Open Access Article
Open Access ArticleDevelopment of a portable lab-on-a-valve device for making primary diagnoses based on gold-nanoparticle aggregation induced by a switchable linker
Jungwoo Hahn†
 b,
Eunghee Kim†
b,
Eunghee Kim† a,
Hyebin Han
a,
Hyebin Han a and
Young Jin Choi
a and
Young Jin Choi *abc
*abc
aDepartment of Agricultural Biotechnology, Seoul National University, 1 Gwanakro, Gwanakgu, Seoul 151-921, Korea. E-mail: choiyj@snu.ac.kr; Tel: +82 2 880 4851
bCenter for Food and Bioconvergence, Seoul National University, 1 Gwanakro, Gwanakgu, Seoul 151-921, Korea
cResearch Institute of Agriculture and Life Sciences, Seoul National University, 1 Gwanakro, Gwanakgu, Seoul 151-921, Korea
First published on 24th August 2020
Abstract
We have developed a low-cost, portable lab-on-a-valve (LOV) integrated microdevice for the detection of pathogens in primary-diagnosis settings. This system was designed for field-based pathogen detection based on the aggregation of gold nanoparticles induced by a switchable linker. A three-way valve, which has attracted much attention as a functional mesofluidic platform for pressure-driven flow, has been designed as a universal reaction platform that combines the functions of fluid flow and a reaction chamber. In addition, we obtain rapid and enhanced visual signals by the use of a syringe filter to remove gold nano-aggregates (Au NAs). Using this device, Salmonella Typhimurium down to 101 CFU mL−1 can be visually detected within 30 min by performing a simple operation that requires no complex equipment. This prototype device has great potential for use in the semi-quantitative and qualitative identification of pathogens in on-site primary diagnoses.
Introduction
Novel bioengineering and diagnostic technologies for the detection of pathogens are expanding their application to a variety of fields, including molecular diagnostics, food-safety testing, medical diagnostics, and environmental monitoring.1–4 The development of such platforms that enable miniaturization, rapid diagnosis, and reduced cost for on-site diagnoses represents an important advance.5–7 To date, standard testing for pathogen detection has been based on traditional cell-culture methods that require at least 48 hours.8 In addition, the enzyme-linked immunosorbent assay (ELISA) is widely used to detect pathogens with high sensitivity and selectivity.9,10 However, these methods are difficult to apply in the field because of their need for complex, labour-intensive processing steps and well-trained operators.11 Therefore, it is essential to develop a diagnostic method that enables quick and inexpensive testing without the need for peripheral devices. Device miniaturization techniques improve portability and minimize the need for bulky laboratory infrastructures in primary-diagnosis settings.12,13 In addition, the development of these compact diagnostic devices widens the range of strategies for disease monitoring and management.14Recently, lab-on-a-valve (LOV) devices have emerged as a new technology for flow control.15–17 LOV devices have the advantage of being directly manipulated by a programmable flow-based platform that can adopt different coupling modes and controlled fluid movement, as compared to the lab-on-a-chip.18 Flow analyses by LOV devices seem to be suitable for the development of portable analysis systems in that this technology reduces instrument size and reagent and energy consumption, which are important benefits for field systems, while simplifying the work required for analysis.19
In this study, we applied flow analysis to switchable linker (SL)-based analysis using 3-way valve chambers (3-VCs). The 3-way valve, which provides a convenient and economical means of selecting various flow rates and directions, plays a crucial role in almost all industrial processes, including the food field.20–22 In addition, the 3-way valve can control the flow of fluid via a simple operation and can be applied to various fluids based on their materials, volume, and ranges of temperature and pressure.23,24 However, a 3-way valve that is sealable, portable, and easy to operate has not yet been developed for use as a reaction chamber for bioassays. In addition, both disposable syringes and syringe filters, which have the advantages of sterilization, low cost, and wide application, can comprise a good biosensing application kit in the field.25–28
Colorimetric detection based on gold-nanoparticle aggregation has attracted interest as a novel alternative for on-site detection due to its simplicity and the fact that it needs no sophisticated equipment or technology.29–32 The gold-nanoparticle aggregation-based colorimetric method has a limitation in that its detection signal is weak when detecting a relatively large target such as a pathogen because the spectral aggregation shift decreases as the distance between gold nanoparticles increases.33 Therefore, in our previous research, we proposed the use of a switchable linker to induce the large-scale aggregation of gold nanoparticles and to enhance the detection signal by sedimentation of the aggregates. This novel colorimetric detection method is expected to increase the capability of on-site detection.
In this study, we first developed a portable kit comprising a 3-way valve for the on-site detection of Salmonella Typhimurium. This kit is based on the use of colorimetric detection by the precipitation of gold nano-aggregates (Au NAs) and a novel switchable linker to amplify the visible signal by the aggregation of gold nanoparticles.34–37 In addition, for quick on-site detection, we introduced filtration into the device as a strategy for improving the visual detection signal.
Experimental
Chemicals, reagents, and instruments
We purchased streptavidin, phosphate-buffered saline (PBS), 1 mM tetrachloroaurate, and bovine serum albumin (BSA) from Sigma-Aldrich (Madison, WI), and tetraborate pH standard solution from Wako Pure Chemicals (Osaka, Japan). We purchased anti-Salmonella polyclonal antibody (biotin) (ab69255) from Abcam (Cambridge, MA) and vials, ethanol, and trisodium citrate from local suppliers.As the buffer in the synthesis of 13 nm Au NPs, we used sterilized distilled water, which we prepared with 1% (w/v) tri-sodium citrate as a reducing agent. The buffers used for the SL-based assay for the detection of streptavidin as a model target and Salmonella Typhimurium were 0.01 mol L−1 PBS (pH 7.4) prepared with 0.1% and 0.5% (w/v) BSA, respectively, for blocking, and 0.01 mol L−1 tetraborate pH standard solution (pH 7.4) for diluting the streptavidin. The bacterial strain used in the assay was Salmonella Typhimurium (ATCC 19585) which was purchased from ATCC.
We performed the SL-based immunoassay in 3-VCs purchased from a local supplier, and filtered the Au NAs using a 50 mL syringe (local supplier) and syringe filters (Hyundai Micro, South Korea). To determine the assay absorbance, we used a UV-1700 spectrophotometer (Shimadzu, Japan). The size (number) of the Au NAs was determined using a Zetasizer (Nano ZS, Malvern Instruments Ltd., Worcestershire, UK).
Preparation of colloidal streptavidin-coated Au NPs
We prepared colloidal Au NPs with an average size of 13 nm by reducing tetrachloroaurate with tri-sodium citrate, which is a well-established citrate reduction method.38 We verified the particle size by transmission electron microscopy (TEM, JEM1010, JEOL, Japan) and UV-Vis spectroscopy (UV-1700 PC, Shimadzu). The synthesized Au NPs had an ultraviolet-visible spectrum peak (λmax) at 520 ± 1.0 nm. Based on the absorbance value (0.4 in a 1/10 dilution sample), the concentration of the Au NPs was estimated to be ∼7 × 1012 particles per mL. The prepared Au NPs were coated with streptavidin for surface functionalization using an electrostatic adsorption procedure.39 To functionalize the prepared Au NPs, we coated 40 μg of streptavidin dissolved in a tetraborate solution (pH 7.4) onto 600 μL of the citrate-stabilized Au NPs with an absorbance of 0.4 (1/10 dilution sample) at 520 ± 1.0 nm. To remove unbound streptavidin, centrifugation was conducted at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min. Then, the supernatant containing the unbound streptavidin was removed and washed off and then mixed with 0.5% (w/v) BSA dissolved in PBS as a blocking buffer. The Au NPs coated with streptavidin molecules (st-Au NPs) exhibited a peak at 531.5 ± 0.5 nm at a concentration with a peak absorbance value of 0.4 (1/10 dilution sample) in their UV-Vis spectra.
000g for 30 min. Then, the supernatant containing the unbound streptavidin was removed and washed off and then mixed with 0.5% (w/v) BSA dissolved in PBS as a blocking buffer. The Au NPs coated with streptavidin molecules (st-Au NPs) exhibited a peak at 531.5 ± 0.5 nm at a concentration with a peak absorbance value of 0.4 (1/10 dilution sample) in their UV-Vis spectra.
Fabrication of the portable kit
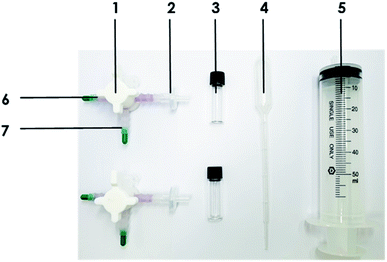
The major objective in developing the prototype kit was to produce a low-cost portable reaction chamber with a 3-way valve to provide an easy-to-use device for primary diagnosis. As shown in Fig. 1, the portable kit consists of a 3-way valve that acts as a reaction chamber, a 50 mL syringe that is used for sample injection and as a filter press, and a syringe filter that filters out Au NAs to obtain a rapid detection signal. To store 200 μL of streptavidin-coated Au NPs (st-Au NPs) and handle the total volume of 400 μL in the reaction, we determined the required pipe length and diameter of the 3-VC to be 16 mm and 4 mm, respectively. To minimize interaction of the device material with proteins, we selected polyvinylidene fluoride (PVDF, hydrophilic) as the 3-VC material.Optimization of the syringe filter
In the SL-based assay, the visible detection signal is amplified by the loss of colour following the precipitation of the large scale-aggregates of Au NPs induced by the SL. In this study, since a filter was introduced to quickly obtain the detection result, experiments were conducted to compare the performances of various filters and determine which was most effective. As shown in Table 1, filters comprising various materials (mixed cellulose ester, polyethersulfone, cellulose acetate, polyvinylidene fluoride, polyamide, and polytetrafluoethylene) and sizes (0.2 μm and 0.45 μm) were used in the filtration of Au NAs in PBS, milk, and 0.1% milk. The total 400 μL volume of the st-Au NP solution was filtered through each syringe filter.| Types | Pore size (μm) | Filtering (PBS) | Filtering (milk) | Filtering (0.1% milk) |
|---|---|---|---|---|
| Mixed cellulose ester (MCE) | 0.2 | Possible | — | — |
| Mixed cellulose ester (MCE) | 0.45 | Possible | Less | Less |
| Polyethersulfone (PES) | 0.2 | Possible | — | — |
| Polyethersulfone (PES) | 0.45 | Possible | Less | Less |
| Cellulose acetate (CA) | 0.45 | Possible | Less | Less |
| Polyvinylidene fluoride (PVDF, hydrophilic) | 0.2 | Possible | — | Less |
| Polyvinylidene fluoride (PVDF, hydrophilic) | 0.45 | Possible | Possible | Possible |
| Polyvinylidene fluoride (PVDF-D, hydrophobic) | 0.2 | Possible | — | Less |
| Polyamide (NYLON) | 0.2 | Possible | — | Less |
| Polyamide (NYLON) | 0.45 | Possible | Less | Possible |
| Polytetrafluoethylene (PTFE-D) | 0.2 | Possible | — | — |
| Polytetrafluoethylene (PTFE-D) | 0.45 | Possible | Less | Possible |
| Polytetrafluoethylene (PTFE-H) | 0.2 | Possible | Less | Possible |
| Polytetrafluoethylene (PTFE-H) | 0.45 | Possible | — | Less |
Detection procedure
The SL-based assay, developed in a previous study, was performed using the portable kit. As shown in Fig. 2C and D, SL-based assays were performed in a total reaction volume of 400 μL containing 200 μL of st-Au NP solution at a fixed concentration, 100 μL of biotinylated polyclonal Salmonella antibody (b-Ab) as the SL, and 100 μL of a test sample (PBS at pH 7.4). First, 100 μL of b-Ab was prepared in a closed reaction chamber of the 3-VC and 200 μL of st-Au NPs was prepared in the closed pipe of the 3-VC. One hundred microliter of the test sample was injected into one pipe of the 3-VC using the syringe, and then this 3-VC mixture was turned and shaken several times. After an adequate reaction time, we opened the valve to allow the mixture to react with the st-Au NPs. After a sufficient reaction time, we filtered the Au NAs through the syringe filter (0.45 μm, PVDF) to obtain a visible detection signal.Prior to performing the SL-based assays for the detection of Salmonella Typhimurium, we had to determine the range of the SL (b-Ab) concentration exhibiting visible colour change (REVC). An assay was performed in PBS at various b-Ab concentrations (10–45 μg mL−1). Based on the REVC results, we selected the concentrations of 13 to 15 μg mL−1 to be the most effective in clearly showing the colour change at the low end of REVC.
Optimization of secondary reaction time for filtration
To filter out the Au NAs, the change in the sizes of the Au NAs over time was measured using a UV-Vis spectrometer and a Zetasizer, respectively. As SLs, 10 and 13 μg mL−1 of biotinylated Salmonella polyclonal antibodies were used as control and aggregates samples, respectively. We added 200 μL of st-Au NP to each test samples and tracked the sizes every 2 min using a Zetasizer.Statistical analysis
The data represent the average of at least three independent experiments or measurements.Results and discussion
Optimization of 3-way valve chamber (3-VC)
To apply the 3-VC to the SL-based assay, it was necessary to optimize the material, the pore and bore sizes of the tubes, the maximum volume, and the loss volume (Table 2). To minimize friction with the solution, among the 3-VC materials considered were polyvinylidiene fluoride (PVDF), polypropylene/polyethylene (PP/PE), and polyvinyl chloride. Ultimately, PVDF was selected due to its low loss volume with the system solution for every pore size. To select the final volume of the 3-way valve, using inner and outer diameters of 4 mm, 6 mm, and 8 mm and 6 mm, 8 mm, and 10 mm, respectively, we considered the compatibility of the syringes and syringe filters and their application to the assay. As they were found to be applicable to the assay under all volume conditions, we selected a valve with a 4 mm inner diameter and 6 mm outer diameter that was compatible with the syringe and syringe filter. Finally, a 3-way valve consisting of PVDF material with an inner diameter of 4 mm and an outer diameter of 6 mm was finally determined to be most suitable as a portable device for field application of the SL-based assay.| Materials | Diameter (pore, mm) | Diameter (bore, mm) | Max volume (μL) | Loss volume (μL) | REVC signal |
|---|---|---|---|---|---|
| PP/PE | 6 | 4 | 500 | <120 | Work |
| 8 | 6 | 1200 | <220 | Work | |
| 10 | 8 | 4000 | <380 | Work | |
| PVDF | 6 | 4 | 500 | <100 | Work |
| 8 | 6 | 1200 | <190 | Work | |
| 10 | 8 | 4000 | <290 | Work |
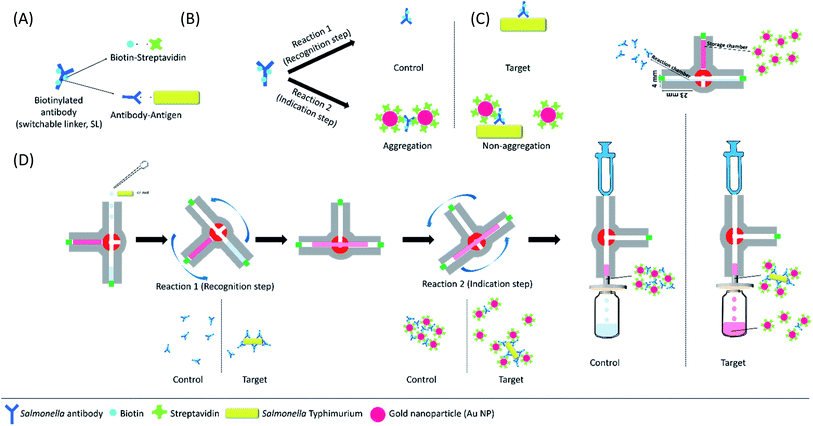
The SL-based immunosensing mechanism by 3-VC
Fig. 2 shows the proposed SL-based immunosensing mechanism for the on-site detection of Salmonella Typhimurium using the developed kit. The SL-based assay can induce the aggregation of st-Au NP with b-Ab and detect the target bacteria in a real matrix. As shown in Fig. 2A and B, we designed the SL (biotinylated antibody) to have two reactions: target recognition (the antibody–antigen immune response) and signal recognition (biotin–streptavidin binding response). Generally, the SL-based assay is performed in two sequential steps. First, the target and the SL undergo a sufficient reaction. Thereafter, depending on the presence or absence of the target, the quantitatively readjusted SLs react with st-Au NPs. When the SLs are bound to their targets, their ability to cross-link with st-Au NP is limited. We refer to this state as the linker being switched-off. Therefore, since the SL is switched off by the target, the degree of aggregation of the st-Au NPs is changed, so the SL-based assay can exhibit a visible colour-fading signal depending on the presence or absence of the target.In this study, a 3-VC was introduced in the proposed kit. As shown in Fig. 2C, the 3-VC consists of a storage chamber and a reaction chamber. The storage chamber contains 200 μL of st-Au NPs solution and the reaction chamber contains 100 μL of biotinylated Salmonella antibody (b-Ab) solution. The amount of SL required to induce aggregation of st-Au NPs depends on the ‘concentration’ of the target in the system. There is also a range of SL concentrations for a given concentration of the target and st-Au NPs that leads to large-scale aggregation and a visible colour change by precipitation of the aggregates, which is known as the REVC. Therefore, the SL can be distinguished as having two regions, i.e., the REVC and the region with no aggregation of st-Au NPs and no colour change (more specifically, the SL concentration range can actually be divided into three parts, which are not explained here). The SL-based assay can identify the target organism based on the REVC shift in response to the ‘switching-off’ of the SL by the target.
System stability of pH and salt conditions
Various and complex components in food are likely to interfere with the detection system. In particular, various pH and high salt concentrations in foods are one of the main causes of biosensor inhibition. In particular, the main factor of Au NP aggregation in this detection system is biotin–streptavidin binding force. In areas where this system will be used, there is no way to remove salt and adjust the pH of the system, so these harsh conditions have to be checked. Streptavidin is a protein as known for its high thermal and mechanical stability. Therefore, stability testing of this system was conducted with emphasis on salt and pH resistance. As shown in Fig. 3, the system confirmed the REVC shift of the control under all pH and salt conditions. As a result, the system was stable under all conditions used in the experiment.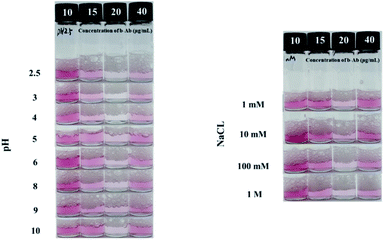 | ||
| Fig. 3 REVC shift of the control using 3-way valve chamber and syringe filter at various system conditions: (A) pH 2.5, 3, 4, 5, 6, 8, 9, 10, (B) 1, 10, 100 mM, and 1 M of NaCL. | ||
The stabilization study of this system was conducted over a wide range of linker concentrations, which can be divided into three regions associated by the size of the aggregates. The size of aggregates was determined by a quantitative correlation between the Au NP and the linker. In the two regions of the linker concentrations of 10 to 15 μg mL−1 and 40 μg mL−1 as shown in Fig. 3, the size of the aggregates of the Au NPs was small so that precipitation did not completely occur. In the case of the linker concentration of 20 μg mL−1, the size of Au NAs was large enough to precipitate completely. This was explained in previous system studies.
There are various components including sugars, proteins, and lipids in real food matrices, and the operation of the system may be inhibited by those things. However, since this system sets a control sample without a target, even if the real food matrix affects the system operation, the REVC in control sample could be reset. Therefore, it can be effectively detected of pathogens if the REVC could be shifted by only a target. In addition, in previous studies, we reported that the SL-based assay could work in serum as well as in various food matrices such as tomato and milk.34–37 Therefore, we dealed with the minimum environmental factors (salt and pH) in this study.
Determination of reaction time (primary reaction time) for targets to crosslink with switchable linkers
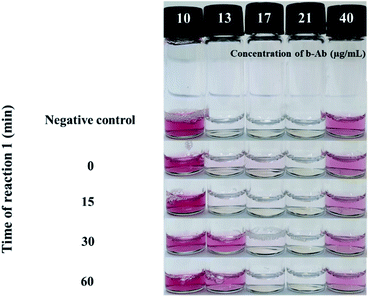
The SL used in this study was designed based on an antibody with biotins, and unlike the ELISA, a specific SL concentration is used in the SL-based assay without any washing step. Therefore, it is necessary to confirm the activity of the SL reacting with the target. In doing so, we tried to optimize the first reaction time of this assay, i.e., the reaction between the SL and the target in the 3-VC. As shown in Fig. 4, 10 μg mL−1 of target Salmonella Typhimurium reacted with 10 μg mL−1 to 40 μg mL−1 of b-Ab as SLs for 0 min, 15 min, 30 min, and 60 min, and then the st-Au NPs were treated to observe the change in the REVC. After 30 min, an REVC shift was observed at a linker concentration of 13 μg mL−1.Determination of reaction time (secondary reaction time) for producing the REVC signal using filter
Switchable linkers can play the roles of both crosslinking the Au NPs and binding the target bacteria, both of which are mutually limited.In other words, in the sequential steps of the SL-based assay, when the SL is combined with the target bacteria, it is switched off and plays no role in inducing aggregation of gold nanoparticles. The SL that is not bound to the target induces large-scale aggregation of gold nanoparticles, which can eventually produce a visual signal whose colour fades as it slowly precipitates out over time (up to 2 h without shaking). The second step of this assay, which determines the REVC, consists of an aggregation reaction of the combination of streptavidin and biotin and a precipitation reaction based on the size of the Au NAs, the latter being the rate-determining step. Therefore, in this study, a filter was applied to the SL-based assay to filter out Au NAs of a certain size to minimize the time required for precipitation and thereby obtain visual signals more quickly.
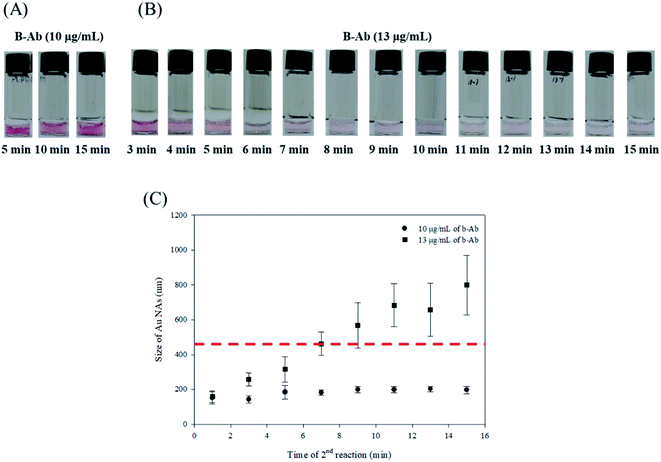
To filter out Au NAs, it was necessary to track the size of the aggregates. As shown in Fig. 5A and B, Au NA was filtered out for 15 min at an SL concentration of 10 μg mL−1 in cases in which no large aggregation was formed and 13 μg mL−1 in cases in which large aggregations were formed. The time to visually recognize that the colour had faded was about 4 min, and after 6 min the solution became clear enough to notice the difference in colour completely. As a result of tracking the size of Au NA for 15 min in 2 min increments by a Zetasizer, it took 7 min for the size of Au NA to grow as large as the size of the filter pore (0.45 μm, the red dash line in Fig. 5C). Taken together, we finally determined that the enough time required for filtration was 7 min. Switchable linkers can crosslink Au NPs to form Au NAs. In the case of the previous SL-based assay, the colour change time can be determined by the sum of the time for forming the aggregate and the time for precipitation, in which the latter takes longer than the former. Therefore, in this study, as a result of introducing the filtration process with the aim of reducing the reaction time of the precipitation and aggregates formation time, the total detection time was greatly reduced by reducing the second step from 2 h to 7 min. This result is expected to introduce a simple syringe filter as part of the kit so that the SL-based assay can detect food-borne pathogens rapidly in the field.
Evaluation experiments
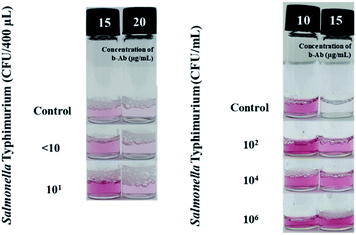
To evaluate the performance of the developed device, we used it to detect Salmonella Typhimurium on the basis of the optimized parameters at various target concentrations. Salmonella Typhimurium, which confirmed the selectivity of SL-based assay in the previous study, was used as a representative strain.35 As shown in Fig. 6, the REVC shift occurred at a linker concentration of 15 μg mL−1, as compared to the control in the tested Salmonella Typhimurium samples (102, 104, and 106 CFU/400 μL). The REVC shift occurred at a linker concentration of 15 μg mL−1, as compared to the control in all the tested Salmonella Typhimurium samples. In addition, to identify an LOD, Salmonella Typhimurium with low level of the concentrations (0, <10, 101 CFU/400 μL) were tested. The assay was evaluated by the naked eye after filtering of Au NAs. As a result, the clear colour change (REVC) in 15 μg mL−1 of b-Ab was seen down to 101 CFU/400 μL of Salmonella Typhimurium. Therefore, the LOD of the BL-based assay using 3-VC was determined to be 101 CFU/400 μL.Since the conventional SL-based assay is based on the precipitation of Au NAs, up to two hours is required to confirm the results. However, in the proposed device, a filter was introduced so that when the Au NAs grow to a certain size that can be filtered, the whole detection time can be reduced by shortening the time required for settling. By applying the filter, the detection result can be quickly confirmed, but we found there to be a volume difference in the filtered system depending on the pressure applied to the filter. In addition, depending on the aggregation mechanism between the target and the gold nanoparticles for each linker concentration, there was a difference in the colour concentration because of the size of the aggregates and the number of gold nanoparticles that did not participate in the reaction. Therefore, when using the proposed apparatus, when selecting a specific linker density, it is necessary to select a section in which the difference in colour density is apparent and the REVC shift is reliably evident. By choosing linker concentrations of 10 μg mL−1 and 15 μg mL−1, Salmonella Typhimurium could be detected down to 101 CFU/400 μL.
This study has suggested two factors to make the SL-based assay more suitable for on-site application. First, a prototype kit that can be easily applied to this assay beyond the laboratory level was proposed. Since the existing method uses reagents, such as buffers, and instruments, such as pipettes, it has application limitations in the field. Second, in order to reduce the detection time, we applied a filter to control the time required for the Au NAs to precipitate. In the existing assay, a shaking method was used to control the time required for precipitation, which required a device such as a vortex, or limiting its use in the field. Therefore, introduction of a 3-way valve-based kit and filter is considered to have upgraded the steps to make the SL-based assay practical.
Conclusions, future directions
Although reliable detection methods for pathogens have been developed for use in the medical, food, and environmental fields, there are many problems to overcome before they can be used in mobile field applications, including the need for miniaturization, rapid diagnosis, and low cost. In this study, we developed a prototype device for making primary diagnoses using the SL-based assay with a 3-VC and a syringe filter for the detection of Salmonella Typhimurium. The use of this assay will be very advantageous in situations where the target bacteria must be identified quickly in the field using a simple experimental procedure and a miniaturized device. It is necessary to solve problems, such as automation of each step's reaction for increasing user convenience using a mechanical and electrical instrument, massification of a wide linker concentration for quantitative detection, and stability of the system through the development of new linker such as an aptamer, in order for this strategy to be used at industrial sites. When these problems are associated with a device and system, they can be solved, and it is expected that targets, such as biological hazards in real food samples can be detected very quickly and easily.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), which is funded by the Ministry of Education (NRF-2013R1A1A2012823 and NRF-2020R1I1A1A0106444511).Notes and references
- K. K. Alharbi and Y. A. Al-sheikh, Saudi J. Biol. Sci., 2014, 21, 109–117 CrossRef PubMed.
- Y.-F. Chen, L. Jiang, M. Mancuso, A. Jain, V. Oncescu and D. Erickson, Nanoscale, 2012, 4, 4839–4857 RSC.
- S. Nayak, N. R. Blumenfeld, T. Laksanasopin and S. K. Sia, Anal. Chem., 2016, 89, 102–123 CrossRef PubMed.
- P. Yager, G. J. Domingo and J. Gerdes, Annu. Rev. Biomed. Eng., 2008, 10, 107–144 CrossRef CAS PubMed.
- M. U. Ahmed, M. M. Hossain, M. Safavieh, Y. L. Wong, I. A. Rahman, M. Zourob and E. Tamiya, Crit. Rev. Biotechnol., 2016, 36, 495–505 Search PubMed.
- R. Dragone, G. Grasso, M. Muccini and S. Toffanin, Frontiers in Public Health, 2017, 5, 80 CrossRef PubMed.
- S. K. Vashist, P. B. Luppa, L. Y. Yeo, A. Ozcan and J. H. Luong, Trends Biotechnol., 2015, 33, 692–705 CrossRef CAS PubMed.
- P. Mandal, A. Biswas, K. Choi and U. Pal, Am. J. Food Technol., 2011, 6, 87–102 CrossRef.
- S.-H. Seo, Y.-R. Lee, J. H. Jeon, Y.-R. Hwang, P.-G. Park, D.-R. Ahn, K.-C. Han, G.-E. Rhie and K.-J. Hong, Biosens. Bioelectron., 2015, 64, 69–73 CrossRef CAS PubMed.
- L. Xiang, H. Wu, Z. Cui and J. Tang, Anal. Lett., 2019, 52, 1966–1975 CrossRef CAS.
- H. Chen, K. Liu, Z. Li and P. Wang, Clin. Chim. Acta, 2019, 493, 138–147 CrossRef CAS PubMed.
- C. D. Chin, T. Laksanasopin, Y. K. Cheung, D. Steinmiller, V. Linder, H. Parsa, J. Wang, H. Moore, R. Rouse and G. Umviligihozo, Nat. Med., 2011, 17, 1015 CrossRef CAS PubMed.
- P. Yager, T. Edwards, E. Fu, K. Helton, K. Nelson, M. R. Tam and B. H. Weigl, Nature, 2006, 442, 412 CrossRef CAS PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368 CrossRef CAS PubMed.
- M. L. de Castro, J. Ruiz-Jiménez and J. Pérez-Serradilla, TrAC, Trends Anal. Chem., 2008, 27, 118–126 CrossRef.
- J. Ruzicka, Analyst, 2000, 125, 1053–1060 RSC.
- J. Wang and E. H. Hansen, TrAC, Trends Anal. Chem., 2003, 22, 225–231 CrossRef CAS.
- M. Miró and E. H. Hansen, Anal. Chim. Acta, 2012, 750, 3–15 CrossRef PubMed.
- E. Mattio, F. Robert-Peillard, L. Vassalo, C. Branger, A. Margaillan, C. Brach-Papa, J. Knoery, J.-L. Boudenne and B. Coulomb, Talanta, 2018, 183, 201–208 CrossRef CAS PubMed.
- P. Athanasatos and T. Costopoulos, Mechanism and Machine Theory, 2012, 50, 64–89 CrossRef.
- M. Giousouf and G. Kovacs, Smart Mater. Struct., 2013, 22, 104010 CrossRef.
- M. L. S. Silva, M. B. Q. Garcia, J. L. Lima and E. Barrado, Talanta, 2007, 72, 282–288 CrossRef CAS PubMed.
- W. J. Kitsch, US Pat., 7 895 850, 2011.
- S. Shoji, in Microsystem technology in chemistry and life science, Springer, 1998, pp. 163–188 Search PubMed.
- J. Fang, Q. Wang and N. Hu, in Micro/Nano Cell and Molecular Sensors, Springer, 2016, pp. 97–123 Search PubMed.
- T. C. Hinkley, S. Singh, S. Garing, A.-L. M. Le, K. P. N. Ny, J. E. Peters, J. N. Talbert and R. Sam, Bioengineering Bacteriophage for the Sensitive & Rapid Detection of Bacteria in Water, 2018, p. 147 Search PubMed.
- H. E. Indyk, E. A. Evans, M. C. B. Caselunghe, B. S. Persson, P. M. Finglas, D. C. Woollard and E. L. Filonzi, J. AOAC Int., 2000, 83, 1141–1148 CrossRef CAS PubMed.
- L. Y. Yeo, H. C. Chang, P. P. Chan and J. R. Friend, Small, 2011, 7, 12–48 CrossRef CAS PubMed.
- G.-H. Chen, W.-Y. Chen, Y.-C. Yen, C.-W. Wang, H.-T. Chang and C.-F. Chen, Anal. Chem., 2014, 86, 6843–6849 CrossRef CAS PubMed.
- Z. Guo, J. Duan, F. Yang, M. Li, T. Hao, S. Wang and D. Wei, Talanta, 2012, 93, 49–54 CrossRef CAS PubMed.
- Y. Luo, J. Xu, Y. Li, H. Gao, J. Guo, F. Shen and C. Sun, Food Control, 2015, 54, 7–15 CrossRef CAS.
- S. E. Magubane, S. Ntlhoro, M. Sabela, S. Kanchi, M. Mlambo, S. C. Onwubu, P. S. Mdluli and A. M. Asiri, J. Taiwan Inst. Chem. Eng., 2019, 101, 159–166 CrossRef CAS.
- I. E. Sendroiu, S. F. Mertens and D. J. Schiffrin, Phys. Chem. Chem. Phys., 2006, 8, 1430–1436 RSC.
- J. Hahn, E. Kim, Y. S. You and Y. J. Choi, Analyst, 2019, 144, 4439–4446 RSC.
- J. Hahn, E. Kim, Y. S. You, S. Gunasekaran, S. Lim and Y. J. Choi, J. Food Sci., 2017, 82, 2321–2328 CrossRef CAS PubMed.
- S. Lim, O. K. Koo, Y. S. You, Y. E. Lee, M.-S. Kim, P.-S. Chang, D. H. Kang, J.-H. Yu, Y. J. Choi and S. Gunasekaran, Sci. Rep., 2012, 2, 456 CrossRef PubMed.
- Y. You, S. Lim, J. Hahn, Y. J. Choi and S. Gunasekaran, Biosens. Bioelectron., 2018, 100, 389–395 CrossRef CAS PubMed.
- G. Frens, Nat. Phys. Sci., 1973, 241, 20 CrossRef CAS.
- K. C. Grabar, R. G. Freeman, M. B. Hommer and M. J. Natan, Anal. Chem., 1995, 67, 735–743 CrossRef CAS.
Footnote |
| † These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2020 |