Open Access Article
Open Access ArticleLabel-free single-molecule identification of telomere G-quadruplexes with a solid-state nanopore sensor†
Sen Wang‡
ab,
Liyuan Liang‡ *ab,
Jing Tangab,
Yao Caiac,
Chuanqi Zhao
*ab,
Jing Tangab,
Yao Caiac,
Chuanqi Zhao bd,
Shaoxi Fangab,
Huabin Wangab,
Ting Wengab,
Liang Wang
bd,
Shaoxi Fangab,
Huabin Wangab,
Ting Wengab,
Liang Wang ab and
Deqiang Wang
ab and
Deqiang Wang *ab
*ab
aChongqing Key Laboratory of Multi-scale Manufacturing Technology, Chongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences, Chongqing 400714, P. R. China. E-mail: liangliyuan@cigit.ac.cn; dqwang@cigit.ac.cn
bChongqing School, University of Chinese Academy of Sciences, Chongqing 400714, P. R. China
cCollege of Instrument Science and Electrical Engineering, Jilin University, Changchun 130016, P. R. China
dLaboratory of Chemical Biology, State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, Jilin 130022, P. R. China
First published on 21st July 2020
Abstract
Telomere sequences can spontaneously form G-quadruplexes (G4) in the presence of some cations. In view of their relevance to genetic processes and potential as therapeutic-targets, hitherto, a wealth of conventional techniques have been reported for interrogation of G4 conformation diversity and corresponding folding kinetics, most of which are limited in precision and sensitivity. This work introduces a label-free solid-state nanopore (SSN) approach for the determination of inter-, intra- and tandem molecular G4 with distinct base permutation in various cation buffers with a tailored aperture and meanwhile captures the single-molecule G4 folding process. SSN translocation properties elucidated that both inter- and intramolecular G4 generated higher current blockage with longer duration than flexible homopolymer nucleotide, and intramolecular G4 are structurally more stable with higher event frequency and longer blockage time than intermolecular ones; base arrangement played weak role in translocation behaviors; the same sequences with one, two and three G4 skeletons displayed an increase in current blockage and a gradual extension in dwell time with the increase of molecule size recorded in the same nanopore. We observed the conformation change of single-molecule G4 which indicated the existence of folding/unfolding equilibration in nanopore, and real-time test suggested a gradual formation of G4 with time. Our results could provide a continued and improved understanding of the underlying relevance of structural stability and G4 folding process by utilizing SSN platform which exhibits strategic potential advances over the other methods with high spatial and temporal resolution.
Introduction
Telomeres are DNA–protein complexes that are located at the end of eukaryotic chromosomes for the inhibition of cellular degradation and maintenance of genome integrity.1,2 Human telomeric DNA comprises hundreds of repetitive 5′-(TTAGGG)n-3′ sequences that are 5000–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 base pairs in full length overhanging at the 3′ end with a single strand of 100–250 bases. This guanine-rich single-stranded DNA is known to form highly ordered secondary structures, called G-quadruplexes (G4) in the presence of mono- or divalent cations. The skeleton of a G4 is composed of stacked G-quartets which contains four planar Gs held together through Hoogsteen hydrogen bonds and further stabilized by intercalating metal ions.3,4 A telomere is prone to shortening along with cell division and is thus closely related to human lifespan; the folded G4 secondary structures at the end of telomeres have been proved to resist the length extension by telomerase and to play a critical role in gene regulation, DNA transcription and replication, and they also exhibit potential in targeted cancer intervention and drug design.5,6 Therefore, investigation of the conformation diversity, relative stability and the folding process of G4 is essential for understanding the modulation mechanism in biological processes.
000 base pairs in full length overhanging at the 3′ end with a single strand of 100–250 bases. This guanine-rich single-stranded DNA is known to form highly ordered secondary structures, called G-quadruplexes (G4) in the presence of mono- or divalent cations. The skeleton of a G4 is composed of stacked G-quartets which contains four planar Gs held together through Hoogsteen hydrogen bonds and further stabilized by intercalating metal ions.3,4 A telomere is prone to shortening along with cell division and is thus closely related to human lifespan; the folded G4 secondary structures at the end of telomeres have been proved to resist the length extension by telomerase and to play a critical role in gene regulation, DNA transcription and replication, and they also exhibit potential in targeted cancer intervention and drug design.5,6 Therefore, investigation of the conformation diversity, relative stability and the folding process of G4 is essential for understanding the modulation mechanism in biological processes.
Computational analysis revealed that there are tens of thousands of sequences that have potential to form G4 structures and they are non-randomly located at functionally important genomic regions. In view of the association of G4 with cell senescence and pathology of cancer, the structural heterogeneity and stability of G4 have aroused broad concern.7,8 Human telomere G4 adopt distinct folds in the presence of different metal ions9,10 and there are a wealth of accessible techniques reported for the investigation of G4 conformation along with the folding dynamics at both single-molecular and single-atomic level, such as X-ray diffraction,11,12 NMR spectroscopy,13 circular dichroism spectroscopy,14 fluorescence resonance energy transfer,15 surface plasmon resonance16 and magnetic tweezers.17,18 However, despite the increased sensitivity, some are limited to solid-phase analysis such as crystallographic methods, some require large quantity of sample with high concentration and purity, such as NMR, and they give average information of molecules. Although the developed single-molecular approaches provide more accurate information, their time resolution may not be enough to capture the quick folding of G4, and the time-consuming and costly labeling or modifications may perturb the G4 folding process.
The nanopore technique with a nano-scaled orifice as sensing platform has emerged to be ultrasensitive for single-molecule identification, including the properties of the size, structure, conformation, charge status and interactions with pores and other probed molecules in real time.19–22 Solid-state nanopore (SSN) fabrication methods are versatile, including ion or electron beam milling, chemical etching and multilevel pulse dielectric breakdown;23–27 one can precisely craft the pore diameter to match the size of analytes and this method is therefore suitable for integrated biological sensors. Due to its high spatial and temporal resolution, size-matched feature, cost-effectiveness and being a label-free process, the nanopore-based sensing platform has attracted great consideration and thereby has been widely and actively used in DNA, RNA, protein, and nanoparticle determination and chirality distinguishing.28–31
At present, there is significant work reported on G4 with biological nanopores including structural identification,32,33 monitoring the folding/unfolding dynamics,34 and investigation of the interplay of G4 with ligands,35,36 while the constricted size of biological nanopores is rather limited for larger-sized G4 translocation except unwinding strands. Also, SSN have been introduced for the investigation of secondary structures of DNA owing to their remarkable thermal and mechanical stability, as well as extraordinary surface, shape and size versatility compared with their biological counterparts. For instance, Long's group has utilized silicon nitride nanopores for ready determination of i-motif structure as well as DNA dynamics;37–39 the employment of tiny SSN for sieving out G4 structure was also reported;40,41 Jiang's group have observed the G4 DNA conformational switching in a responsive nanochannel;42,43 i-motif folding dynamics44 and G4 formation with DNA carrier in SSN was also reported.45 However, inter- and intramolecular and consecutive G4 have not been detected with SSN since inter- and intramolecular DNA with distinct base permutation and stability may perform differently in organisms, and consecutive G4 units are the dominating factor for diseases.
In this work, we have realized the application of label-free SSN for the identification of human telomeric G4 with different strand length and base arrangement in diverse ionic electrolytes and decoded the relative stability of G4 structures under various conditions. Moreover, we have captured the folding process of single-molecule G4 and monitored the G4 formation in temporal dimension under diverse conditions which turned out to be a moderate tendency. Our work provided structural differentiation of inter-, intra- and tandem molecular G4 under distinct conditions and monitored their folding and splitting process at single-molecule level, which helps to understand the modulation mechanism of DNA secondary structures in biological processes in vivo.
Experimental section
Nanopore fabrication
The details of nanopore fabrication with SiNx nanochips via dielectric breakdown were reported previously23–25 and described briefly in ESI.† The diameter of the nanopore is estimated based on a theoretical derivation under nanopore quasi-1D resistance approximation plus access resistances23,46–48 shown below:| G = σ[(4L/πd2) + (1/d)]−1 | (1) |
G4 sensing principle
The details of nanopore sensing of G4 are illustrated in ESI,† and a schematic setup of the nanopore sensing system is exhibited in Scheme 1. As for G4 translocation process, 0.1 μM of the pre-dissolved telomere DNA sequences in 1 M K+ or Na+ buffer solution were heated at 95 °C for 5 minutes before cooling down to room temperature and incubated for one hour which allows the accomplishment of G4 folding equilibration before loading for nanopore testing with 200 mV bias voltage. Each setup was repeated at least two or three times and the resultant data were statistically averaged.Monitoring single-molecule G4 formation with label-free SSN sensor
G4 formation assays were monitored with 10 nM aqueous solution of telomere DNA that was heated at 95 °C for 5 minutes and stored immediately at 0 °C to avoid the formation of any secondary structures before testing. The pre-melted DNA aqueous solution was injected into a flow cell and mixed immediately after the blank recording with a buffer solution containing 1 M KCl or NaCl and 10 mM Tris, 1 mM EDTA at pH 7.4 under 100 mV and 200 mV external voltages in a nanopore of 3.5 nm (for DNA1 and DNA2) or 4.2 nm (for DNA6) in diameter.Results and discussion
Single-molecule resolution of telomeric inter- and intramolecular G4 folding under diverse conditions
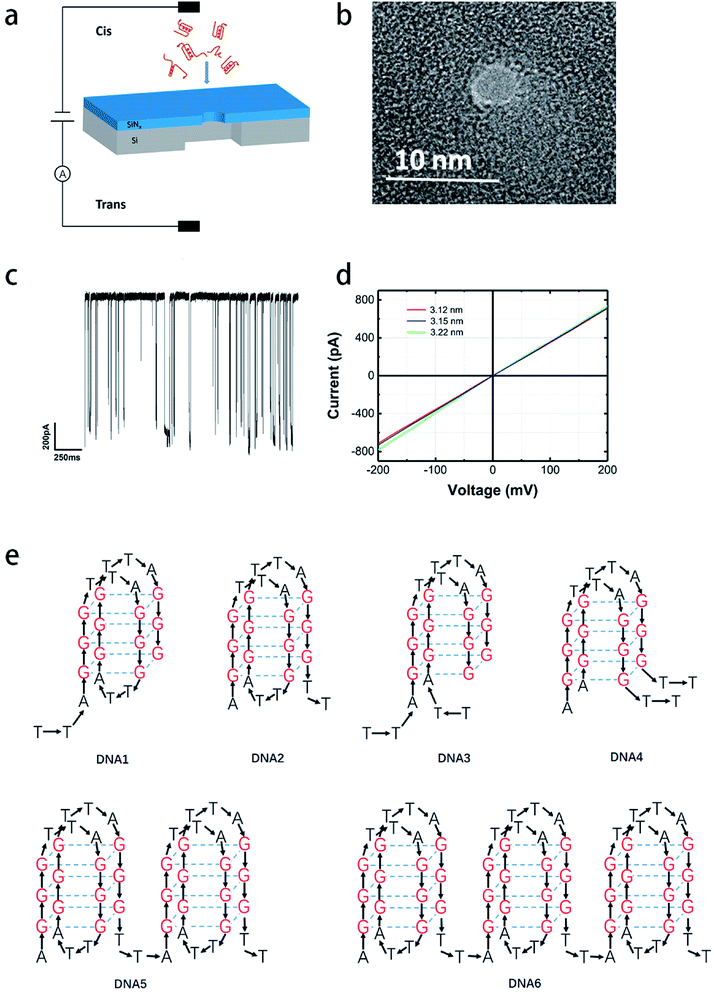
A nanopore-based sensing platform has been employed in this work to identify DNA secondary structures folded with distinct strands in length and base permutation under diverse metal ion induced conditions. Scheme 1a and c demonstrate schematically the nanopore detection system and representative current traces produced by G4 translocation. Scheme 1b is a TEM image of a 3.5 nm nanopore generated via dielectric current breakdown approach. Scheme 1d shows the I–V curves of three nanopores recorded on patch clamp. The pore size for testing is around 3.2 nm in diameter, which is appropriate for the capture of G4 in nanopores; in order to minimize the error arising in nanopores of different size, current blockages were normalized as ΔI/I0, where I0 represents the open pore current and ΔI represents the translocation current blockage produced by the analytes. In this work, four telomeric G4 (structural skeletons are shown in Scheme 1e) were designed to investigate the correlation between the length, base arrangement of the sequence and the folding size and structural stability. K+- and Na+-containing buffers were utilized as they are common and important in vivo and are reported to well stabilize G4 skeleton via intercalation.9,10 100 nM DNA1–4 was melted and cooled down before incubation for one hour in 1 M KCl or NaCl, 10 mM Tris, 1 mM EDTA, pH 7.4 to ensure the completion of the folding process before subjecting to nanopore measurement under 200 mV in a nanopore of 3.2 nm in diameter. In order to corroborate the formation of G4 in the indicated buffers, CD characterization has been implemented. The spectra gave a representative absorption at around 290 nm for all telomeric sequences in both 1 M KCl and NaCl, and absorption density for DNA1 and DNA2 is apparently higher than that for DNA3 and DNA4 (Fig. 1). G4 folded in other cation buffers are also exhibited in Fig. S1.† In comparison, homopolymer nucleotide polyA20 that is unable to fold into secondary structure in buffer does not present an absorption at 290 nm in its CD spectrum, this further proving the formation of G4 structure under our indicated buffer conditions.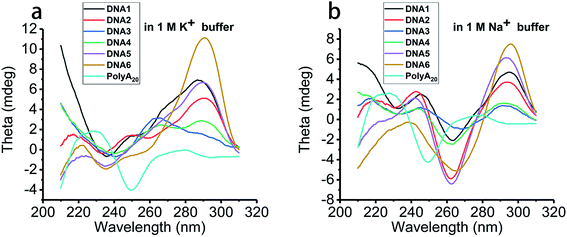 | ||
| Fig. 1 CD spectra of 10 μM human telomeric G4 prepared in 1 M (a) KCl and (b) NaCl buffer solution composed of 1 mM EDTA, 10 mM Tris with pH 7.4 at 22 °C. | ||
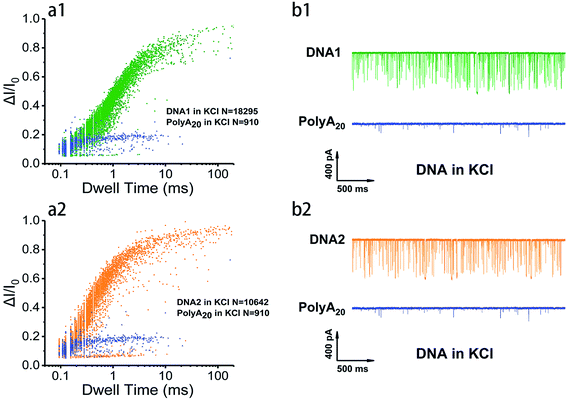
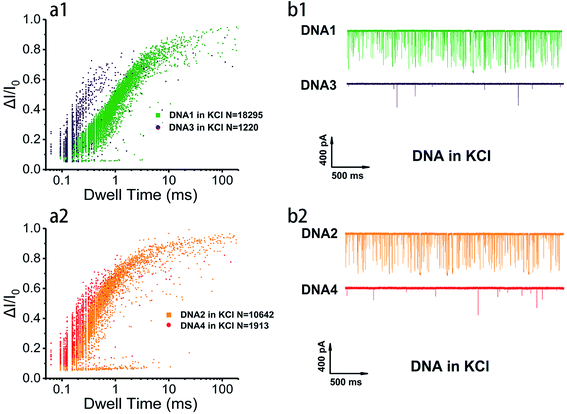
In order to survey the structural stability and compare the translocation properties among the four inter- and intramolecular telomeric G4 and polyA20 in both 1 M K+- and 1 M Na+-containing buffers, parallel assays were carried out with 100 nM of telomere DNA and current traces were collected under 200 mV in the same nanopore. Superimposed scatter plots illustrated that G4 folded with intramolecular DNA1 and DNA2 (Fig. 2 and S2†) and intermolecular DNA3 and DNA4 (data not shown) produced much higher current blockage with longer duration than the flexible homopolymer polyA20 due to their larger physical dimension. This has further verified that our prescient designed four sequences have folded into nanopore size-matched G4 and have performed characteristically based on the SSN approach. In addition, the intramolecular G4 folded with DNA1 and DNA2 threaded through the nanopore with longer duration and higher event frequency than that of intermolecular G4 folded with DNA3 and DNA4 in both Na+ and K+ buffers (Fig. 3a, and S3a†), which indicated that the intramolecular G4 is more stable. The representative current traces also demonstrated a clear distinction in translocation behaviors among inter- and intramolecular G4 and labile sequences (Fig. 3b, and S3b†). Much fewer events were collected for polyA20 and intermolecular G4 due to the larger pore size (3.2 nm) compared with the DNA geometry, and intermolecular G4 do produce spikes with higher current amplitude but much less than that of intramolecular G4, owning to the instability and unwinding during the translocation process. Therefore intermolecular G4 produce events with different levels of blockage amplitude and event frequency was even less than that of polyA20 due to the shorter DNA sequence.
The details of nanopore translocation properties of DNA1–4 and polyA20 are displayed in Fig. S4–S6† and the statistics are collected in ESI Table 1.† It is clear that both current blockage and dwell time of intermolecular G4 folded with DNA3 and DNA4 are quite similar in KCl and NaCl. This implies that the base arrangement is not the determinant factor that impacts the structural difference of intermolecular G4 based on the SSN approach; while intramolecular G4 exhibited distinct properties as shown in Fig. S4.† In 1 M NaCl, the normalized current blockage ΔI/I0 exhibited two Gaussian distributions with a fitting value near 0.2 and 0.7. We proposed that ΔI/I0 lower than 0.2 could be assigned to bumping and the free sequences translocation, which could also be proved by the statistical data in ESI Table 1† and translocation behavior of polyA20, threading through the nanopore without folding to G4 (Fig. S6†); while ΔI/I0 above 0.7 should be mainly contributed by G4 translocation. However, in 1 M KCl, the distribution of current blockage produced by intramolecular G4 is more concentrated. Several repetitions (data not shown) ended up with similar phenomena and ΔI/I0 focused on the low value owing to significant probability of bumping events generated by intramolecular G4 with nanopore which has a highly closed size.
Discrimination of G4 with the same base arrangement while distinct in length
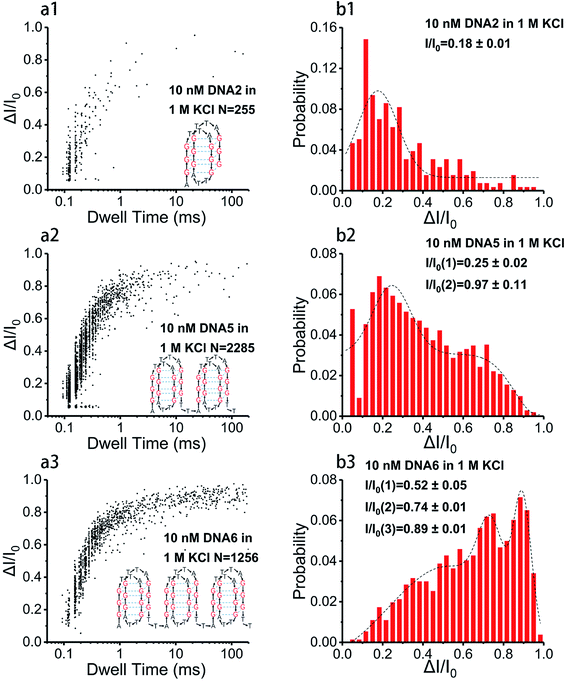
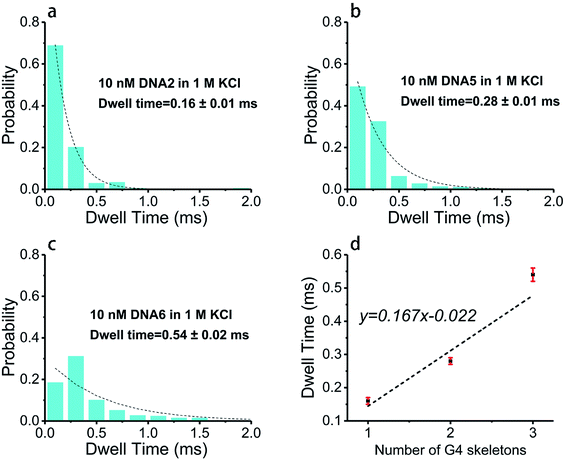
Base arrangement does not significantly influence the translocation behavior of G4 folded in a sequence of the same length as illustrated above. Here we want to further determine if the G4 folded with the same sequence but different in length performed characteristically in the SSN system, as the number of consecutive G4 units in a DNA chain is the dominating factor for diseases. DNA2, DNA5 and DNA6 were designed to share the same sequence but altered in length, the skeletons of which are presented in Scheme 1e. 10 nM of DNA2, DNA5 and DNA6 were melted by heating at 95 °C for 5 minutes before cooling down to room temperature and incubating for one hour in 1 M KCl or NaCl, 10 mM Tris, 1 mM EDTA, pH 7.4 for G4 formation; the prepared G4 samples were characterized with CD spectra (Fig. 1) before subjection to nanopore detection under 200 mV in a nanopore of 4.2 nm. Fig. 4 and 5 demonstrate the translocation properties of the three G4 samples. The event frequency, higher normalized current blockage amplitude ΔI/I0 and blockage duration increase along with the increase of the sequence length as expected; the translocation characteristics of DNA2, DNA5 and DNA6 in NaCl are similar to those in KCl, as exhibited in Fig. S7 and S8.† Statistical analysis in ESI Table 2† illustrated that both higher current blockage ratio and dwell time increase along with prolonging of the length of DNA, which is in agreement with the physical size of G4 skeletons; and the line fitting graphs of blockage duration in Fig. 5d and S8d† suggest an approximate linear relation with the number of G4 skeletons.Exploration of the folding process of G4 under distinct conditions
In order to decipher the folding and splitting process of distinct G4 under varied conditions, we performed the setups by monitoring the produced current traces of G4 translocation immediately after the addition of the melted DNA solution into test buffer in a flow cell. We have captured quite a few representative translocation traces of single-molecule G4 folded with 10 nM DNA2 in 1 M KCl (Fig. S9†). The analysis of all point histograms for these typical single-molecule DNA events suggested that events with two or more distributions represent two or more conformations being involved during the translocation process. Single-molecule G4 could change their conformation in a confined space, and they might involve various intermediates (hair pins or triplexes)49,50 according to the spikes in Fig. S9a.† Some single-molecule signals hold two levels of blockage in which the higher blockage could be a G4 structure and the lower one might be any partly folded intermediate involved during the threading process. Some translocation current traces of long-strand DNA6 and their all point histograms are provided in Fig. S10.† They also gave a good sense of the captured conformation change of G4 in SSN assays. Some other sorts of events (Fig. S11†) also indicated a complicated mixture of G4 system in translocation process.To reveal the G4 folding process in temporal dimension, firstly we attempted to survey the influence of the concentration of metal ions on G4 folding rate by analyzing migration of the current blockage amplitude with increasing time, since the formation and stability of G4 highly rely on the type and concentration of intercalated cations. We performed the assay in 1 M K+ or Na+ buffer solution (the optimized concentration of 1 M buffer is illustrated in ESI†) to ensure a good signal to noise ratio in nanopores and recorded the current traces with time for inter-, intra- and tandem molecular telomeric G4 (due to the poor stability of intermolecular G4, the tendency of the translocation data is not shown). We analyzed the current blockage in the first tens of seconds, as many works have proved that the time scale of the G4 folding process is of a few seconds or even microseconds.45,51 10 nM DNA1, DNA2 and long-sequence DNA6 were subjected to the SSN system with a pore size of 3.5 nm (for DNA1 and DNA2) and 4.2 nm (for DNA6) in both 1 M K+ and Na+ for folding/unfolding process investigation. Fig. S12 and S13† demonstrate a part of the typical translocation behavior of DNA1 every 10 seconds. We monitored the first 40 seconds of translocation behaviors. The histograms of normalized current blockage (Fig. S12b, and S13b†) and line graph (Fig. S12c, and S13c†) gave a slight tendency of which the ratio of lower current blockage (ΔI/I0 = 0–0.2) that is proposed to be produced by labile sequence decrease, and the ratio of higher current blockage (ΔI/I0 = 0.7–1) that represents the G4 translocation increase with time. Although the first point was anomalous with longer duration which might due to the access resistance at the beginning of translocation test, the blockage duration presented an apparent increase during our indicated time period (Fig. S12d, S13d, and S14†), as G4 with a larger size than labile sequence will block the nanopore to a larger extent, thereby the dwell time increases along with the augmentation of the ratio of G4 in the translocation analytes. This is in line with our expectation and it is also matched with our statistical analysis for current blockage during the full-set translocation of the first 40 seconds and the first 3 minutes of measurement (Fig. S15†).
We further monitored G4 formation under various conditions with different DNA sequences. All the statistical line graphs are shown in Fig. S16.† Several replicates of DNA1 in 1 M KCl were surveyed (Fig. S16a–d†) and they all demonstrated an increase of ratio in higher current blockage (ΔI/I0 = 0.7–1) that signified an increase of G4 formation with time; the folding dynamics of DNA6 in both 1 M KCl and NaCl were also examined under 100 mV and 200 mV, the line graphs presenting a moderate increased ratio of G4 with an increase of time (Fig. S16g–j†). It is worth mentioning that in the first tens of seconds, the current traces with higher blockage ratio were greater than those with lower blockage ratio for both DNA 1 and DNA2 (Fig. S16a–f†) while the opposite was the case for DNA6 (Fig. S16g–j†), which signified a slower folding process of DNA6 than DNA1 and DNA2 that have smaller molecular size; and the variation of the applied voltages (200 mV in Fig. S16g and h† and 100 mV in Fig. S16i and j†) did not influence the tendency. Even the line graphs are not regular and perfect, but all the examples in Fig. S16† display a tendency of an increase ratio of G4 with increasing time during the first tens of seconds or first few minutes. This confirmed that we could realize real-time monitoring of G4 formation with our nanopore platform despite this process being exceedingly fast and complicated.
Conclusions
We have realized the determination of intermolecular, intramolecular and tandem telomeric G4 with altered base permutation in the presence of distinct cations based on a SSN sensing platform. The translocation properties revealed that both inter- and intramolecular G4 produced higher event frequency with larger current blockage and longer translocation time than labile polymer nucleotide; in addition, intramolecular G4 is apparently structurally more stable which produced more events with longer blockage duration than intermolecular G4 in both 1 M KCl and NaCl buffer solutions, and intramolecular G4 performed distinctly in K+ and Na+ due to the disparity of physical size and stability of G4 in the two cation buffers. Base alteration does not contribute to evident discrepancy in translocation behaviors. Furthermore, the same sequences with consecutive G4 units exhibited an increase in the higher current blockage and a gradual extension in dwell time recorded in the same nanopore. A wealth of representative translocation events and corresponding distribution profiles illustrated a dynamic G4 conformation change during nanopore measurement, signifying that a folding/unfolding dynamic process was captured with our SSN system. In addition, the exploration of the G4 formation process performed with 10 nM of different telomeric sequences exhibited a moderate tendency of which the distribution of higher current blockage that represents the G4 translocation augments along with time during the formation of G4 for all the assays. Compared with biological nanopores, the SSN approach with tailored size is more favorable for the discrimination of G4 molecules with intrinsic conformation intricacies and diversity and also real-time monitoring the G4 formation by SSN under diverse conditions. Our work provides a mechanistic insight into the monitoring and interpretation in single-molecule reactions including intermediate capture based on the SSN technique. Further study will be on the immobilization of telomeric sequences in SSN and monitoring the folding kinetics by the variation of cations and loop permutation.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank Mr Jianqiang Fu for help with TEM characterization. This work was supported by the Natural Science Foundation of Chongqing, China (grant no. cstc2017jcyjB0105), the Youth Innovation Promotion Association (2017392), the Pioneer Hundred Talents Program of the Chinese Academy of Sciences (to Liang Wang), the Instrumentation Development Program of the Chinese Academy of Sciences (YZ201568) and the University of Chinese Academy of Sciences.References
- L. I. Jansson, J. Hentschel, J. W. Parks, T. R. Chang, C. Lu, R. Baral, C. R. Bagshaw and M. D. Stone, Telomere DNA G-quadruplex folding within actively extending human telomerase, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 9350–9359 CrossRef CAS PubMed.
- H. Izumi and K. Funa, Telomere Function and the G-Quadruplex Formation are Regulated by hnRNP U, Cells, 2019, 8, 390–400 CrossRef CAS PubMed.
- J. L. Mergny and D. Sen, DNA Quadruple Helices in Nanotechnology, Chem. Rev., 2019, 119, 6290–6325 CrossRef CAS.
- H. L. Bao, H. S. Liu and Y. Xu, Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells, Nucleic Acids Res., 2019, 47, 4940–4947 CrossRef CAS PubMed.
- Z. Yu, K. D. Fenk, D. Huang, S. Sen and J. A. Cowan, Rapid Telomere Reduction in Cancer Cells Induced by G-Quadruplex-Targeting Copper Complexes, J. Med. Chem., 2019, 62, 5040–5048 CrossRef CAS PubMed.
- D. Rhodes and H. J. Lipps, G-quadruplexes and their regulatory roles in biology, Nucleic Acids Res., 2015, 43(18), 8627–8637 CrossRef CAS PubMed.
- X. Zhang, D. Zhang, C. Zhao, K. Tian, R. Shi, X. Du, A. J. Burcke, J. Wang, S. J. Chen and L. Q. Gu, Nanopore electric snapshots of an RNA tertiary folding pathway, Nat. Commun., 2017, 8, 1458–1468 CrossRef PubMed.
- J. Shim and L. Q. Gu, Single-molecule investigation of G-quadruplex using a nanopore sensor, Methods, 2012, 57, 40–46 CrossRef CAS PubMed.
- N. An, A. M. Fleming and C. J. Burrows, Interactions of the human telomere sequence with the nanocavity of the alpha-hemolysin ion channel reveal structure-dependent electrical signatures for hybrid folds, J. Am. Chem. Soc., 2013, 135, 8562–8570 CrossRef CAS PubMed.
- N. An, A. M. Fleming, E. G. Middleton and C. J. Burrows, Single-molecule investigation of G-quadruplex folds of the human telomere sequence in a protein nanocavity, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 14325–14331 CrossRef CAS PubMed.
- Y. He, Y. Zhang, L. Wojtas, N. G. Akhmedov, D. Thai, H. Wang, X. Li, H. Guo and X. Shi, Construction of a cross-layer linked G-octamer via conformational control: a stable G-quadruplex in H-bond competitive solvents, Chem. Sci., 2019, 10, 4192–4199 RSC.
- B. Bakalar, B. Heddi, E. Schmitt, Y. Mechulam and A. T. Phan, A Minimal Sequence for Left-Handed G-Quadruplex Formation, Angew. Chem., Int. Ed., 2019, 58, 2331–2335 CrossRef CAS PubMed.
- H. L. Bao and Y. Xu, Investigation of higher-order RNA G-quadruplex structures in vitro and in living cells by (19)F NMR spectroscopy, Nat. Protoc., 2018, 13, 652–665 CrossRef CAS PubMed.
- G. Tao, Y. Chen, R. Lin, J. Zhou, X. Pei, F. Liu and N. Li, How G-quadruplex topology and loop sequences affect optical properties of DNA-templated silver nanoclusters, Nano Res., 2018, 11, 2237–2247 CrossRef CAS.
- D. Panda, P. Saha, T. Das and J. Dash, Target guided synthesis using DNA nano-templates for selectively assembling a G-quadruplex binding c-MYC inhibitor, Nat. Commun., 2017, 8, 16103–16113 CrossRef CAS PubMed.
- Y. Zhao, Z.-Y. Kan, Z.-X. Zeng, Y.-H. Hao, H. Chen and Z. Tan, Determining the Folding and Unfolding Rate Constants of Nucleic Acids by Biosensor. Application to Telomere G-Quadruplex, J. Am. Chem. Soc., 2004, 126, 13255–13264 CrossRef CAS PubMed.
- P. Shrestha, S. Jonchhe, T. Emura, K. Hidaka, M. Endo, H. Sugiyama and H. Mao, Confined space facilitates G-quadruplex formation, Nat. Nanotechnol., 2017, 12, 582–588 CrossRef CAS PubMed.
- S. Selvam, S. Mandal and H. Mao, Quantification of Chemical and Mechanical Effects on the Formation of the G-Quadruplex and i-Motif in Duplex DNA, Biochemistry, 2017, 56, 4616–4625 CrossRef CAS.
- M. van den Hout, A. R. Hall, M. Y. Wu, H. W. Zandbergen, C. Dekker and N. H. Dekker, Controlling nanopore size, shape and stability, Nanotechnology, 2010, 21, 115304–115309 CrossRef PubMed.
- J. Houghtaling, C. Ying, O. M. Eggenberger, A. Fennouri, S. Nandivada, M. Acharjee, J. Li, A. R. Hall and M. Mayer, Estimation of Shape, Volume, and Dipole Moment of Individual Proteins Freely Transiting a Synthetic Nanopore, ACS Nano, 2019, 13, 5231–5242 CrossRef CAS PubMed.
- J. D. Spitzberg, A. Zrehen, X. F. van Kooten and A. Meller, Plasmonic-Nanopore Biosensors for Superior Single-Molecule Detection, Adv. Mater., 2019, 31, 1900422–1900439 CrossRef PubMed.
- Z. Long, S. Zhan, P. Gao, Y. Wang, X. Lou and F. Xia, Recent Advances in Solid Nanopore/Channel Analysis, Anal. Chem., 2018, 90, 577–588 CrossRef CAS PubMed.
- B. Yin, W. Xie, L. Liang, Y. Deng, S. He, F. He, D. Zhou, C. Tlili and D. Wang, Covalent Modification of Silicon Nitride Nanopore by Amphoteric Polylysine for Short DNA Detection, ACS Omega, 2017, 2, 7127–7135 CrossRef CAS PubMed.
- H. Tian, W. Xie, S. He, D. Zhou, S. Fang, L. Liang and D. Wang, Investigation of the adsorption behavior of BSA with tethered lipid layer-modified solid-state nanopores in a wide pH range, RSC Adv., 2019, 9, 15431–15436 RSC.
- F. He, L. Liang, S. Zhou, W. Xie, S. He, Y. Wang, C. Tlili, S. Tong and D. Wang, Label-Free Sensitive Detection of Microcystin-LR via Aptamer-Conjugated Gold Nanoparticles Based on Solid-State Nanopores, Langmuir, 2018, 34, 14825–14833 CrossRef CAS PubMed.
- Y. Deng, Q. Huang, Y. Zhao, D. Zhou, C. Ying and D. Wang, Precise fabrication of a 5 nm graphene nanopore with a helium ion microscope for biomolecule detection, Nanotechnology, 2017, 28, 045302–045307 CrossRef.
- R. Gao, Y. Lin, Y. L. Ying, Y. X. Hu, S. W. Xu, L. Q. Ruan, R. J. Yu, Y. J. Li, H. W. Li, L. F. Cui and Y. T. Long, Wireless nanopore electrodes for analysis of single entities, Nat. Protoc., 2019, 14, 2015–2035 CrossRef CAS.
- R. J. Yu, Y. L. Ying, R. Gao and Y. T. Long, Confined Nanopipette Sensing: From Single Molecules, Single Nanoparticles, Angew. Chem., Int. Ed., 2019, 58, 3706–3714 CrossRef CAS.
- J. Beaulaurier, E. E. Schadt and G. Fang, Deciphering bacterial epigenomes using modern sequencing technologies, Nat. Rev. Genet., 2019, 20, 157–172 CrossRef CAS PubMed.
- S. Cai, J. Y. Y. Sze, A. P. Ivanov and J. B. Edel, Small molecule electro-optical binding assay using nanopores, Nat. Commun., 2019, 10, 1797–1805 CrossRef PubMed.
- X. Zhao, R. Ma, Y. Hu, X. Chen, R. Dou, K. Liu, C. Cui, H. Liu, Q. Li, D. Pan, X. Shan, L. Wang, C. Fan and X. Lu, Translocation of tetrahedral DNA nanostructures through a solid-state nanopore, Nanoscale, 2019, 11, 6263–6269 RSC.
- N. An, A. M. Fleming, H. S. White and C. J. Burrows, Nanopore Detection of 8-Oxoguanine in the Human Telomere Repeat Sequence, ACS Nano, 2015, 9, 4296–4307 CrossRef CAS PubMed.
- J. W. Shim, Q. Tan and L. Q. Gu, Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity, Nucleic Acids Res., 2009, 37, 972–982 CrossRef CAS PubMed.
- Y. Ding, A. M. Fleming, L. He and C. J. Burrows, Unfolding Kinetics of the Human Telomere i-Motif Under a 10 pN Force Imposed by the alpha-Hemolysin Nanopore Identify Transient Folded-State Lifetimes at Physiological pH, J. Am. Chem. Soc., 2015, 137, 9053–9060 CrossRef CAS PubMed.
- A. Zhao, S. E. Howson, C. Zhao, J. Ren, P. Scott, C. Wang and X. Qu, Chiral metallohelices enantioselectively target hybrid human telomeric G-quadruplex DNA, Nucleic Acids Res., 2017, 45, 5026–5035 CrossRef CAS.
- C. Zhao, L. Wu, J. Ren, Y. Xu and X. Qu, Targeting human telomeric higher-order DNA: dimeric G-quadruplex units serve as preferred binding site, J. Am. Chem. Soc., 2013, 135, 18786–18789 CrossRef CAS PubMed.
- S. C. Liu, Q. Li, Y. L. Ying and Y. T. Long, Detection of structured single-strand DNA via solid-state nanopore, Electrophoresis, 2019, 40, 2112–2116 CrossRef CAS PubMed.
- X. Shi, Q. Li, R. Gao, W. Si, S. C. Liu, A. Aksimentiev and Y. T. Long, Single-molecule analysis in an electrochemical confined space, J. Phys. Chem. Lett., 2018, 9, 4686–4694 CrossRef CAS PubMed.
- W. Si, H. Yang, J. Sha, Y. Zhang and Y. Chen, Dynamics of a Molecular Plug Docked onto a Solid-State Nanopore, Electrophoresis, 2019, 14, 2117–2124 CrossRef PubMed.
- W. Si, H. Yang, J. Sha, Y. Zhang and Y. Chen, Discrimination of single-stranded DNA homopolymers by sieving out G-quadruplex using tiny solid-state nanopores, Electrophoresis, 2019, 14, 2117–2124 CrossRef PubMed.
- Y. Goto, I. Yanagi, K. Matsui, T. Yokoi and K. I. Takeda, Identification of four single-stranded DNA homopolymers with a solid-state nanopore in alkaline CsCl solution, Nanoscale, 2018, 10, 20844–20850 RSC.
- M. Liu, H. Zhang, K. Li, L. Heng, S. Wang, Y. Tian and L. Jiang, A Bio-inspired Potassium and pH Responsive Double-gated Nanochannel, Adv. Funct. Mater., 2015, 25, 421–426 CrossRef CAS.
- J. Wang, R. Fang, J. Hou, H. Zhang, Y. Tian, H. Wang and L. Jiang, Oscillatory Reaction Induced Periodic C-Quadruplex DNA Gating of Artificial Ion Channels, ACS Nano, 2017, 11, 3022–3029 CrossRef CAS PubMed.
- A. Megalathan, B. D. Cox, P. D. Wilkerson, A. Kaur, K. Sapkota, J. E. Reiner and S. Dhakal, Single-molecule analysis of i-motif within self-assembled DNA duplexes and nanocircles, Nucleic Acids Res., 2019, 47, 7199–7212 CrossRef CAS PubMed.
- F. Boskovic, J. Zhu, K. Chen and U. F. Keyser, Monitoring G-Quadruplex Formation with DNA Carriers and Solid-State Nanopores, Nano Lett., 2019, 19(11), 7996–8001 CrossRef CAS PubMed.
- S. W. Kowalczyk, A. Y. Grosberg, Y. Rabin and C. Dekker, Modeling the conductance and DNA blockade of solid-state nanopores, Nanotechnology, 2011, 22, 315101–315105 CrossRef PubMed.
- M. Chinappi, T. Luchian and F. Cecconi, Nanopore tweezers: Voltage-controlled trapping and releasing of analytes, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2015, 92, 032714–032722 CrossRef PubMed.
- M. Wanunu, W. Morrison, Y. Rabin, A. Y. Grosberg and A. Meller, Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient, Nat. Nanotechnol., 2010, 5, 160–165 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka and H. Sugiyama, Direct and single-molecule visualization of the solution-state structures of G-hairpin and G-triplex intermediate, Angew. Chem., Int. Ed., 2014, 53, 4107–4112 CrossRef CAS PubMed.
- W. Li, X.-M. Hou, P.-Y. Wang, X.-G. Xi and M. Li, Direct measurement of sequential folding pathway and energy landscape of human telomeric G-quadruplex structures, J. Am. Chem. Soc., 2013, 135, 6423–6426 CrossRef CAS PubMed.
- R. D. Gray and J. B. Chaires, Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures, Nucleic Acids Res., 2008, 36, 4191–4203 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of the characterization, the translocation properties of all the telomere G4 samples and polyA20 under different conditions are available from the corresponding authors on request. See DOI: 10.1039/d0ra05083k |
| ‡ These authors contributed equally to the paper. |
| This journal is © The Royal Society of Chemistry 2020 |