Open Access Article
Open Access ArticleThe simultaneous detection of the squamous cell carcinoma antigen and cancer antigen 125 in the cervical cancer serum using nano-Ag polydopamine nanospheres in an SERS-based lateral flow immunoassay†
Ji Xiaab,
Yifan Liuab,
Menglin Ranab,
Wenbo Lu d,
Liyan Bie,
Qian Wanga,
Dan Lu*abf and
Xiaowei Cao
d,
Liyan Bie,
Qian Wanga,
Dan Lu*abf and
Xiaowei Cao *abcdf
*abcdf
aDepartment of Obstetrics and Gynecology, College of Clinical Medicine, Yangzhou University, Yangzhou, PR China. E-mail: ludan1968@126.com; cxw19861121@163.com
bThe First Clinical College, Dalian Medical University, Dalian, PR China
cInstitute of Translational Medicine, Medical College, Yangzhou University, Yangzhou, PR China
dKey Laboratory of Magnetic Molecules and Magnetic Information Materials (Ministry of Education), School of Chemistry and Material Science, Shanxi Normal University, Linfen, 041004, PR China
eTransformative Otology and Neuroscience Center, College of Special Education, Binzhou Medical University, Yantai 264003, PR China
fJiangsu Key Laboratory of Integrated Traditional Chinese and Western Medicine for Prevention and Treatment of Senile Diseases, Yangzhou University, Yangzhou, PR China
First published on 6th August 2020
Abstract
The accurate analysis of tumor related biomarkers is extremely critical in the diagnosis of the early stage cervical cancer. Herein, we designed a novel and inexpensive surface-enhanced Raman scattering-based lateral flow assay (SERS-based LFA) strip with a single test line, which was applied for the rapid and sensitive quantitative simultaneous analysis of SCCA and CA125 in serum samples from patients with cervical cancer. In the presence of target antigens, the monoclonal antibody-coupled and Raman reporter-labeled nano-Ag polydopamine nanospheres (PDA@Ag-NPs) aggregated on the test line modified by the polyclonal antibody to form a double-antibody sandwich structure. The finite difference time domain simulation demonstrated that large number of “hot spots” was generated among the nanogaps of aggregated PDA@AgNPs, which resulted in a huge enhancement of the signal of the Raman reporters. Accordingly, the limit of detection was determined to be 7.156 pg mL−1 for SCCA and 7.182 pg mL−1 for CA125 in phosphate buffer and 8.093 pg mL−1 for SCCA and 7.370 pg mL−1 for CA125 in human serum, revealing high sensitivity of this SERS-based LFA strip. Significantly, the detection of SCCA and CA125 using the SERS-based LFA was observed to have high specificity and reproducibility, and the whole detection was completed within 20 min. Furthermore, the SERS-based LFA and enzyme-linked immunosorbent assay were also employed in serum samples obtained from patients with cervical cancer, cervical intraepithelial neoplasia and healthy subjects, and perfect agreement existed between both the methods. Thus, clinically, the developed SERS-based LFA strip has strong potential for the simultaneous detection of multiple cancer biomarkers in serum.
Introduction
Cervical cancer is one of the most prevalent gynecological malignancies worldwide, which is inferior only to breast cancer.1 Also, developing countries bear the major portion of the disease burden.2 The current detection methods for the initial assessment of cervical cancer include the thinprep cytology test (TCT) and human papillomavirus (HPV) test and have low specificity and sensitivity. Additionally, colposcopy and cervical biopsy are the invasive tests, which make them difficult for patients to accept. The early clinical manifestations of cervical cancer are atypical and non-specific, and the detection rate of existing cervical cancer screening methods is not very stable.3 Consequently, it is urgent to establish simple, noninvasive, sensitive and accurate methods for the early screening and clinical diagnosis of cervical cancer. With the rapid development of oncomolecularbiology, scientists have discovered that tumor biomarkers are indicative of a particular disease process and highly important for early diagnosis, attracting enormous interest in clinical viewpoints.4,5 However, parallel single biomarker detection is inadequate in the complex pathological process because of its low specificity and sensitivity. Thus, the simultaneous detection of multiple biomarkers can improve the efficiency and accuracy and is becoming essential in contemporary cancer diagnosis.6 Squamous cell carcinoma antigen (SCCA) is isolated from squamous cell carcinoma of the uterine cervix and is characterized as a glycoprotein with a molecular mass of 48 kDa.7,8 SCCA is one of the auxiliary indicators for cervical cancer diagnosis, condition detection and prognosis judgment.9 Cancer antigen 125 (CA125) is a common marker in the detection of female reproductive system tumors, where the level of CA125 is elevated significantly in the serum of patients with cervical cancer.10,11 Therefore, SCCA and CA125, as cancer biomarkers of cervical cancer universally, are used in routine cancer diagnosis. Only trace amounts of biomarkers are present in a patient's serum in the early stages of the disease, which requires screening experiments with high reliability and sensitivity. Presently, the methods for the detection of SCCA and CA125 mainly include enzyme-linked immunosorbent assay (ELISA), fluorescence immunoassay, radioimmunoassay and electrochemical immunoassay.12–15 Although these methods are relatively reliable, they limited in diagnosis due to several disadvantages such as photobleaching, time-consuming operation process, large sample volume and use of expensive instruments.Accordingly, the membrane-based lateral flow assay (LFA) has developed rapidly because of its advantages of low cost, rapid analysis, user-friendly operation, and long-term stability.16 Thus, over the past few years, it has become one of the most successful commercial point-of-care-testing (POCT) detection products.17,18 Based on chromatographic separation, the LFA test generally uses gold nanoparticles as markers to display signals. However, the drawbacks of the LFA test such as limited sensitivity, lack of quantification and interference in the sample matrix hinder its further applications for cancer diagnosis.19,20 Accordingly, to overcome these disadvantages, much effort has been focused on improving the performance of the LFA test, where the SERS-based LFA test is one outcome. Surface-enhanced Raman scattering (SERS), as an ultrasensitive analytical method to quantify analytes, has attracted increasing attention.21 SERS can enhance the Raman signal of analytes located near the surface of Ag or Au metallic nanostructures by as much as 6 to 14 orders of magnitude, which is associated with the electromagnetic coupling (namely “hot spots”) at the gaps between nanoparticles or rough surfaces of the metallic nanoparticles.22–26 The fingerprint characteristic of SERS causes it to have better detection accuracy for biomolecule samples.27 SERS also has the advantages of a narrow spectral band, non-destructive, and weak Raman signal of water.28–30 Therefore, SERS has been considered to be a potential candidate method for the analysis of fluid samples in vitro. SERS-based LFA tests, which combine the advantages of SERS and LFA, have been established and widely employed to detect proteins, chemical component and DNA in vitro. For example, Lin et al. reported the detection of bisphenol A using gold nanostars in an SERS-improved lateral flow immunochromatographic assay.31 Wang et al. developed an SERS-based lateral flow assay biosensor using AuNPs as SERS tags for the quantitative and ultrasensitive detection of interleukin-6 in unprocessed whole blood.32 Wang et al. reported the simultaneous detection of dual nucleic acids using an SERS-based LFA biosensor with double test lines, which utilized Au NPs as the SERS nanotags.33 However, compared to the single T line, SERS-based LFA tests with multiple lines require a longer operation process and analytical time, which limit their wider application in the field of multiplex analyte detection. In addition, with multiple T lines on one strip, the performance of SERS-based LFA may be influenced by the location of the T lines.34
Polydopamine (PDA) nanospheres have received widespread attention due to their abundant surface active groups, large surface area and strong adhesion property, which are beneficial to facilitate the robust binding of nanoparticles and antibodies onto PDA nanospheres.35 Besides, the spherical morphology and good hydrophilicity of PDA nanospheres allow them to easily diffuse in water and rapidly migrate under the action of capillary chromatography.36,37 In weakly alkaline solution, PDA nanospheres can be synthesized via the self-polymerization of dopamine with low environmental requirement and controllable morphology.38 Thus, based on these attributes, PDA nanospheres are expected to act as novel carriers for nanoparticles. Ag particles are evenly distributed on resin nanospheres, which not only prevents the oxidation of the Ag particles, but also the formation of a large number of “hot spots” through the junction between nanoparticles. Based on the above advantages, PDA@Ag-NPs can be used as a new nanomaterial for SERS detection.
Herein, we propose an ultrasensitive SERS-based LFA integration strip for the analysis of two specific cervical cancer biomarkers including SCCA and CA125 in a single test line on an NC membrane. Compared to multiple test lines, a single test line significantly reduces the preparation time, analysis time and reagent consumed. In this system, two SERS immunoprobes were prepared via the surface modification of PDA@Ag-NPs with different Raman reporters (4-ATP and DTNB) and target-specific monoclonal antibodies. The experimental conditions were optimized for the fabrication of a special integration strip, including the amount of monoclonal antibodies, and the concentration of antigens and SERS immunoprobes. The selectivity, sensitivity and reproducibility were assessed to confirm the performance of this design. Ultimately, the simultaneous detection of the concentrations of SCCA and CA125 in serum from healthy, cervical cancer and cervical intraepithelial neoplasia (CIN) subjects was measured via the SERS-based LFA and ELISA methods to verify its feasibility in practical application. To the best of our knowledge, there are few reports on the simultaneous detection of cancer markers using SERS-based LFA strips with a single T line, which is promising and has good potential to be employed for diagnosis in the early stage of cervical cancer.
Materials and methods
Materials
Silver nitrate (AgNO3), sodium borohydride (NaBH4), dopamine hydrochloride (DA), ethanol (CH3CH2OH), ammonia (NH4OH), 4-aminothiophenol (4-ATP) and 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) were purchased from Noah Chemical Inc. (Yangzhou, China). Bovine serum albumin (BSA), Tris–HCl buffer solution, Tween 20, Triton X-100, phosphate buffer saline (PBS, pH 7.4, 0.01 M), N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) were purchased from Younuo Chemicals Inc. (Yangzhou, China). Squamous cell carcinoma antigen (SCCA), SCCA monoclonal antibody (labeling), SCCA polyclonal antibody (coating), cancer antigen 125 (CA125), CA125 monoclonal antibody (labeling) and CA125 polyclonal antibody (coating) were purchased from Linc-Bio Science Inc. (Shanghai, China) and were stored at −20 °C. Ultra-pure water prepared on a Milli-Q (Millipore, USA, resistivity > 18 M) purifier was used for all experiments. All glassware and Teflon-coated magnetic stirring bars were immersed in aqua regia (HCl/HNO3, volume ratio 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and rinsed with ultra-pure water and dried in an oven prior to each examination. All other reagents were from commercial sources and used without further purification.
1) and rinsed with ultra-pure water and dried in an oven prior to each examination. All other reagents were from commercial sources and used without further purification.
The processing and storage of blood specimens
Peripheral blood specimens were obtained from 30 healthy subjects, 30 CINI subjects, 30 CINII subjects, 30 CINIII subjects and 30 cervical cancer subjects from the College of Clinical Medicine of Yangzhou University. Consent documents were also obtained from all donors. All peripheral blood specimens were collected on the day of diagnosis. After centrifugation at 2500 rpm for 15 min at 4 °C, the serum specimens were collected in EP tubes, labeled according to their classification and stored at −80 °C before analysis. Table 1 summarizes the age and histopathologic stage details.| Group | Healthy | CINI | CINII | CINIII | Cervical cancer |
| Average age | 25 | 33 | 39 | 44 | 54 |
| Sample | 30 | 30 | 30 | 30 | 30 |
Synthesis of monodisperse polydopamine nanospheres
In a typical synthesis, PDA nanospheres were prepared via the sol–gel method according to previously reported methods with some modifications.36,40 Typically, 30 mL of ethanol, 60 mL of ultra-pure water and 1.5 mL of ammonia solution (25–28%) were added to a conical flask and stirred vigorously for 30 min at room temperature for 30 min to obtain a uniform mixture. Subsequently, 6 mL of dopamine hydrochloride solution (0.05 g mL−1) was dropped in the alkaline water–ethanol mixture and it was observed that the color of the mixture gradually turned from colorless and transparent to pale brown and deepened gradually, finally becoming dark brown under vigorous stirring, which indicated the formation of PDA nanospheres. This reaction was allowed to proceed for 40 h at room temperature before completion. The resulting PDA nanospheres were washed thoroughly with ethanol and ultra-pure water by centrifugation (5000 rpm, 10 min) three times. Finally, the sediment was collected by centrifugation and re-dispersed in 50 mL ultra-pure water.Synthesis of the nano-Ag polydopamine composite nanospheres
For the synthesis of PDA@Ag-NPs, 180 mL of AgNO3 (25 mmol L−1) solution was added dropwise to the 20 mL PDA nanosphere solution. After stirring for 30 min, Ag+ was sufficiently adsorbed on the surface of the PDA nanospheres. Subsequently, 20 μL of NaBH4 solution (0.15 mol L−1) was added and the resulting mixture was stirred for 1 h at room temperature. It was observed that the solution gradually changed from brown to dark gray. The PDA@Ag-NPs composite nanospheres were washed with ultra-pure water by centrifugation (5000 rpm, 10 min) three times and re-dispersed in 20 mL ultra-pure water.Preparation of Raman reporter-labeled PDA@Ag-NPs and antibody conjugation
For the preparation of the SERS immunoprobes, PDA@Ag-NPs were labeled with 4-ATP and DTNB to generate SERS signals, followed by coupling with monoclonal antibodies though the stable amide bond. The carboxyl group (–COOH) on the surface of PDA@Ag-NPs was activated by adding EDC and NHS.41,42 Briefly, 5 mL as-prepared PDA@Ag-NP suspension was co-incubated with 50 μL of 10−3 mol L−1 4-ATP (or 10−5 mol L−1 DTNB) ethanol solution at room temperature for 30 min. Then, the separated 4-ATP/DTNB-labeled PDA@Ag-NPs from the solution were obtained by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) to ensure that the unconjugated 4-ATP (or DTNB) was removed completely. Then the sediment was re-dispersed in 1 mL PBS buffer. Subsequently, 20 μL of 150 mM EDC and 20 μL of 30 mM NHS were added and incubated for 1 h. In the following step, 4 μL of 0.2 mg mL−1 SCCA monoclonal antibody (or 3 μL of 0.2 mg mL−1 CA125 monoclonal antibody) was added to the suspension of PDA@Ag-NPs@4-ATP (or PDA@Ag-NPs@DTNB) to prepare the PDA@Ag-NPs–Raman reporter–antibody conjugates (denoted as PDA@Ag-NPs@4-ATP@anti-SCCA and PDA@Ag-NPs@DTNB@anti-CA125), and incubated at room temperature for 1 h with mild agitation. Subsequently, the redundant binding sites of PDA@Ag-NPs were blocked by BSA (1 wt%) for an additional hour to eliminate the effect of nonspecific binding. The unreacted reagent was removed by centrifugation (10
000 rpm, 10 min) to ensure that the unconjugated 4-ATP (or DTNB) was removed completely. Then the sediment was re-dispersed in 1 mL PBS buffer. Subsequently, 20 μL of 150 mM EDC and 20 μL of 30 mM NHS were added and incubated for 1 h. In the following step, 4 μL of 0.2 mg mL−1 SCCA monoclonal antibody (or 3 μL of 0.2 mg mL−1 CA125 monoclonal antibody) was added to the suspension of PDA@Ag-NPs@4-ATP (or PDA@Ag-NPs@DTNB) to prepare the PDA@Ag-NPs–Raman reporter–antibody conjugates (denoted as PDA@Ag-NPs@4-ATP@anti-SCCA and PDA@Ag-NPs@DTNB@anti-CA125), and incubated at room temperature for 1 h with mild agitation. Subsequently, the redundant binding sites of PDA@Ag-NPs were blocked by BSA (1 wt%) for an additional hour to eliminate the effect of nonspecific binding. The unreacted reagent was removed by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) and the precipitate was re-suspended in 1 mL PBS buffer containing 1% BSA. The conjugates were stored at 4 °C prior to use.
000 rpm, 10 min) and the precipitate was re-suspended in 1 mL PBS buffer containing 1% BSA. The conjugates were stored at 4 °C prior to use.
Fabrication of the SERS-based LFA strip
The components of the SERS-based LFA strip included a sample pad, conjugate pad, NC membrane (test and control lines), and absorption pad. In advance, the sample pad was prepared by saturating it with 50 mM Tris–HCl (pH 8.0) containing 150 mM NaCl and 0.25% Triton® X-100, followed by drying at 37 °C for 2 h to enhance its surface activity and hydrophilicity. The conjugate pad was pretreated with 10 mM PBS buffer solution containing 0.2% Tween-20, and 2% sucrose and dried at room temperature. Subsequently, 5 μL of the SERS immunoprobes (PDA@Ag-NPs@4-ATP@anti-SCCA and PDA@Ag-NPs@DTNB@anti-CA125 mixed in the proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were pipetted onto the conjugate pad and the as-prepared conjugate pads were dried at 37 °C for 2 h. Subsequently, 5 μL of SCCA polyclonal antibody (0.2 mg mL−1) and CA125 polyclonal antibody (0.2 mg mL−1) in solution as capture agents and 5 μL of anti-human IgG solution (0.2 mg mL−1) were sprayed at a dispensing rate of 0.6 μL cm−1 onto the virtual test line and the virtual control line of the NC membrane via a commercial line dispensing system, respectively, leaving about a 2 cm gap between both lines. The width of the test line was 1.5–2 mm. The NC membrane was dried at 37 °C for 3 h before being blocked with PBS buffer containing 1% BSA. To fabricate the LFA strip, all components were assembled sequentially with a 2 mm overlap to ensure that the sample solution could migrate through the whole LFA strip. The integrated strips were cut into individual 1 cm-wide strips using a paper cutter and stored at 4 °C in a sealed bag in a dry state for further detection.
1) were pipetted onto the conjugate pad and the as-prepared conjugate pads were dried at 37 °C for 2 h. Subsequently, 5 μL of SCCA polyclonal antibody (0.2 mg mL−1) and CA125 polyclonal antibody (0.2 mg mL−1) in solution as capture agents and 5 μL of anti-human IgG solution (0.2 mg mL−1) were sprayed at a dispensing rate of 0.6 μL cm−1 onto the virtual test line and the virtual control line of the NC membrane via a commercial line dispensing system, respectively, leaving about a 2 cm gap between both lines. The width of the test line was 1.5–2 mm. The NC membrane was dried at 37 °C for 3 h before being blocked with PBS buffer containing 1% BSA. To fabricate the LFA strip, all components were assembled sequentially with a 2 mm overlap to ensure that the sample solution could migrate through the whole LFA strip. The integrated strips were cut into individual 1 cm-wide strips using a paper cutter and stored at 4 °C in a sealed bag in a dry state for further detection.
SERS measurement
During the test, 100 μL of sample solution was dropped onto the sample pad 0.5 cm away from the end. The solution could flow toward the direction of absorbent paper together with the SERS immunoprobes due to capillary force. During the flow process, gray bands visible to the naked eye gradually appeared on the T line and C line. The entire LFA process was completed within 20 min. The SERS spectra of 4-ATP and DTNB on the T line were measured using a Renishaw inVia Raman microscope system (50× objective lens) with a power of 5 mW and exposure time of 1 s, with a laser operating at λ = 785 nm as the excitation source. The width of the laser spot for the 50× objective lens was 2 μm. All the Raman spectra reported in this study were collected in the range of 400–1800 cm−1 in continuous mode. The average SERS spectrum measured from 10 different spots along the T line was employed to quantify SCCA and CA125 in the sample solution, which ensured the authenticity and rationality of the data.Instruments
An ultraviolet absorption spectrometer (Cary 5000, Agilent, USA) was used to acquire ultraviolet-visible-near infrared (UV-vis-NIR) absorption spectra in the wavelength range of 400–1800 nm. Transmission electron microscopy (TEM) images were obtained on a transmission electron microscope (Tecnai 12, Philips, Netherlands) with an accelerating voltage of 60 kV. An S-4800II field-emission scanning electron microscope (Gemini SEM 300, Carl Zeiss, Germany) at an accelerating voltage of 3.0 kV was applied to gain scanning electron microscopy (SEM) images. High-resolution TEM (HRTEM) and energy dispersive X-ray (EDX) mapping were carried out on a field-emission-transmission electron microscope (Tecnai G2F30 S-TWIN, FEI, USA) operating at 20 kV. All experiments were carried out at room temperature.Results and discussion
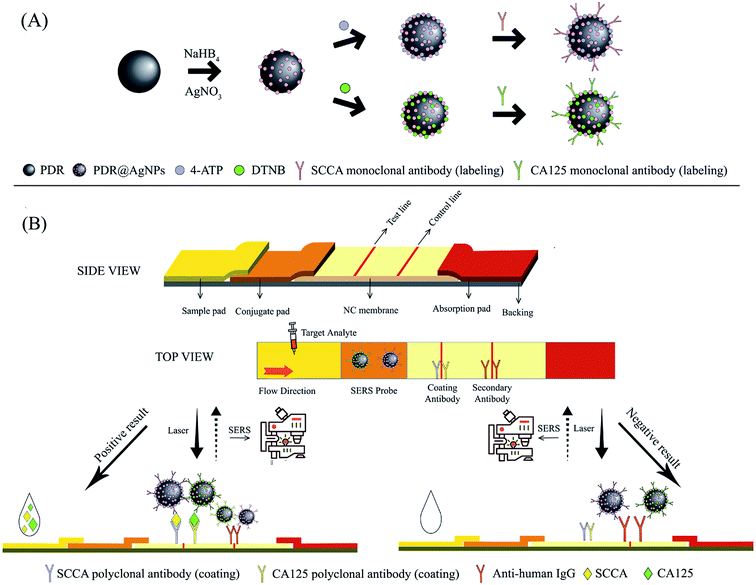
Principle of the SERS-based LFA strip
The working principle of the developed integration strip is schematically illustrated in Scheme 1. As mentioned above, the strip consisted of four parts. The sample pad carries the sample solution, while ensuring that the sample solution is sufficiently distributed to the downstream area. The conjugate pad closely adjacent to the sample pad maintains the stability of the SERS immunoprobes and controls their release. The NC membrane is the area where the immunoreaction takes place, including one test line (T line) and one control line (C line). The SERS-based LFA strip can be considered effective only when the C line is colored. The absorption pad enhances the capillary force and absorbs all redundant substances.16,18 In the detection process, the sample solution containing SCCA and CA125 is applied on the sample pad and gradually migrates in the longitudinal direction due to capillary action. When the sample solution migrates through the conjugate pad, SCCA and CA125 can specifically bind to corresponding SERS immunoprobes (PDA@Ag-NPs@4-ATP@anti-SCCA and PDA@Ag-NPs@DTNB@anti-CA125) immobilized on the conjugate pad to form an antigen-SERS immunoprobe complex. When the complex reaches the NC membrane, it can be captured by the polyclonal antibodies of SCCA and CA125 pre-immobilized on the T line, leading to the formation of a sandwich-structure complex. Subsequently, excessive complex continues to migrate through the C line and the anti-human IgG pre-immobilized on the C line captures the complex. The SERS spectra of the samples at the T line were measured using a Renishaw Raman microscope. Due to the distinguishable characteristic peaks of 4-ATP and DTNB, the concentrations of SCCA and CA125 in the samples could be calculated according to the intensities of the peaks.Characterization of the PDA@Ag-NPs modified with Raman reporters and antibodies
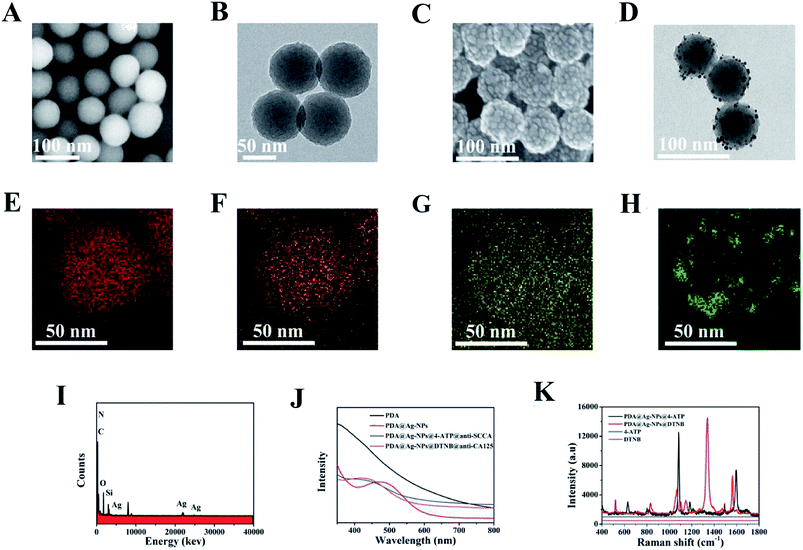
SEM images and TEM images were employed to characterize the synthesized PDA nanospheres. As shown in Fig. 1A and B, PDA nanospheres were obtained with a uniform and regular spherical morphology on a large scale. It is displayed more clearly in Fig. 1B that the average diameter of the PDA nanospheres was approximately 70 nm. The PDA nanospheres had excellent dispersion in aqueous solution, and there was no aggregation between the nanospheres. A large number of amino and carboxylic groups was exposed on the surface of the PDA nanospheres in the process of polymerization.39,40 After adding AgNO3 solution, the negatively charged carboxylic groups (–COOH) were successfully functionalized with Ag+ through ion-pairing and the Ag nanoparticles were reduced in situ on the surface of PDA nanospheres under the reducing action of NaBH4.35,40 Fig. 1C displays the SEM images of PDA@Ag-NPs, which has a relatively rough surface compared with that of the PDA nanospheres. This variation indicates that dense Ag nanoparticles were modified on the surface of the PDA nanospheres. It can be observed in Fig. 1D that the diameter of the Ag nanoparticles is about 1–3 nm, and they were uniformly attached to the surface of the nanospheres. There were gaps between the Ag nanoparticles, which allowed the active groups between the gaps to combine with antibodies.40PDA@Ag-NPs were characterized via energy dispersion X-ray spectroscopy (EDX) element mapping to display the spatial distributions of carbon (Fig. 1E), nitrogen (Fig. 1F), oxygen (Fig. 1G) and silver (Fig. 1H). The results exhibited that the elemental carbon, nitrogen and oxygen were evenly distributed in PDA@Ag-NPs, while elemental silver was evenly distributed on the surface of PDA@Ag-NPs, which indicates that the Ag nanoparticles were successfully immobilized on the PDA nanospheres. The element distribution of the material can also be seen in the EDX image (Fig. 1I), which further confirms that the Ag nanoparticles were reduced on the surface of PDA. The preparation process of the SERS immunoprobes is illustrated by UV-vis-NIR absorption spectroscopy (Fig. 1J). For the PDA nanospheres, there was no obvious absorption peak in the wavelength range of 400–800 nm. After the immobilization of Ag nanoparticles, a peak appeared near 475 nm due to the surface plasmon resonance of the Ag nanoparticles.43,44 The blue shift in this peak from 475 nm to 438 nm and 440 nm demonstrates that the Raman reporters and antibodies were modified successfully on the PDA@Ag-NPs.45,46
Fig. 1K presents the Raman spectra of the two Raman reporters and 4-ATP/DTNB-labeled PDA@Ag-NPs. 4-ATP and DTNB were labeled on PDA@Ag-NPs via the sulfhydryl group (–SH). Two main Raman peaks at 1083 cm−1 (related to the C–S stretching vibrational mode) and 1584 cm−1 (related to the C–C stretching vibrational mode) were observed in SERS spectra of both 4-ATP and DTNB. In addition, for DTNB, a characteristic peak was also observed at around 1330 cm−1 (related to the C–N stretching vibrational mode).16,47,48 The Raman signals of 10−2 M 4-ATP and DTNB were both weak, whereas strong SERS signals were detected for 4-ATP/DTNB-labeled PDA@Ag-NPs. The analytical enhancement factor (AEF) for PDA@Ag-NPs was calculated using the equation AEF = (ISERS/CSERS)/(IRS/CRS), where C and I denote the concentration and intensity, and SERS and RS denote the SERS conditions and non-SERS conditions, respectively.24 When CSERS = 1 × 10−6 M and CRS = 10−2 M, with the intensities at 1330 cm−1 measured in Fig. 1K, the AEF of PDA@Ag-NPs was calculated to be 2.13 × 105. Using the measured intensities at 1083 cm−1 in Fig. 1K, AEF = 2.33 × 105 was obtained. Thus, the results demonstrate that PDA@Ag-NPs had a prominent SERS enhancement effect.
FDTD simulation of different aggregations of PDA@Ag-NPs
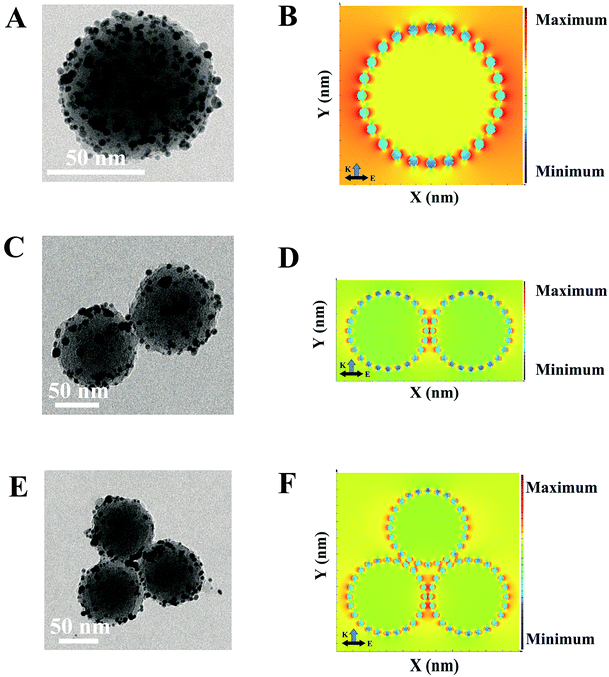
To understand the SERS behaviors of the SERS-based LFA, a series of numerical calculations were carried out to study the nanostructures-dependent electric field enhancements.49 The models of PDA@Ag-NPs with different aggregations were constructed by employing the nanostructure parameters extracted from the TEM images. The TEM images in Fig. 2A, C and E clearly indicate that the average diameter of the PDA nanospheres is approximately 70 nm and the diameter of the Ag nanoparticles is about 1–3 nm. The spacing gap between each PDA@Ag-NPs was approximately 0.5 nm. Fig. 2B, D and F show the simulated local electric field distribution of PDA@Ag-NPs with different aggregations. Fig. 2B demonstrates that the enhanced electric field distribution of a single PDA@Ag-NP is not uniform, but rather areas of the most “hot spots” with high electric field intensity among the gaps of the Ag nanoparticles. In Fig. 2D and F, the hot spots are mainly located in the gaps among the dimers and trimers of PDA@Ag-NPs and that of the Ag nanoparticles on the surface of PDA. The electric field intensity near the gap among PDA@Ag-NPs was much stronger than that among Ag nanoparticles, which reveals that the PDA@Ag-NP polymer generated more “hot spots” and induced strong SERS signals. Thus, the results theoretically prove that the PDA@Ag-NP aggregations caused by immunochromatography can be used as an excellent SERS sensor for detecting biomolecules.Optimization of SERS-based LFA parameters
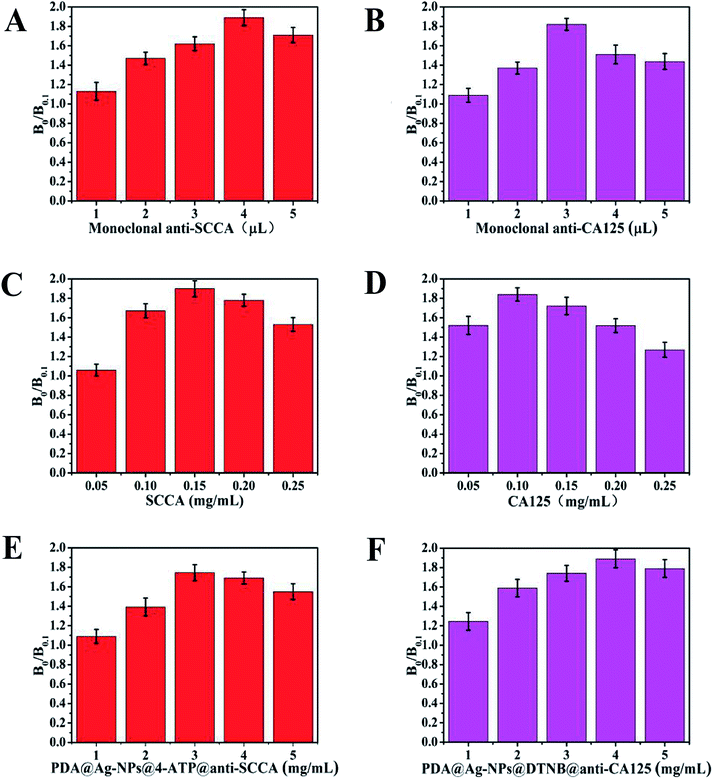
The amount of monoclonal antibodies, and the concentration of antigens and SERS immunoprobes strongly affect the sensitivity of the detection. Thus, these experimental factors were investigated to achieve the optimal analytical performance. The inhibition rate (B0/B0.1) was used as the index parameter for the optimization result, where B0 and B0.1 are the Raman intensities of the highest peak when the concentration of the sample was 0 ng mL−1 and 0.1 ng mL−1, respectively.50 The sensitivity of the detection was positively correlated with the inhibition rate. Different amounts of 0.2 mg mL−1 monoclonal antibodies (ranging from 1 mL to 5 mL) were added during the preparation of the SERS immunoprobes. As shown in Fig. 3A and B, the maximum inhibition rate was obtained with the addition of 4 μL of monoclonal anti-SCCA and 3 μL of monoclonal anti-SCCA, respectively. Herein, the most suitable amount of monoclonal anti-SCCA was 4 μL and the optimum amount of monoclonal anti-CA125 was 3 μL. For the antigens applied on the sample pad, SCCA and CA125 with concentrations ranging from 0.05 mg mL−1 to 0.25 mg mL−1 were investigated (Fig. 3C and D). The highest inhibition rate was achieved at 0.15 mg mL−1 SCCA and 0.10 mg mL−1 CA125, respectively. Thus, the optimal concentration of SCCA was 0.15 mg mL−1, and the optimal concentration of CA125 was 0.10 mg mL−1. Fig. 3E and F show the variation in inhibition rate as a function of the concentration of PDA@Ag-NPs@4-ATP@anti-SCCA and PDA@Ag-NPs@DTNB@anti-CA125 (ranging from 1 μM to 5 μM), respectively. Therefore, a concentration of 3 μM was estimated to be the optimum concentration for PDA@Ag-NPs@4-ATP@anti-SCCA and a concentration of 4 μM was chosen as the optimum concentration for PDA@Ag-NPs@DTNB@anti-CA125.Qualitative analysis of SCCA and CA125 using SERS-based LFA strip
Under the optimal experimental conditions, a qualitative test was performed to analyze the absence or presence of SCCA (1 μg mL−1) and CA125 (1 μg mL−1) in PBS buffer. As illustrated in Fig. 4A, when SCCA and CA125 were present in PBS buffer, gray bands appeared on the T line and C line. As expected from the original design, a cluster of SERS immunoprobes were captured on the T line by the immunoreaction, which was confirmed by SEM imaging. Meanwhile, the SERS peaks at 1083 cm−1 and 1584 cm−1 from 4-ATP and 1330 cm−1 from DTNB appeared in the corresponding SERS spectrum. In the absence of SCCA and CA125 (Fig. 4B), no color change was observed on the T line, and thus only one gray band appeared on the C line, which indicated that the strip was working properly. In this case, no obvious Raman signal was shown and no SERS immunoprobe aggregation occurred on the T line. These results demonstrate that the SERS-based LFA strip could efficiently identify SCCA and CA125.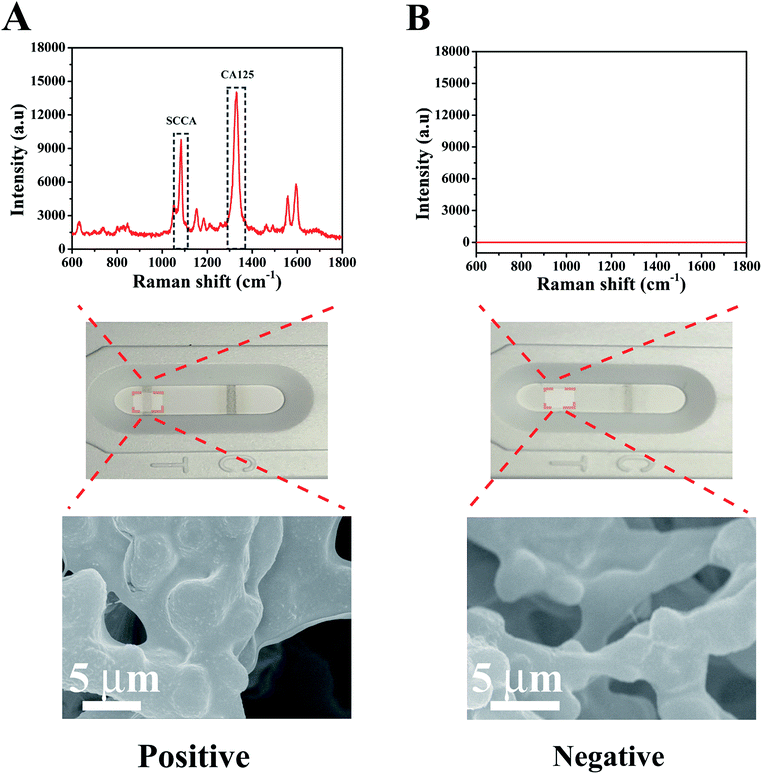 | ||
| Fig. 4 SERS spectra of the T line from a (A) positive result and (B) negative result together with the corresponding optical images and SEM images. | ||
Specificity and reproducibility of SERS-based LFA for SCCA and CA125
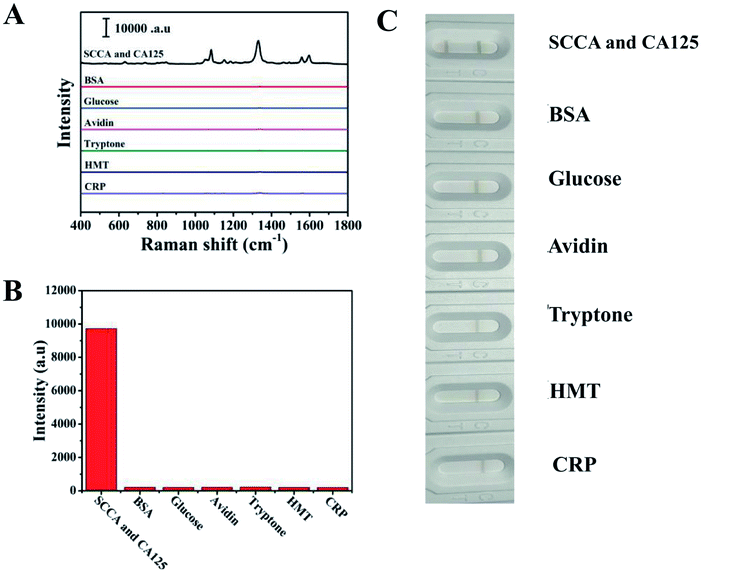
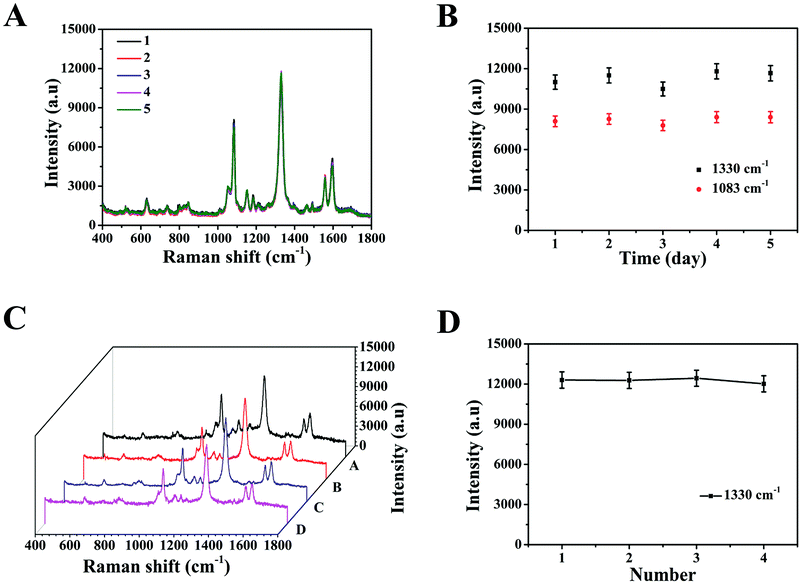
Specificity is considered a crucial parameter for the practical application of the SERS-based LFA strip. Thus, to investigate the specificity of this special integration strip, specific biomarkers (SCCA and CA125) and nonspecific biomarker or protein (BSA, glucose, avidin, tryptone, HMT and CRP) with the same concentration (5 μg mL−1) in PBS buffer were detected, respectively. The intensities of the peaks at 1083 cm−1 and 1330 cm−1 were chosen to trace SCCA and CA125, respectively. According to the SERS spectra shown in Fig. 5A, it can be seen that SERS intensity of SCCA and CA125 was much higher than that of the interferants. The histogram in Fig. 5B shows the results more clearly using the peak intensity at 1330 cm−1. As illustrated in Fig. 5C, two gray bands appeared in the presence of SCCA and CA125, whereas only one pale gray band was observed on the C line in the presence of the nonspecific biomarker and protein, which is consistent with SERS results.The reproducibility of the SERS-based LFA strip as another crucial parameter was investigated under the optimal conditions. The SERS spectra of five different SERS-based LFA strips prepared at different times are shown in Fig. 6A. These SERS spectra were almost no difference. The scattergram of the peak intensity at 1083 cm−1 and 1330 cm−1 is displayed in Fig. 6B, where the deviation in the peak intensity at 1083 cm−1 and 1330 cm−1 is 5.6% and 6.9%, respectively. Fig. 6C exhibits the SERS spectra of four individual SERS-based LFA strips prepared simultaneously. According to the scattergram shown in Fig. 6D, the deviation in the peak intensity at 1330 cm−1 was calculated to be 6.3%. These results demonstrate the high precision of the SERS-based LFA strip. Therefore, the SERS-based LFA strip possessed good specificity and reproducibility, which demonstrate its high precision for the detection for SCCA and CA125 in clinical fluid samples.
Quantitative detection of SCCA and CA125 in PBS buffer and human serum
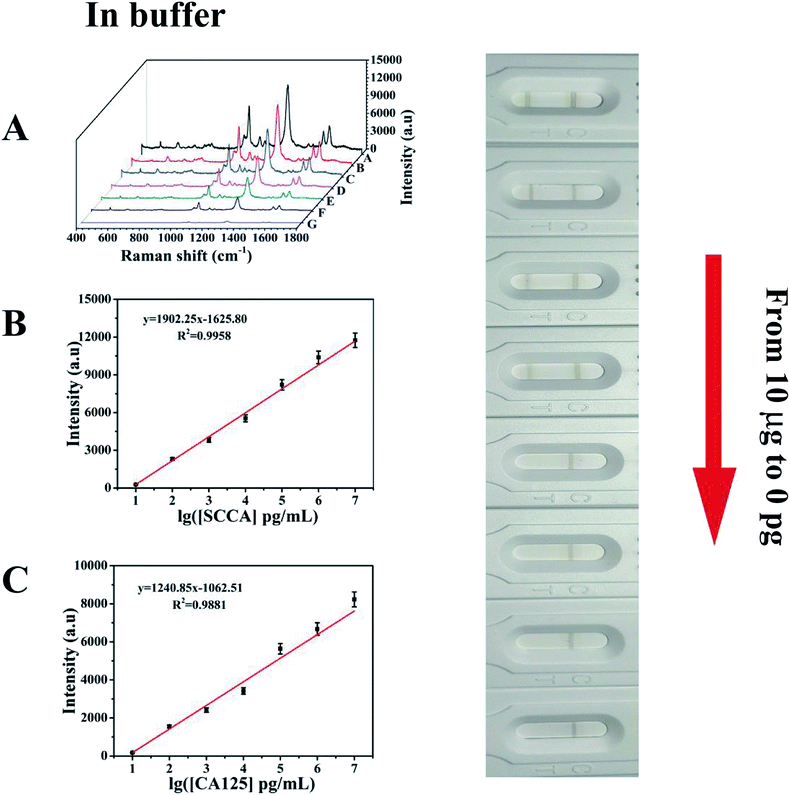
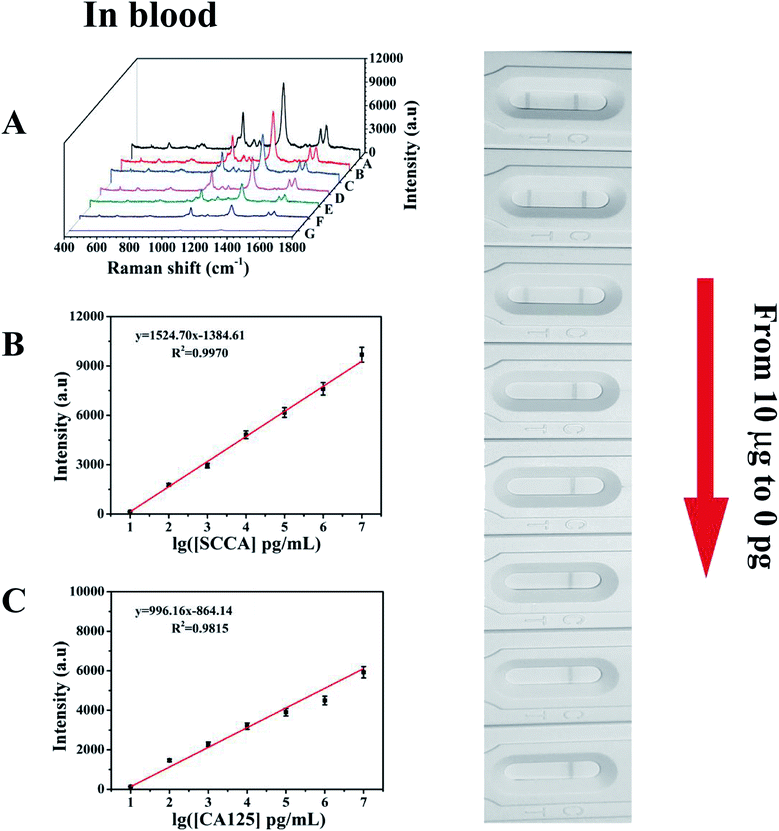
Under the above optimized conditions, the ability of the developed SERS-based LFA strip for the quantitative analysis of SCCA and CA125 was evaluated. SCCA and CA125 were dissolved together in PBS buffer to obtain the same final concentrations, which ranged from 10 pg mL−1 to 10 μg mL−1. With an increase in the concentration of SCCA and CA125, more double-antibody sandwich immunocomplexes were formed, which means that more SERS immunoprobes were captured on the T line, thus resulting in the color on the T line becoming darker accordingly. Consequently, the SERS intensity concomitantly increased with an increase in the concentration of SCCA and CA125 (Fig. 7A). The peak intensity of 4-ATP at 1083 cm−1 was measured and employed for the quantitative evaluation of the SCCA concentration. In the range of 10 pg mL−1 to 10 μg mL−1, the SERS intensity was almost linear with the logarithm of SCCA concentration (Fig. 7B) and the linear regression equation was y = 1902.25x − 1625.80 (where the SERS intensity of the band at 1083 cm−1 is represented by y and the logarithm of SCCA concentration is represented by x) with a linear regression coefficient of R2 = 0.9958. Consequently, the limit of detection (LOD) of the SCCA immunoassay using the SERS-based LFA strip was estimated to be 7.156 pg mL−1 in PBS buffer. For the quantitative analysis of the CA125 detection, the peak of DTNB at 1330 cm−1 was monitored for the construction of a calibration curve for CA125, as displayed in Fig. 7C, which indicated that the SERS intensity showed a good linear response to the logarithm of CA125 concentration. The linear regression equation was y = 1240.85x − 1062.51 (R2 = 0.9881) and the LOD was as low as 7.182 pg mL−1 in PBS buffer.To evaluate the feasibility of this special integration strip for the analysis of human serum, various concentrations of SCCA and CA125 (10 pg mL−1 to 10 μg mL−1) were mixed in human serum and detected by the SERS-based LFA strip. It can be seen from Fig. 8A that the group of SERS spectra corresponding to different SCCA and CA125 concentrations exhibit an increase as the concentration increased, accompanied by the corresponding darkening of the T line color. As displayed in Fig. 8B, the calibration curve showed that the SERS intensities at 1083 cm−1 (4-ATP) increased with the concentration of SCCA. The linear regression equation and the linear regression coefficient were y = 1524.70x − 1384.61 and 0.9970, and the LOD was as low as 8.093 pg mL−1 for SCCA in human serum. Fig. 8C illustrates the calibration curve obtained by plotting the intensity of DTNB at 1330 cm−1 versus the logarithm of SCCA concentration with a linear regression equation of y = 996.16x − 864.14 and a linear regression coefficient of R2 = 0.9815, achieving an LOD of 7.370 pg mL−1 for CA125 in human serum. Thus, base on these low LOD values, this method is suitable for practical clinical applications.
Clinical serum samples analysis
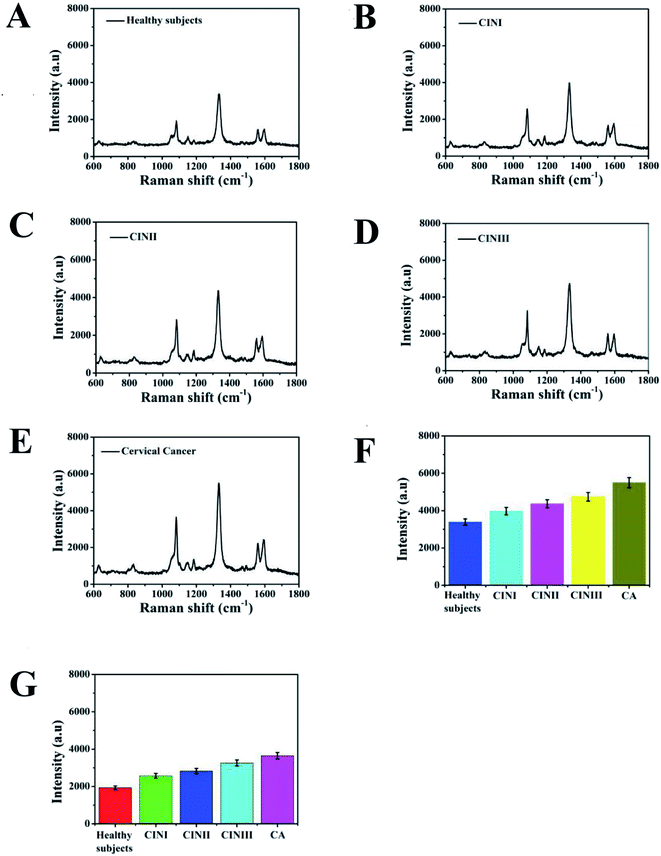
To demonstrate the detection accuracy and clinical practicability of the proposed SERS-based LFA strip for simultaneously identifying SCCA and CA125, clinical serum samples from healthy, CINI, CINII, CINIII and cervical cancer subjects were studied. The whole measurement process could be completed within 20 min. Fig. 9A–E reveal the average SERS spectra of the clinical serum samples. Each spectrum is the result of averaging 30 different serum samples. The SERS signals of SCCA and CA125 in the human serum from the cervical cancer subjects were higher than that from the CIN (I, II and III) patients and healthy subjects. Fig. 9F and G illustrate the histograms of the peak intensities at 1083 cm−1 and 1330 cm−1. As the disease progressed, the SERS intensities at 1083 cm−1 and 1330 cm−1 were both gradually augmented. ELISA, as a routine detection method for biomarkers, was used to verify the accuracy of the SERS-based LFA.12 The concentrations of SCCA and CA125 in the clinical serum samples measured by the SERS-based LFA and ELISA methods are compared in Table 2. The concentrations of SCCA measured by the SERS-based LFA were 1.355 ng mL−1, 3.277 ng mL−1, 5.916 ng mL−1, 10.421 ng mL−1 and 32.771 ng mL−1, while that of SCCA measured by ELISA were 1.291 ng mL−1, 3.441 ng mL−1, 5.733 ng mL−1, 9.982 ng mL−1 and 34.019 ng mL−1. Compared with ELISA, the relative error of the SERS-based LFA was 4.723%, −4.766%, 3.093%, 6.212% and −3.667%, respectively. The relative standard deviations of the two methods were 4.723%, −4.766%, 3.093%, 6.212% and −3.667%, respectively. The concentrations of CA125 measured by SERS-based LFA were 0.639 ng mL−1, 2.814 ng mL−1, 5.502 ng mL−1, 13.801 ng mL−1 and 33.812 ng mL−1, while that of CA125 measured by ELISA were 0.617 ng mL−1, 2.719 ng mL−1, 5.379 ng mL−1, 13.174 ng mL−1 and 35.447 ng mL−1. The relative error of the two methods was calculated to be 3.443%, 7.376%, 2.236%, 4.543% and −4.612%, respectively. There was no significant difference between the results given by these two methods. The results confirmed the detection accuracy and practicability of this special integration strip.| Sample | SERS-based LFA strip (ng mL−1) | ELISA (ng mL−1) | Relative error [%] | |||
|---|---|---|---|---|---|---|
| SCCA | CA125 | SCCA | CA125 | SCCA | CA125 | |
| Healthy subjects | 1.355 | 0.639 | 1.291 | 0.617 | 4.723 | 3.443 |
| CINI | 3.277 | 2.814 | 3.441 | 2.606 | −4.766 | 7.376 |
| CINII | 5.916 | 5.502 | 5.733 | 5.379 | 3.093 | 2.236 |
| CINIII | 10.421 | 13.801 | 9.982 | 12.944 | 6.212 | 4.543 |
| Cervical cancer | 32.771 | 33.812 | 34.019 | 35.447 | −3.667 | −4.612 |
Conclusions
In conclusion, a novel SERS-based LFA strip for the ultrasensitive simultaneous detection of SCCA and CA125 was developed, which was based on the formation of PDA@Ag-NP aggregations by immunoreaction on a single T line. PDA@Ag-NPs exhibited a promising SERS enhancement effect, which was certified by FDTD simulation, and modified with Raman reporters and antibodies to be employed as SERS immunoprobes. By measuring the peak intensity of 4-ATP at 1083 cm−1 and DTNB at 1330 cm−1, the concentrations of SCCA and CA125 in the samples were quantitatively analyzed. In the concentration ranged from 10 pg mL−1 to 10 μg mL−1, and the LOD of SCCA and CA125 was 7.16 pg mL−1 and 7.18 pg mL−1 in PBS buffer, respectively. The detection ability of practical clinical applications was investigated by analyzing SCCA and CA125 in human serum, with the LOD of 8.03 pg mL−1 for SCCA and 7.37 pg mL−1 for CA125. This established that the SERS-based LFA strip also has high specificity and reproducibility. Clinical serum samples collected from healthy, CINI, CINII, CINIII and cervical cancer subjects were detected using the SERS-based LFA strip and the detection was completed within 20 min. The results of the SCCA and CA125 concentrations were consistent with ELISA method, which demonstrated the accuracy of this strip for practical use. Thus, considering these merits, the SERS-based LFA strip can be employed as an inexpensive POCT detection for analyzing multiple biomarkers in the early stage of cervical cancer.Ethical statement
During the experiment, the Ethical Guidelines issued by the Council for International Organizations of Medical Sciences were strictly followed and informed consent was signed by donors. This work was approved by the Ethics Committee of the College of Clinical Medicine of Yangzhou University and followed the Guidelines of the Declaration of Helsinki.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81701825), the Social Development Foundation of Jiangsu (No. BE2018684), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 17KJB416012), the Chinese Medicine Science and Technology Development Plan Project of Jiangsu (No. YB201972), the Research Project of Maternal and Child Health of Jiangsu (No. F201809).References
- W. Small, M. A. Bacon, A. Bajaj, L. T. Chuang, B. J. Fisher, M. M. Harkenrider, A. Jhingran, H. C. Kitchener, L. R. Mileshkin, A. N. Viswanathan and D. K. Gaffney, Cancer, 2017, 123, 2404–2412 CrossRef.
- N. Vergara, M. Balanda, W. Hidalgo, H. S. Martín, A. Aceituno, F. Roldán, T. Villalón, M. Hott, G. Espinoza, A. Quiero, M. T. Valenzuela and E. Ramírez, Med. Microbiol. Immunol., 2018, 207, 95–103 CrossRef CAS.
- S. Y. Feng, D. Lin, J. Q. Lin, B. H. Li, Z. F. Huang, G. N. Chen, W. Zhang, L. Wang, J. J. Pan, R. Chen and H. S. Zeng, Analyst, 2013, 138, 3967–3974 RSC.
- A. Mahasneh, F. Al-Shaheriv and E. Jamal, Exp. Mol. Pathol., 2017, 102, 475–483 CrossRef CAS.
- M. Bhardwaj, V. Erben, P. Schrotz-King and H. Brenner, Cancers, 2017, 9, 156–178 CrossRef.
- Z. Crannell, A. Castellanos-Gonzalez, G. Nair, R. Mejia, A. C. White and R. Richards-Kortum, Anal. Chem., 2016, 88, 1610–1616 CrossRef CAS.
- A. A. Tikhonov, M. V. Tsybulskaya, Ve. I. Butvilovskaya, E. N. Savvateeva, P. V. Belousov, D. V. Kuprash, O. N. Solopova, M. A. Chernichenko, M. M. Filushin and A. Y. Rubina, Anal. Methods, 2016, 8, 7920–7928 RSC.
- M. D. Esajas, J. M. Duk, H. W. De Bruijn, J. G. Aalders, P. H. Willemse, W. Sluiter, B. Pras, K. Ten Hoor, H. Hollema and A. G. van der Zee, J. Clin. Oncol., 2001, 19, 3960–3966 CrossRef CAS.
- X. Li, J. Zhou, K. C. Huang, F. Tang, H. Zhou, S. Wang, Y. Jia, H. Sun, D. Ma and S. Li, PLoS One, 2015, 10, 4–16 Search PubMed.
- D. P. Bender, J. I. Sorosky, R. E. Bulle and A. K. Sood, Am. J. Obstet. Gynecol., 2003, 189, 113–117 CrossRef CAS.
- B. Kotowicz, J. Kaminska, M. Fuksiewicz, M. Kowalska, J. Jonska-Gmyrek, K. Gawrychowski, J. obotkowski, M. Skrzypczak, J. Starzewski and M. Bidzinski, Int. J. Gynecol. Canc., 2010, 20, 588–592 CrossRef.
- D. Zhang, W. Li, Z. Ma and H. Han, Biosens. Bioelectron., 2019, 126, 800–805 CrossRef CAS.
- S. Çataltepe, C. Schick, C. J. Luke, S. C. O. Pak, D. Goldfarb, P. Chen, M. J. Tanasiyevic, M. R. Posner and G. A. Silvermana, Clin. Chim. Acta, 2000, 295, 107–127 CrossRef.
- P. Samadi, H. Ghanbari, R. Saber and Y. Omidi, Biosens. Bioelectron., 2018, 122, 68–74 CrossRef.
- Q. Mao, X. H. Liu, C. C. Chen, J. W. Ye, H. K. Liang, B. X. Li, X. Y. Sun, L. Sun, Y. M. Sun, X. M. Mu and L. Q. Li, Biotechnol. Appl. Biochem., 2018, 65, 816–821 CrossRef CAS.
- M. Xiao, K. X. Xie, X. H. Dong, L. Wang, C. H. Huang, F. Xu, W. Xiao, M. L. Jin, B. Y. Huang and Y. Tang, Anal. Chim. Acta, 2019, 1053, 139–147 CrossRef CAS.
- W. C. Mak, V. Beni and A. P. F. Turner, Trends Anal. Chem., 2016, 79, 297–305 CrossRef CAS.
- E. B. Bahadır and M. K. Sezgintürk, Trends Anal. Chem., 2016, 82, 286–306 CrossRef.
- W. Ren, S. I. Mohammed, S. Wereley and J. Irudayaraj, Anal. Chem., 2019, 91, 2876–2884 CrossRef CAS.
- D. Quesada-gonzález and A. Merkoci, Biosens. Bioelectron., 2015, 73, 47–63 CrossRef.
- J. P. Camden, J. A. Dieringer, J. Zhao and R. P. V. Duyne, Accounts Chem. Res., 2008, 41, 1653–1661 CrossRef CAS.
- S. Y. Ding, E. M. You, Z. Q. Tian and M. Moskovits, Chem. Soc. Rev., 2017, 46, 4042–4076 RSC.
- Y. Wang, L. J. Tang and J. H. Jiang, Anal. Chem., 2013, 85, 9213–9220 CrossRef CAS.
- B. B. Shan, Y. H. Pu, Y. F. Chen, M. L. Liao and M. Li, Coord. Chem. Rev., 2018, 371, 11–37 CrossRef CAS.
- S. Nie and S. R Emory, Science, 1997, 275, 1102–1106 CrossRef CAS.
- N. A. Hatab, C. H. Hsueh, A. L. Gaddis, S. T. Retterer, J. H. Li, G. Eres, Z. Y. Zhang and B. H. Gu, Nano Lett., 2010, 10, 4952–4955 CrossRef CAS.
- S. Bamrungsap, A. Treetong, C. Apiwat, T. Wuttikhun and T. Dharakul, Microchim. Acta, 2016, 183, 249–256 CrossRef CAS.
- R. A. Halvorson and P. J. Vikesland, Environ. Sci. Technol., 2010, 44, 7749–7755 CrossRef CAS.
- K. Xu, Z. Y. Wang, C. F. Tan, N. Kang, L. W. Chen, L. Ren and E. S. Thian, ACS Appl. Mater. Inter., 2017, 9, 26341–26349 CrossRef CAS.
- X. F. Jia, C. W. Wang, Z. Rong, J. Li, K. L. Wang, Z. W. Qie, R. Xiao and S. Q. Wang, RSC Adv., 2018, 8, 21243–21251 RSC.
- L. K. Lin and L. A. Stanciu, Sens. Actuators, B, 2018, 276, 222–229 CrossRef CAS.
- Y. Wang, J. Y. Sun, Y. J. Hou, C. Zhang, D. W. Li, H. X. Li, M. F. Yang, C. D. Fan and B. L. Sun, Biosens. Bioelectron., 2019, 141, 111432 CrossRef CAS.
- X. K. Wang, N. Choi, Z. Y. Cheng, J. H. Ko, L. X. Chen and J. Choo, Anal. Chem., 2017, 89, 1163–1169 CrossRef CAS.
- Z. Z. Wu, Food Anal. Methods, 2019, 12, 1086–1091 CrossRef.
- C. J. Wu, G. X. Zhang, T. Xia, Z. N. Li, K. Zhao, Z. W. Deng, D. Z. Guo and B. Peng, Mater. Sci. Eng., C, 2015, 55, 155–165 CrossRef CAS.
- Y. L. Liu, K. L. Ai, J. H. Liu, M. Deng, Y. Y. He and L. H. Lu, Adv. Mater., 2013, 25, 1353–1359 CrossRef CAS.
- H. B. Wen, P. C. Jiang, Y. L. Hu and G. K. Li, Microchim. Acta, 2018, 185, 353–361 CrossRef.
- X. Xu, L. Sun, B. Bai, H. L. Wang and Y. R. Suo, Sci. Total Environ., 2019, 665, 113–141 CrossRef.
- J. Yan, L. Yang, M. F. Lin, J. Ma, X. Lu and P. S. Lee, Small, 2013, 9, 596–603 CrossRef CAS.
- W. B. Lu, H. Y. Wang, M. Wang, Y. Wang, L. Tao and W. P. Qian, RSC Adv., 2015, 5, 24615–24624 RSC.
- D. Sun, F. H. Cao, W. Q. Xu, Q. D. Chen, W. Shi and S. Q. Xu, Anal. Chem., 2019, 91, 2551–2558 CrossRef CAS.
- Y. Su, S. T. Xu, J. N. Zhang, X. J. Chen, L. P. Jiang, T. T. Zheng and J. J. Zhu, Anal. Chem., 2019, 91, 864–872 CrossRef CAS.
- W. B. Lu, L. Tao, Y. Wang, J. Ge, J. Dong and W. P. Qian, Microchim. Acta, 2015, 182, 479–486 CrossRef CAS.
- F. Wang, R. Han, G. T. Liu, H. F. Chen, T. R. Ren, H. F. Yang and Y. Wen, J. Electroanal. Chem., 2013, 706, 102–107 CrossRef CAS.
- D. Xie, W. F. Zhu, H. Cheng, Z. Y. Yao, M. Li and Y. L. Zhao, Phys. Chem. Chem. Phys., 2018, 20, 8881–8886 RSC.
- A. J. Wang, W. D. Ruan, W. Song, L. Chen, B. Zhao, Y. M. Jung and X. Wang, J. Raman Spectrosc., 2013, 44, 1649–1653 CrossRef CAS.
- W. Y. Lim, C. H. Goh, T. M. Thevarajah, B. T. Goh and S. M. Khor, Biosens. Bioelectron., 2020, 147, 111792 CrossRef CAS.
- D. Li, L. M. Jiang, J. A. Piper, I. S. Maksymov, A. D. Greentree, E. Wang and Y. Wang, ACS Sens., 2019, 4, 2507–2514 CrossRef CAS.
- S. Lin, X. Lin, S. Han, L. He, H. Y. Zhao, J. Zhang, W. Hasi and L. Wang, J. Alloys Compd., 2019, 805, 318–326 CrossRef CAS.
- X. Z. Li, T. Y. Yang, Y. T. Song, J. H. Zhu, D. L. Wang and W. Li, Sens. Actuators, B, 2019, 283, 230–238 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05207h |
| This journal is © The Royal Society of Chemistry 2020 |