Force Microscopy (AFM) analysis suggest that adsorption of the initial layer of proteins is followed by the embedding of the second layer of protein which in turn results in a formation of multilayered aggregate. The aggregated complex (transient complex) finally exposes the initially adsorbed proteins which desorb from the NPs surface into the solution leaving the NPs with the proteins.23 A better understanding of the evolution of protein corona is explained by the dynamic density functional theory (DDFT) which combines the steric and electrostatic interactions. In addition to the classical diffusion-based Vroman effect, the non-monotonic adsorption pattern of corona formation is rather dependent on parameters such as adsorption energy, bulk density, and charge of the protein. It also suggests that inter-particle interaction can promote cooperative binding of proteins on the NPs surface and not just competitive binding.24 However, other studies do not limit the formation of protein corona only to the Vroman effect.25–28 The difference in the opinions is possibly due to the diversity of the experimental protocols in terms of variation in NPs, analytical methods, and choice of protein source. The principle of competitive displacement of proteins was not applicable and a stable protein composition throughout an extended period of 4 h was observed in the case of metal oxide NPs.29 Label-free liquid chromatography-mass spectrometry (LC-MS) studies conducted by Tenzer et al. also revealed that protein corona formation does not solely depend on the principles of the Vroman effect but instead complex interplay of multiple parameters determine the presence of any particular protein on the NPs surface.30 The Vroman effect of time-bound protein displacement also does not apply to solid lipid nanoparticle (SLN) incubated with dilute human plasma.31 Oil-in-water emulsions and iron oxide NPs have also shown similar trends.25,32 Table 2 summarizes a few of the studies wherein theories of protein coronation varied for different NPs.
| Nanoparticle type | Protein source | Mechanism of protein corona formation | Ref. |
|---|---|---|---|
| Sulfonate and carboxyl-polystyrene NPs | Transferrin and blood plasma | Exchange of transferrin from the soft corona layer in the presence of different concentration of blood plasma | 22 |
| Polystyrene NPs | Cellulase enzyme mixture | Vroman effect showing displacement of proteins along with transient complex formation | 23 |
| Oil-in-water emulsion | Human plasma | No displacement of proteins was observed. Apolipoproteins were dominantly present in the corona component | 25 |
| Citrate capped gold NPs | Ubiquitin | A combinatorial effect of NPs electrostatic potential, surface coating and slow reorientation of the NPs to find ubiquitin's specific binding site promotes corona formation | 27 |
| Citrate-capped gold NPs with PEG coating | Human plasma | No competitive displacement of proteins were observed. Quantitative variation in bound protein due to surface coating was observed | 28 |
| Metal oxide NPs (TiO2, SiO2, ZnO) | Human plasma | No typical Vroman effect was observed. Shape of the NPs played important role in adsorption of particular class of proteins | 29 |
| Ultrasmall supermagnetic iron oxide nanoparticles (USPIO) | Plasma | No typical Vroman effect was observed. An increase in the amount of immunoglobin and fibrinogen was observed for longer incubation time points | 32 |
Reporting a discrete mechanism of protein corona formation is a challenging task. A case-by-case approach is thus required to address the questions related to the formation of corona on NPs. The understanding of the interactive process would enable us to answer the vital questions regarding the biological response of these coronated NPs. Considering the inevitable nanobio-interactions it is necessary to tactfully work on the architectural framework of the NPs. Several reviews have summarized and categorized the varying nature of protein corona formation over a plethora of NPs and how it influences the cellular interactions.8,17,33 Though several research groups have collectively provided new insights about protein coronation there are few pivotal aspects regarding corona formation that are unanswered
• What role does protein corona play in a drug delivery system?
• Can artificial engineering of the protein corona help NPs attain a better disease targeting efficiency?
• How does the protein corona affect the sustainability of therapeutic NPs?
• Can we extend the concept of protein corona for sensing and/or diagnostic applications?
3. Morphological variations in NPs affecting protein corona
The type and quantity of corona formation on NPs is dependent on the intrinsic property of the NPs. Size, shape, surface charge, and hydrophobicity can effectively modulate the corona formation. The physicochemical property of the NPs not only alter the identity of the adsorbed protein but also induce conformational change. Reports suggest that smaller sized NPs due to a higher degree of surface curvature generally tend to adsorb less protein and also the protein adsorbed forms a thick layer whereas, particles with relatively large size generally adsorb more proteins and the proteins are more evenly layered.34 In a combination study, the effect of surface charge and size of polystyrene NPs was studied. It was observed that both the parameters simultaneously influence protein coronation.6,35 It has been reported that rod-like structure tends to adsorb more proteins as compared spherical particles.36 The surface charge or zeta potential of the NPs have a significant impact on the electrostatic interactions. The positive and negative amino acid residues play a significant role in binding to charged particles. Reports suggest that positively charged particle generally tends to attach to proteins that have a pI < 5.5 whereas negatively charged particles bind to protein with pI > 5.5.37 Adsorption of human serum albumin (HSA) which is predominantly negatively charged at physiological pH has shown higher binding affinities towards cetyltrimethylammonium bromide (CTAB)-capped selenium NPs as compared IgG which is positively charged.15 The repulsive forces along with other parameters play a critical role in the enrichment of the corona proteins. Study shows that the increase in the hydrodynamic size of positively charged magnetic nanoparticles (MNPs) (3000 nm) following incubation in serum decreases its cellular uptake as compared to negatively charged MNPs which adsorbs less protein.38 The degree of protein conformational loss follows the pattern positive > negative > neutral.39 In the case of the nature of the material i.e. hydrophobic/hydrophilic, it is observed that hydrophobic NPs tend to adsorb more proteins as compared to hydrophilic NPs. Reports suggest that hydrophobic carbon NPs show considerably higher protein adsorption as compared to hydrophilic metal NPs.40 It is suggested that the stabilizing interactions such as hydrogen bonding, hydrophobic and van der Waals are generally disrupted when protein comes in contact with a hydrophobic NPs. During the attachment of proteins to the NPs, new secondary bonds are established along with the breakdown of the old stabilizing bonds resulting in conformational loss.414. Extrinsic parameters influencing protein corona formation
The physiological barriers that the NPs composite encounters upon entering the biological system such as a change in pH and temperature, change in the composition of the biological fluids, and several immune cells significantly impact the overall performance of the NP system. Optimizing the crucial factors for the formulation of more efficient and stable nanomedicine thus remains a challenging concern. Smart modification and alterations in the architectural framework of the study can enhance the understanding of NPs–cellular interactions. The complex process of nanobio-interactions is also dependent on the blood flow dynamics and the presence of various other primary cells in the human whole blood.42,43 In this regards, the alterations can be introduced into the NPs system by either be modifying the material composition (metal, semiconducting, magnetic), morphology (size and shape), surface functionalization (positive/negative or neutral charge); or by engineering several parameters related to the interacting analyte. While we, aim to focus mainly on protein's interaction with the NPs in this review, it is necessary to highlight some of the studies wherein modifying functional moieties, or changing the type and concentration of the protein affected the corona formation as well as impacted cellular interactions.4.1. Protein corona upon varying the concentration and composition of serum
In an attempt to understand the biomolecular corona, several studies have been reported wherein, the concentration of the protein in the medium was varied. It was suggested that exposure of NPs at a low protein concentration allows relatively few proteins to adhere and distribute on the NPs surface forming a stable NP–protein complex. The protein layers experience a higher degree of conformational changes while masking the NPs surface. However, when the exposure media contains a high concentration of proteins, dense layering occurs and in this case, the majority of the interactions are governed by the adjacent polypeptide fragments. Comparatively lesser conformational changes are noticed since the contact between the NPs surface and the proteins are minimized.44,45Polystyrene- and silica-NPs (200 nm) demonstrated varied patterns of bound corona upon incubation with an increasing concentration of plasma (3–80%). Densitometric analysis showed that for polystyrene NPs increasing the protein concentration directly increased the amount of bound corona, whereas for silica NPs the amount of protein decreased. SDS-PAGE and LC-MS/MS analysis suggested that in a multiprotein system such a blood plasma, high binding proteins which are present in smaller amounts such as histidine-rich glycoprotein tends to replace low-binding affinity protein such as fibrinogen and proteins with a molecular weight in the 50–70 kDa range (present in comparatively much higher amounts in the blood plasma).44 Similarly, for MNPs, for increasing serum concentration the apolipoprotein E-1 (ApOE-1) protein component significantly increased as compared to albumin and transferrin.46 Analysis of potential protein adsorptive property of poly(lactic-co-glycolic acid) nanoparticles (PLGA-NPs) demonstrated that variation in the protein source such as fetal bovine serum (FBS) or human serum, can impact corona formation in terms of both quantity and quality. For 0.08 m2 area of PLGA-NPs incubated with 50 μL human serum or FBS, the bound protein was observed to be 160.91 μg and 23.08 μg respectively. Apart from this, LC-MS/MS showed that PLGA-NPs incubated with human serum tended to adsorb opsonins, components of proteases C1r, and other cationic proteins whereas that with FCS adsorbed hemoglobin subunits.47 Modifying the composition of the incubating media such as complement depletion by heat inactivation was seen to alter the cellular uptake of coronated-polystyrene particles in A549 cells.48 In another study, HSA was modified by succinylation and arylation. It was observed that succinylated-HSA had a weaker binding affinity towards dihydrolipoic acid-coated quantum dots (DHLA-QDs) and formed a thick clustered layer on the NPs surface as compared to its counterpart. Internalization of such modified-protein-coronated QDs in HeLa cells were seen to be influenced by the varying pattern of coronation.49
4.2. Effect of incubation time, temperature, shear stress and pH of the media
Several external factors such as incubation time, temperature, pH, and shear stress can either be co-dependent on each other or function individually while forming protein corona on NPs. Considering the several forces involved in the formation of the biomolecular corona, studies have reported that adsorption of proteins onto the NPs surface occurs rapidly mostly within 30 s of incubation. In an attempt to understand the combined effects of incubation time and temperature, MNPs were incubated with fetal calf serum (FCS) for different time points (1, 5, 10, and 20 min). SDS-PAGE analysis revealed an increase in protein content on the NPs at 25 °C and 37 °C. However, the NPs incubated at higher temperatures such as at 50 °C and 70 °C demonstrated accumulation of the denatured polypeptides in the range of 25 to 75 kDa on NPs surface.50 In another study 30 nm polyethylene glycol (PEG)-coated gold NPs were incubated with pooled plasma samples at room 37 °C wherein it was observed that the total amount of protein decreased with time from 5 to 60 min, but there was no dramatic change in the composition of the protein corona.28External factors such as temperature and pH can change the binding parameters of proteins to NPs. Reports have shown enthalpy-driven binding of bovine serum albumin (BSA) to QDs, wherein at high temperature the BSA binding was lowered.51 The normal body temperature of an individual varies depending on the physiological conditions. Reports on magnetic and copper NPs have shown that change in temperature can control both the pattern and quantity of corona formation respectively.52,53
FCS demonstrated that the binding nature of BSA to QDs altered at different pH (6, 7.4, and 9). The weaker binding was observed due to an increase in repulsive forces at a lower pH, whereas at pH-9, due to loss in rigidity and tertiary contact, BSA acquired a more flexible conformation which in turn led to stronger interactions.51 Different pH inside the cell (lysosomal—5.3; endosomal—4.6) can trigger similar reactions wherein, the conformational stability and biological activity of the coronated proteins may be altered.16
Another factor, such as shear stress imparted by the blood in the blood vessels, has also been reported to alter corona formation on PEGylated-liposomes. Analysis via dynamic light scattering (DLS), nanoLC-MS/MS has proved an increase in the size of the NPs following incubation in FBS. Nearly 108 proteins were identified on the liposome surface under both dynamic and static reaction conditions. Enrichment of complement proteins was observed in case of dynamic incubation whereas albumin, transferrin, and alpha-2-HS-glycoprotein appeared more abundantly in the case of static incubation.54 A schematic representation summarizing the overall factors that influence protein corona formation is given below (Scheme 2).
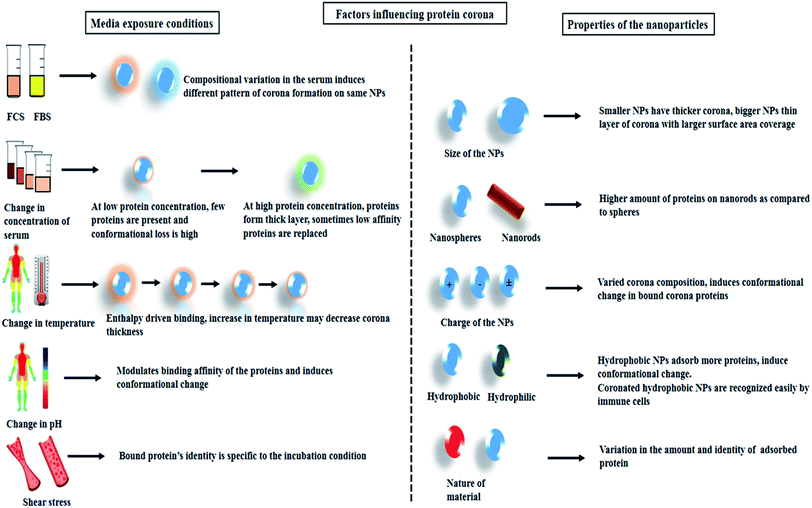 | ||
| Scheme 2 Schematic representation showing the external media conditions and intrinsic property of the NPs that influence protein corona formation. | ||
5. Role of protein corona in nanomedicine
5.1. Employing protein corona for drug loading
The nano-size property of the NPs makes them efficient as drug delivery agents as it enables them to escape the immune system and reach the targeting site. Apart from this the leaky vasculature of the blood vessels readily helps the NPs to penetrate and accumulate via enhanced permeability and retention (EPR) effect.55,56 The composition, shape, size, and surface properties determine the half-life, rate of uptake, toxicity, and accumulation rate within the targeted cells/tissues.56 From the parameters described in the above sections, it can be suggested that several modifications in the architectural design of the nano-composite or the incubating medium can help us generate effective NPs-based DDS. In a few of the recent reports, several research groups have rather utilized the corona components as such for the delivery of the desired drug. Ideally, albumin which contributes nearly 50–60% of the total plasma proteins acts as an effective transportation molecule. It has the potential to hold onto several payloads. Shahabi et al. have mentioned “sponge” like properties of the corona that can hold onto several charged molecules. Silica NPs incubated in 10% FCS have thus been utilized for loading doxorubicin or meloxicam simultaneously. The drug-loaded composite had excellent anti-proliferative properties.57 Considering the tunable physicochemical properties of gold nanorods (GNRs) several studies have combined the benefits of protein corona on GNRs for designing effective nano-DDS. Protein coronated CTAB-capped gold nanorods (GNRs) were used to load doxorubicin (dox) and DNA.58 The passive release of the payload was optimized by tuning the hard and soft corona composition around gold-nanorods, nanobones, and carbon nanotubes.59 Few other studies have incorporated the same strategy for multimodal cancer therapy and combined photodynamic therapy (PDT) and photothermal therapy (PTT).60,61 Apart from this, prodrug nanoassemblies with exposed surface maleimide groups could effectively recruit albumin corona upon incubation in rat plasma. The presence of albumin enhanced their accumulation in tumor cells and also was showed better activity as compared to commercially available Abraxane®.625.2. Protein corona and its effect on drug release kinetics
The release kinetics of drugs is an important parameter that needs to be considered while designing a targeted DDS. The presence of protein corona has shown a significant reduction in the burst release of drugs. Reports suggest that a sustained release profile was observed for commercially available Abraxane®.63 Similarly the release of camptothecin from silica NPs was also slowed in the presence of protein corona.64 A controlled release of the payload was obtained by using ultrafast laser excitation of the GNRs at longitudinal surface plasmon resonance (LSPR). Such an approach dismantles the corona layer and disrupts the binding between the proteins and the payload, finally releasing it at the desired site.58 CTAB-capped GNRs along with polystyrene sulfonate- (PSS) and PEG-capped GNRs were similarly employed for drug loading and pH-triggered release. The study demonstrated the combined effect of exposed surface ligand along with protein corona upon loading and sustained release. Owing to partial protein coverage of NRs surface, the exposed PSS ligand could additionally hold an excess of dox payload and also comparatively had slower release under different pH-trigger.14 In another similar approach sustained dox release from selenium NPs showed a direct relation with the adsorbed albumin corona on the NPs surface.16 These studies reveal that the corona can tactfully hold and release drugs in a controlled fashion. However, reports also suggest that protein corona tend to disturb the integrity and leakage of dox from commercially available Doxove drug.655.3. Protein corona and disease targeting
Efficient disease targeting is one of the crucial parameters that need to be considered while designing therapeutic nano-formulations. Under all the circumstances, it is necessary to analyze the change in the functional properties of the composite material after its interaction with the physiological milieu. Several studies have been reported wherein attachment of targeting ligands have improved the efficiency of NPs-based DDS.66 Pristine NPs coated with peptide fragments and layers of proteins are what the cell sees. Hence, under the ideal in vivo situation, the identity and the performance of the targeting NPs are significantly altered and the desired performance is also compromised. It has been reported that transferrin coated NPs are masked due to protein adsorption layer which in turn results in a loss in active targeting ability of the NPs.7 In another interesting study, IgG pre-coated NPs that was designed for opsonization mediate-uptake by the macrophage cells (RAW 264.7) did not significantly increase NPs cellular uptake due to protein coronation.67 Selenium NPs with different surface functionalization CTAB, polyoxyethylene-(20)-cetyl ether (Brij-58 and SDS) showed reduced cellular uptake upon individual corona formation with HSA and IgG, however, the presence of transferrin corona on positively charged CTAB-capped selenium NPs enhanced its uptake and cellular toxicity in MDA-MB-231 cell lines.15 There are other studies wherein, pre directing protein corona formation can maintain the targeting ability of the NPs system. It was reported that apolipoprotein E4 adsorption on polysorbate 80 stabilized NPs enhanced in targeting ability towards cerebrovascular endothelium and promoted a 3-fold higher accumulation in the brain parenchyma as compared to bare NPs system.68 Apart from pre-directed corona formation with desired proteins, another approach is to design a biomimetic cell-membrane coated NPs system which successfully prevents protein coronation.69 Considering the inherent problems associated with cancer cell targeting, researchers have partially mitigated corona formation by altering the surface properties of the NPs. Modulating the NPs surface with PEG helps the targeting ligand maintain its specific binding properties. The steric hindrance created by the linear chain prevents the accumulation of proteins over the NPs surface. The strategy of pegylation can significantly circumvent opsonin adsorption. Due to lesser protein adsorption, smaller particle size will be maintained resulting in decreased recognition/clearance by the immune cells and higher cellular uptake. However, the PEG chain length must be selected such that it does not interference ligand linker.70 It must be noted that there are limitations associated with the use of PEGylated NPs. The immunogenic response generated due to the presence of anti-PEG antibodies promotes faster clearance of PEG-based drugs which in turn lowers the desired efficacy of the delivery system. Hypersensitive reactions caused due to the immunogenic response can further impart life-threatening side effects.715.4. Sustainability of protein coronated nanomedicine
For nano-formulation to function efficiently, it is necessary that the NPs remain in their active form and is also not eliminated from the system too soon. It was observed that myeloperoxidase (MPO) release and hypochlorite (OCl-) production enhanced the degradation of HSA attached to single-wall carbon nanotubes (SWCNTs) in the neutrophils.72 Also the protein corona initiated a sulfidation-mediated decrease in the toxicity of silver NPs.73 PEG as mentioned previously reduces the process of coronation and prevents the premature clearance of the NPs from the body. Apart from this, the usage of hydrophilic polymers such as polyvinylpyrrolidone (PVP) has also been reported. However, NPs coated with PVP had lower retention time due to the adsorption of a higher amount of opsonins as compared to PEG-coated NPs.74 Reports also suggest that engineering the corona on the NPs surface by incorporating dysopsonins (albumin or apolipoproteins) tend to increase their circulation time. It has been shown that albumin forms protein corona around poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) (PHBHHx) NPs and decreases complement activation.75 Apart from that to prevent macrophage-based clearance of large protein coronated particles use of poly(2-ethyl-2-oxazoline) has also shown good results.76 Other antifouling agents that promote longer stay of NPs in the physiological system includes brushed phosphorylcholine and zwitterionic coatings.77,78Scheme 3 collectively summarizes the role of protein corona in nanomedicine. It can be suggested that while designing novel techniques for corona-based drug delivery systems one should consider the following factors:
• Elucidating the relative affinity of the corona proteins for particular drug payload so that the corona components can itself be utilized for controlled loading and release.
• The predetermination of the plasma concentration at which the corona is enriched with targeting proteins e.g. at 50% plasma concentration liposomes was enriched with vitronectin which can be used further for targeting several tumor cells.79
• Use of hydrophilic surface or anti-fouling polymers so that the protein adsorption is minimized.
• Promoting the binding of dysopsonins on the NPs so that the retention time of the NPs is increased.
6. Protein corona and its diagnostic applications
Apart from using the corona components only for therapeutics, it has been suggested that they can even be exploited for disease diagnostics. The concept of personalized protein corona gives rise to the idea that NPs interacted with serum obtained from individuals having different pathological conditions would behave differently and would acquire a unique identity that is particular to the diseased condition the individual is suffering from.80 Recent patent by the inventors O. C. Farokhzad and M. Mahmoudi claim the development of liposome-based sensor array that has the potential to detect five different types of cancers (lung, pancreas, myeloma, meningioma, or glioblastoma) based on the varying pattern of protein corona fingerprint.81 Few of the other patents related to protein corona based disease diagnosis,82,83 biomarker analysis,84 and therapeutic response,85 has been summarized in Table 3. Zheng et al. exploited the concept of the dynamic nature of protein corona and developed a protein corona based assay system specific for prostate cancer.86 Misfolded or denatured proteins occurring during several pathological conditions such as amyloidosis, Parkinson's disease can coronate differently on the surface of the NPs as compared to the normal protein which has its native conformation intact.87,88 Deng et al. in a study demonstrated the protein corona containing fibrinogen could attach itself to gold NPs and could unfold itself, the unfolded protein can further interact with leucocyte receptors and generate an inflammatory response, however, normal fibrinogen structure could not do the same. The approach can readily act as a biosensing platform for disease diagnosis.89 Another interesting lung cancer detection strategy utilized the principles of the Vroman effect. The competitive replacement of low molecular weight cells by high weight cells facilitated a change in the refractive index which further introduced wavelength shift in the multilayered grated surface plasmon resonance (SPR) sensor.90 In an attempt to extend the application of protein corona based detection beyond cancer diagnostics, Hajipour et al. analyzed the components of hard corona formed on graphene oxide sheets. The serum samples used for the study included healthy individuals along with patients with cardiovascular disease, diabetes, thalassemia, rheumatism, etc. The hard corona components analyzed via gel electrophoresis showed the appearance of disease-specific bands that acted as fingerprints.91 Sensing systems that could identify the probable composition of protein corona under ideal in vivo situation would ease the task of designing an effective NPs system. Surface immobilized liposomes were used as a multi-parametric SPR system to determine the composition of protein without interfering with its natural dynamics.92 It can be suggested that such detailed work protocols can not only help us understand the nano-biointerface better but also pave new ways to resolve several problems associated with the elimination of NPs in clinical trials.| Inventors | Title | Application no. | Ref. |
|---|---|---|---|
| O. C. Farokhzad, M. Mahmoudi | System and method for protein corona sensor array for early detection of diseases | WO2018112460A1 | 81 |
| Q. Huo | Methods for biomolecule and biomolecule complex (bmc) detection and analysis and the use of such for research and medical diagnosis | WO2011088128A2 | 82 |
| K. Dawson, L. Lynch, M. Lundqvist, T. Cedervall | A method for the selective concentration of a specific low abundance biomolecule | WO2010097785A1 | 83 |
| K. Kostarelos, M. Hadjidemetriou | Detection of cancer biomarkers using nanoparticles | WO2018046542A1 | 84 |
| Q. Huo | Biomolecular interactions and interaction products as biomarkers for detection, diagnosis, prognosis and predicting therapeutic responses of human diseases | US20130058923A1 | 85 |
7. Summary and future perspectives
Instead of several research outputs over the decade, there are very few nanomedicines that practically reach clinical trials. The limitations and inherent problems that inhibit the progression and utility of such NPs lies in the fact that several pivotal nano-bio interactions are generally overlooked during the study. Thorough knowledge regarding the varying pattern of corona formation is very important as under ideal in vivo conditions the true identity, fate, and performance of the desired NPs depend on the adsorbed biomolecules. The toxicity, drug loading efficiency, sustainable release, biodistribution, and accumulation can be altered based on the protein-adsorptive behavior of the NPs. Critical aspects such as mapping the protein binding sites and evaluating the spatial organization have technically improved the understanding of the NPs–protein interaction.93 The details of protein coronation at a molecular level still needs more work. Additionally, advanced analytical instruments such as super-resolution fluorescence microscopy and Forster resonance energy transfer spectroscopy that can analyze/visualize real-time protein coronation would potentially add interesting insights. Information about protein corona evolution based on the individual's physiological condition, age and lifestyle can be used as a potential disease monitoring tool. To practically employ such applications, all the parameters that determine protein coronation should be studied in a simulated biological system. Several groups have identified different physicochemical properties of corona proteins; however, sometimes results from different labs were contradicting in nature even when the investigating NPs were the same.94,95 Hence, the interaction of the NPs with the proteins based on the circadian rhythm, body temperature, pH, and shear-stress from the fluids in the presence of other major biomolecules such as lipids, carbohydrates, immune cells, degrading enzymes, should be collectively analyzed. Apart from the immune cells, the presence of white blood cells (WBCs) and coagulation factors can also influence the interaction of NPs with the proteins. Experimental setups thus can include the use of whole blood to understand the impact of such interfering factors. Also to better understand the scenario of corona formation it is necessary to take into consideration the hemodynamics of the blood in the vascular system.Reports have demonstrated that in the case of NPs that are designed for cancer targeting the architectural framework wherein, targeting ligands that are embedded onto the surface of the NPs, gets masked due to corona formation.7 Under such circumstance careful modification in the surface chemistry (incorporation of anti-fouling polymer) or topography of the NPs can partially help in maintaining the pristine properties of the NPs; however since complete elimination of corona formation is not possible, the corona can itself be engineered to achieve active targeting and prolonged circulation. The properties of the corona proteins to further hold onto several charged payload can also be exploited and the coronated-NPs can be used as nano-carriers. While such strategies of preformed corona are put into use, it must be kept in mind that changes in the protein structure can in itself generate an immunogenic response, the overall complexities both after interaction with NPs and payload individually and in combination must be carefully analyzed.
In summary, studies that enhance the in-depth understanding of the adsorptive behavior of the proteins can open up new nanoplatforms that would enable screening, diagnosis, and therapy of several diseases. Efficient overall planning thus would ease the inherent problems associated with the in vitro–in vivo transitions often faced by researchers.
Conflicts of interest
There are no conflicts to declare.References
- S. B. Khan, M. Faisal, M. M. Rahman and A. Jamal, Sci. Total Environ., 2011, 409, 2987–2992 CrossRef CAS PubMed.
- C. M. Rico, M. I. Morales, A. C. Barrios, R. McCreary, J. Hong, W.-Y. Lee, J. Nunez, J. R. Peralta-Videa and J. L. Gardea-Torresdey, J. Agric. Food Chem., 2013, 61, 11278–11285 CrossRef CAS PubMed.
- K. T. Nguyen, J. Nanomed. Nanotechnol., 2011, 2, 103e Search PubMed.
- K. Schmid and M. Riediker, Environ. Sci. Technol., 2008, 42(7), 2253–2260 CrossRef CAS PubMed.
- I. Khan, K. Saeed and I. Khan, Arab. J. Chem., 2017, 12, 908–931 CrossRef.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- A. Salvati, A. S. Pitek, M. P. Monopoli, K. Prapainop, F. B. Bombelli, D. R. Hristov, P. M. Kelly, C. Åberg, E. Mahon and K. A. Dawson, Nat. Nanotechnol., 2013, 8, 137 CrossRef CAS PubMed.
- N. Durán, C. P. Silveira, M. Durán and D. S. T. Martinez, J. Nanobiotechnol., 2015, 13, 55 CrossRef PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. E. Toledano Furman, F. Salvatore and E. Tasciotti, Nanomedicine, 2016, 11, 81–100 CrossRef CAS PubMed.
- S. Winzen, S. Schoettler, G. Baier, C. Rosenauer, V. Mailaender, K. Landfester and K. Mohr, Nanoscale, 2015, 7, 2992–3001 RSC.
- C. Carrillo-Carrion, M. Carril and W. J. Parak, Curr. Opin. Biotechnol., 2017, 46, 106–113 CrossRef CAS PubMed.
- S. A. Alex, N. Chandrasekaran and A. Mukherjee, J. Mol. Liq., 2017, 248, 703–712 CrossRef CAS.
- D. Chakraborty, L. Mohan, S. A. Alex, N. Chandrasekaran and A. Mukherjee, Biomater. Sci., 2019, 7, 63–75 RSC.
- D. Chakraborty, S. Tripathi, K. Ethiraj, N. Chandrasekaran and A. Mukherjee, New J. Chem., 2018, 42, 16555–16563 RSC.
- D. Chakraborty, P. Chauhan, S. A. Alex, S. Chaudhary, K. Ethiraj, N. Chandrasekaran and A. Mukherjee, J. Mol. Liq., 2018, 268, 335–342 CrossRef CAS.
- D. Chakraborty, P. Chauhan, S. Kumar, S. Chaudhary, N. Chandrasekaran, A. Mukherjee and K. Ethiraj, J. Mol. Liq., 2019, 296, 111864 CrossRef CAS.
- C. Ge, J. Tian, Y. Zhao, C. Chen, R. Zhou and Z. Chai, Arch. Toxicol., 2015, 89, 519–539 CrossRef CAS PubMed.
- L. Treuel, D. Docter, M. Maskos and R. H. Stauber, Beilstein J. Nanotechnol., 2015, 6, 857–873 CrossRef CAS PubMed.
- P. Del Pino, B. Pelaz, Q. Zhang, P. Maffre, G. U. Nienhaus and W. J. Parak, Mater. Horiz., 2014, 1, 301–313 RSC.
- W. Liu, J. Rose, S. Plantevin, M. Auffan, J.-Y. Bottero and C. Vidaud, Nanoscale, 2013, 5, 1658–1668 RSC.
- R. A. Latour, G. L. Bowlin and G. Wnek, Encyclopedia of Biomaterials and Biomedical Engineering, 2005, p. 270 Search PubMed.
- S. Milani, F. Baldelli Bombelli, A. S. Pitek, K. A. Dawson and J. Radler, ACS Nano, 2012, 6, 2532–2541 CrossRef CAS PubMed.
- S. L. Hirsh, D. R. McKenzie, N. J. Nosworthy, J. A. Denman, O. U. Sezerman and M. M. Bilek, Colloids Surf. B Biointerfaces, 2013, 103, 395–404 CrossRef CAS PubMed.
- S. Angioletti-Uberti, M. Ballauff and J. Dzubiella, Mol. Phys., 2018, 116, 3154–3163 CrossRef CAS.
- S. Harnisch and R. H. Müller, Eur. J. Pharm. Biopharm., 2000, 49, 41–46 CrossRef CAS PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779 CrossRef CAS PubMed.
- F. Tavanti, A. Pedone and M. C. Menziani, New J. Chem., 2015, 39, 2474–2482 RSC.
- M. A. Dobrovolskaia, B. W. Neun, S. Man, X. Ye, M. Hansen, A. K. Patri, R. M. Crist and S. E. McNeil, Nanomed. Nanotechnol. Biol. Med., 2014, 10, 1453–1463 CrossRef CAS PubMed.
- Z. J. Deng, G. Mortimer, T. Schiller, A. Musumeci, D. Martin and R. F. Minchin, Nanotechnology, 2009, 20, 455101 CrossRef PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi and C. Reinhardt, Nat. Nanotechnol., 2013, 8, 772 CrossRef CAS PubMed.
- T. Göppert and R. Müller, Int. J. Pharm., 2005, 302, 172–186 CrossRef PubMed.
- M. Jansch, P. Stumpf, C. Graf, E. Rühl and R. Müller, Int. J. Pharm., 2012, 428, 125–133 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, F. Song, R. Liu, H. Guo, D. W. H. Olsen, Y. Cohen, A. Emili and W. C. Chan, ACS Nano, 2014, 8, 2439–2455 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- S. Lindman, I. Lynch, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Nano Lett., 2007, 7, 914–920 CrossRef CAS PubMed.
- J. E. Gagner, M. D. Lopez, J. S. Dordick and R. W. Siegel, Biomaterials, 2011, 32, 7241–7252 CrossRef CAS PubMed.
- P. Aggarwal, J. B. Hall, C. B. McLeland, M. A. Dobrovolskaia and S. E. McNeil, Adv. Drug Deliv. Rev., 2009, 61, 428–437 CrossRef CAS PubMed.
- M. P. Calatayud, B. Sanz, V. Raffa, C. Riggio, M. R. Ibarra and G. F. Goya, Biomaterials, 2014, 35, 6389–6399 CrossRef CAS PubMed.
- B.-J. L. Van Hong Nguyen, Int. J. Nanomed., 2017, 12, 3137 CrossRef PubMed.
- H. Ruh, B. Kühl, G. Brenner-Weiss, C. Hopf, S. Diabaté and C. Weiss, Toxicol. Lett., 2012, 208, 41–50 CrossRef CAS PubMed.
- Q. Peng and H. Mu, J. Controlled Release, 2016, 225, 121–132 CrossRef CAS PubMed.
- S. Y. Khor, M. N. Vu, E. H. Pilkington, A. P. Johnston, M. R. Whittaker, J. F. Quinn, N. P. Truong and T. P. Davis, Small, 2018, 14, 1801702 CrossRef PubMed.
- M. N. Vu, H. G. Kelly, A. K. Wheatley, S. Peng, E. H. Pilkington, N. A. Veldhuis, T. P. Davis, S. J. Kent and N. P. Truong, Small, 2020, 2002861 CrossRef PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- C. Gräfe, A. Weidner, M. vd Lühe, C. Bergemann, F. H. Schacher, J. H. Clement and S. Dutz, Int. J. Biochem. Cell Biol., 2016, 75, 196–202 CrossRef PubMed.
- M. M. Yallapu, N. Chauhan, S. F. Othman, V. Khalilzad-Sharghi, M. C. Ebeling, S. Khan, M. Jaggi and S. C. Chauhan, Biomaterials, 2015, 46, 1–12 CrossRef CAS PubMed.
- K. Partikel, R. Korte, D. Mulac, H.-U. Humpf and K. Langer, Beilstein J. Nanotechnol., 2019, 10, 1002–1015 CrossRef CAS PubMed.
- A. Lesniak, A. Campbell, M. P. Monopoli, I. Lynch, A. Salvati and K. A. Dawson, Biomaterials, 2010, 31, 9511–9518 CrossRef CAS PubMed.
- L. Treuel, S. Brandholt, P. Maffre, S. Wiegele, L. Shang and G. U. Nienhaus, ACS Nano, 2014, 8, 503–513 CrossRef CAS PubMed.
- A. Weidner, C. Gräfe, M. von der Lühe, H. Remmer, J. H. Clement, D. Eberbeck, F. Ludwig, R. Müller, F. H. Schacher and S. Dutz, Nanoscale Res. Lett., 2015, 10, 282 CrossRef PubMed.
- Z. Wang, Q. Zhao, M. Cui, S. Pang, J. Wang, Y. Liu and L. Xie, Nanomaterials, 2017, 7, 93 CrossRef PubMed.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz, M. R. Ejtehadi, R. Hartmann, K. Kantner, U. Linne and P. Maffre, ACS Nano, 2013, 7, 6555–6562 CrossRef CAS PubMed.
- A. Bhogale, N. Patel, J. Mariam, P. Dongre, A. Miotello and D. Kothari, Colloids Surf. B Biointerfaces, 2014, 113, 276–284 CrossRef CAS PubMed.
- S. Palchetti, V. Colapicchioni, L. Digiacomo, G. Caracciolo, D. Pozzi, A. L. Capriotti, G. La Barbera and A. Laganà, Biochim Biophys Acta Biomembr., 2016, 1858, 189–196 CrossRef CAS PubMed.
- H. Maeda, Adv. Drug Deliv. Rev., 2015, 91, 3–6 CrossRef CAS PubMed.
- J. Shi, P. W. Kantoff, R. Wooster and O. C. Farokhzad, Nat. Rev. Cancer, 2017, 17, 20 CrossRef CAS PubMed.
- S. Shahabi, L. Treccani, R. Dringen and K. Rezwan, Nanoscale, 2015, 7, 16251–16265 RSC.
- J. C. Y. Kah, J. Chen, A. Zubieta and K. Hamad-Schifferli, ACS Nano, 2012, 6, 6730–6740 CrossRef CAS PubMed.
- A. Cifuentes-Rius, H. de Puig, J. C. Y. Kah, S. Borros and K. Hamad-Schifferli, ACS Nano, 2013, 7, 10066–10074 CrossRef CAS PubMed.
- E. L. L. Yeo, J. U.-J. Cheah, B. Y. Lim, P. S. P. Thong, K. C. Soo and J. C. Y. Kah, ACS Biomater. Sci. Eng., 2017, 3, 1039–1050 CrossRef CAS.
- E. L. L. Yeo, U. Joshua, J. Cheah, D. J. H. Neo, W. I. Goh, P. Kanchanawong, K. C. Soo, P. S. P. Thong and J. C. Y. Kah, J. Mater. Chem. B, 2017, 5, 254–268 RSC.
- D. Zhang, J. Yang, J. Guan, B. Yang, S. Zhang, M. Sun, R. Yang, T. Zhang, R. Zhang and Q. Kan, Biomater. Sci., 2018, 6, 2360–2374 RSC.
- S. Behzadi, V. Serpooshan, R. Sakhtianchi, B. Müller, K. Landfester, D. Crespy and M. Mahmoudi, Colloids Surf. B Biointerfaces, 2014, 123, 143–149 CrossRef CAS PubMed.
- A. J. Paula, R. T. Araujo Junior, D. S. f. T. Martinez, E. J. Paredes-Gamero, H. B. Nader, N. Durán, G. Z. Justo and O. L. Alves, ACS Appl. Mater. Interfaces, 2013, 5, 8387–8393 CrossRef CAS PubMed.
- G. Caracciolo, S. Palchetti, L. Digiacomo, R. Z. Chiozzi, A. L. Capriotti, H. Amenitsch, P. M. Tentori, V. Palmieri, M. Papi and F. Cardarelli, ACS Appl. Mater. Interfaces, 2018, 10, 22951–22962 CrossRef CAS PubMed.
- K. Cho, X. Wang, S. Nie and D. M. Shin, Clin. Canc. Res., 2008, 14, 1310–1316 CrossRef CAS.
- V. Mirshafiee, R. Kim, S. Park, M. Mahmoudi and M. L. Kraft, Biomaterials, 2016, 75, 295–304 CrossRef CAS PubMed.
- R. Dal Magro, B. Albertini, S. Beretta, R. Rigolio, E. Donzelli, A. Chiorazzi, M. Ricci, P. Blasi and G. Sancini, Nanomed. Nanotechnol. Biol. Med., 2018, 14, 429–438 CrossRef CAS PubMed.
- L. Rao, Q.-F. Meng, L.-L. Bu, B. Cai, Q. Huang, Z.-J. Sun, W.-F. Zhang, A. Li, S.-S. Guo and W. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 2159–2168 CrossRef CAS PubMed.
- Q. Dai, C. Walkey and W. C. Chan, Angew. Chem., Int. Ed., 2014, 53, 5093–5096 CrossRef CAS PubMed.
- T. T. Hoang Thi, E. H. Pilkington, D. H. Nguyen, J. S. Lee, K. D. Park and N. P. Truong, Polymers, 2020, 12, 298 CrossRef.
- N. Lu, J. Li, R. Tian and Y.-Y. Peng, Chem. Res. Toxicol., 2014, 27, 1070–1077 Search PubMed.
- T. Miclăuş, C. Beer, J. Chevallier, C. Scavenius, V. E. Bochenkov, J. J. Enghild and D. S. Sutherland, Nat. Commun., 2016, 7, 11770 Search PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Nat. Nanotechnol., 2016, 11, 372 CrossRef PubMed.
- Q. Peng, S. Zhang, Q. Yang, T. Zhang, X.-Q. Wei, L. Jiang, C.-L. Zhang, Q.-M. Chen, Z.-R. Zhang and Y.-F. Lin, Biomaterials, 2013, 34, 8521–8530 CrossRef CAS PubMed.
- O. Koshkina, D. Westmeier, T. Lang, C. Bantz, A. Hahlbrock, C. Würth, U. Resch-Genger, U. Braun, R. Thiermann and C. Weise, Macromol. Biosci., 2016, 16, 1287–1300 CrossRef CAS PubMed.
- B. Wang, T. Blin, A. Käkinen, X. Ge, E. H. Pilkington, J. F. Quinn, M. R. Whittaker, T. P. Davis, P. C. Ke and F. Ding, Polym. Chem., 2016, 7, 6875–6879 RSC.
- H. Ou, T. Cheng, Y. Zhang, J. Liu, Y. Ding, J. Zhen, W. Shen, Y. Xu, W. Yang and P. Niu, Acta Biomater., 2018, 65, 339–348 CrossRef CAS PubMed.
- G. Caracciolo, D. Pozzi, A. Capriotti, C. Cavaliere, F. Cardarelli, A. Bifone and A. Laganà, Nanotechnology, 2012, 3, 354–357 CAS.
- C. Corbo, R. Molinaro, M. Tabatabaei, O. C. Farokhzad and M. Mahmoudi, Biomater. Sci., 2017, 5, 378–387 RSC.
- O. C. Farokhzad and M. Mahmoudi, System and method for protein corona sensor array for early detection of diseases, WO2018112460A1, 2018.
- Q. Huo, Methods for biomolecule and biomolecule complex (bmc) detection and analysis and the use of such for research and medical diagnosis, WO2011088128A2, 2011.
- K. Dawson, L. Lynch, M. Lundqvist and T. Cedervall, A method for the selective concentration of a specific low abundance biomolecule, WO2010097785A1, 2010.
- K. Kostarelos and M. Hadjidemetriou, Detection of cancer biomarkers using nanoparticles, WO2018046542A1, 2018.
- Q. Huo, Biomolecular interactions and interaction products as biomarkers for detection, diagnosis, prognosis and predicting therapeutic responses of human diseases, US Pat., US20130058923A1, 2013.
- T. Zheng, N. Pierre-Pierre, X. Yan, Q. Huo, A. J. Almodovar, F. Valerio, I. Rivera-Ramirez, E. Griffith, D. D. Decker and S. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 6819–6827 CrossRef CAS PubMed.
- K. A. Conway, J. D. Harper and P. T. Lansbury, Biochemistry, 2000, 39, 2552–2563 CrossRef CAS PubMed.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2006, 75, 333–366 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39 CrossRef CAS PubMed.
- P. K. Teotia and R. Kaler, Opt. Commun., 2018, 406, 188–191 CrossRef CAS.
- M. J. Hajipour, J. Raheb, O. Akhavan, S. Arjmand, O. Mashinchian, M. Rahman, M. Abdolahad, V. Serpooshan, S. Laurent and M. Mahmoudi, Nanoscale, 2015, 7, 8978–8994 RSC.
- O. K. Kari, T. Rojalin, S. Salmaso, M. Barattin, H. Jarva, S. Meri, M. Yliperttula, T. Viitala and A. Urtti, Drug Deliv. Transl. Res., 2017, 7, 228–240 CrossRef CAS PubMed.
- P. M. Kelly, C. Åberg, E. Polo, A. O'connell, J. Cookman, J. Fallon, Ž. Krpetić and K. A. Dawson, Nat. Nanotechnol., 2015, 10, 472 CrossRef CAS PubMed.
- A. M. Alkilany, N. N. Mahmoud, F. Hashemi, M. J. Hajipour, F. Farvadi and M. Mahmoudi, Chem. Res. Toxicol., 2016, 29, 943–948 Search PubMed.
- M. Azhdarzadeh, A. A. Saei, S. Sharifi, M. J. Hajipour, A. M. Alkilany, M. Sharifzadeh, F. Ramazani, S. Laurent, A. Mashaghi and M. Mahmoudi, Nanomedicine, 2015, 10, 2931–2952 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |