Open Access Article
Open Access ArticlePoly(3-hydroxybutyrate)/poly(amine)-coated nickel oxide nanoparticles for norfloxacin delivery: antibacterial and cytotoxicity efficiency†
Nehal
Salahuddin
 *,
Mohamed
Gaber
,
Maie
Mousa
and
Mohamed A.
Abdelwahab
*,
Mohamed
Gaber
,
Maie
Mousa
and
Mohamed A.
Abdelwahab
 *
*
Chemistry Department, Faculty of Science, Tanta University, Tanta, 31527 Egypt. E-mail: nehal.attaf@science.tanta.edu.eg; mohamed.abdelwahab@science.tanta.edu.eg
First published on 17th September 2020
Abstract
Sustained release dosage forms enable prolonged and continuous release of a drug in the gastrointestinal tract for medication characterized by a short half lifetime. In this study, the effect of blending polyamine on poly(3-hydroxybutyrate) (PHB) as a carrier for norfloxacin (NF) was studied. The prepared blend was mixed with different amounts of NiO nanoparticles and characterized using FTIR analysis, X-ray diffraction analysis, thermogravimetric analysis, dynamic light scattering, transmission electron microscopy and scanning electron microscopy. It was found that the drug released from the nanocomposite has a slow rate in comparison with NiO, PHB, and PHB/polyamine blend. The highest ratio of NiO content to the matrix (highest NF loading), leads to a slower rate of drug release. The release from the nanocomposites showed a faster rate at pH = 2 than that at pH = 7.4. The mechanisms of NF adsorption and release were studied on PHB/polyamine–3% NiO nanocomposite. In addition, the antimicrobial efficacy of nanocomposites loaded with the drug was determined and compared with the free drug. Inclusion of NiO into PHB/polyamine showed a higher efficacy against Streptococcus pyogenes and Pseudomonas aeruginosa than the free NF. Moreover, the cytotoxicity of PHB/polyamine–3% NiO against HePG-2 cells was investigated and compared with PHB and PHB/polyamine loaded with the drug. The most efficient IC50 was found for NF@PHB/polyamine–3% NiO (29.67 μg mL−1). No effect on cell proliferation against the normal human cell line (WISH) was observed and IC50 was detected to be 44.95 and 70 μg mL−1 for NiO nanoparticles and the PHB/polyamine–3% NiO nanocomposite, respectively indicating a selectivity of action towards tumor cells coupled with a lack of cytotoxicity towards normal cells.
1 Introduction
Cancer is a major problem around the globe and is the second leading cause of death. Cancer was originally thought to be a genetic disease. However, recent studies have revealed the connection between bacterial infections and growth of different types of cancer.1 Although the idea of bacteria causing different types of cancer exploded about a century ago, the potential mechanisms of carcinogenesis are still not well established. Various targeted proteins may interfere with the normal growth behaviour of host cells, thus altering the outcome of programmed cell death.2Colorectal cancer (CRC) is a heterogeneous disease and considered as one of the foremost reasons for mortality and morbidity in the world.3 Various factors have been associated with the growth of colon cancer including mutation, bacterial infection and irradiation. It was predicted that several proteins of M. hominis may be targeted to the host cell ER, and possibly alter the normal pattern of protein folding. These predicted proteins can modify the normal function of the host cell. Thus, the intercellular infection of M. hominis in host cells may assist as a potential issue in prostate cancer etiology.1
Hyperthermia therapy is a type of medical treatment in which the body tissue is exposed to a higher temperature (up to 45 °C) to treat and destroy cancer cells or shrink tumors. The damage to cells during hyperthermia can make them more sensitive to the effects of chemotherapy and radiation therapy so these treatments work better. Among hyperthermia therapy methods; magnetic hyperthermia is well known as the one that produces a controllable heat inside the body. Magnetic hyperthermia therapy (MHT) involves heat generation using magnetic nanoparticles (MNPs) in response to an externally applied alternating current magnetic field. These MNPs can be specifically targeted to the tumor site for homogenous heating. In recent years, there is an emerging interest to synthesize nanoparticles of Fe, Co, and Ni due to their superior properties and application in various fields like sensors,4 memory storage devices,5 drug delivery,6 catalysis,7 magnetic resonance imaging8 and very recently in the treatment of cancer cells9 due to its modest magnetic susceptibility.
Studies showed that various types of NPs employed for the analysis of anticancer potential against HT-29 and other colon cancer cell line. Various metal oxide NPs, including silver oxide (Ag2O), cobalt oxide (Co3O4), manganese oxide (Mn3O4), titanium dioxide (TiO2), and zinc oxide (ZnO), have been investigated for their anticancer activities.10–13 Recently, NiO-NP with irregular spherical shape (20–25 nm) significantly diminished the viability of HT-29 and SW620 human colon cancer cells and can be participating as a treatment option in colorectal cancer patients.10 The synthesized NiO-NPs yielded IC50 values of 13.72 and 394.41 μg mL−1 for HT-29 and SW620 cells, respectively. These consequences demonstrate that NiO-NPs are more cytotoxic to HT-29 cells at lower doses, indicating therapeutic potential.10
Gupta et al.14 reported that NiO nanoparticles and MnO2/NiO nanocomposites are promising for targeted caffeic acid phenethyl ester anticancer drug delivery applications. Different conjugates NiO nanoparticles erythromycin drug showed different antimicrobial activities and could be applied to overcome drug resistant.15 Several attempts were done to mix the anti-cancer drugs with the biodegradable polymer during the delivery process.16,17 The nanoparticles were coated by polymer, which releases the drug by controlled diffusion or erosion from the core across the polymeric membrane or matrix.
Poly(3-hydroxybutyrate) (PHB) are the most thoroughly studied forms of biopolymer polyesters family for biomedical applications due to their biocompatibility, biodegradability, nontoxic, and adjustable mechanical properties.18,19 These properties make PHB as an ideal candidate in medicine, surgery, pharmacology and in tissue engineering scaffolds. It was reported that PHB nanoparticles acted as an efficient carrier vehicle for doxorubicin drug and delayed the release for 5–7 days.20 In another publication,21 microspheres prepared from a microbial biodegradable polyester, i.e. PHB was used as a potential chemoembolization agent.21 Drug release rates were very rapid and almost 90% of the drug-loaded was released in about 24 h. Both the size and drug content of PHB microspheres were found to be effective in controlling the drug release from these microspheres. The retention of potent antineoplastic drugs within the NPs is crucial for the clinical application. The liberation of docetaxel from poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) NPs in PBS pH 7.4 at 25 °C demonstrated minimal drug release for 3–4 days period. This less liberation of drug provided a window of toxicity protection during circulation and allowing specificity to leaky blood vasculature of tumors by the enhanced permeability and retention effect (EPR). The negatively charged zeta potential and a low in vitro drug release rate ensure safety in the biological environment toward premature degradation and cytotoxic capacity. Therefore, PHB NPs could be considered as a promising vehicle for hydrophobic drug administration for better treatment of breast cancer.22
Polyamines are abundantly available in the liver of poultry, fermented soybeans and mushrooms. Polyamines have various advantages such as high-water solubility, absorptivity, high cationic charge density, and high reactivity. It has been reported that polyamine prevents arteriosclerosis23,24 and promotes hair growth25 due to its anti-inflammatory properties and cell proliferative effect. Polyamines are essential components in cell growth and cell differential process. They have a role in the regulation and stimulation of DNA, RNA and synthesis of protein.26 In addition polyamines affect on tumor growth and carcinogenesis which reflect their multifunctional character during growth.
Norfloxacin (NF) belongs to the group of fluoroquinolone antibiotics that depict a broad spectrum of antimicrobial activity, relatively low incidence of adverse and toxic effects as well as an excellent safety profile.27,28 These quinolones showed a potentially important source of new anticancer agents. These agents work by inhibiting the two enzymes namely DNA gyrase and topoisomerase IV that are essential for DNA replication as well as transcription leading to cell death.29 NF has a short half-life time (3–4 h) and high doses of antibiotics causes severe side effect.30–32 Copper II complex of N-propyl-norfloxacin (Hpr-nor) and 1, 10-phenanthroline (phen) showed dramatically decreased cell viability in time-dependent manner with higher in vitro cytotoxicity on leukemia cells in comparison with CuCl2, Hpr-nor-Cu(op)2 and phen due to synergistic contribution of its components.33 Recently, Abdel-Aziz el al.,34 synthesized 7-(4-substituted piperazin-1-yl)-4-oxoquinolines based on ciprofloxacin and NF scaffolds. Derivatives based on NF scaffold were more active against cancer cell lines in comparison with ciprofloxacin. Interestingly, these compounds showed high efficiency compared with 5-fluorouracil.34 Vivek et al.35 reported that lipid–polymer hybrid nanoparticles were prepared for topical and site targeting the delivery of NF by emulsification solvent evaporation method (ESE). The results justified the formulation of lipid–polymer hybrid nanoparticles (LPNPs) containing NF is a good alternative and suitable carrier for the treatment of burn bacterial infection. Ellipticine (EPT) encapsulated polyhydroxyalkanoates nanoparticles demonstrated higher inhibition of cancer cell line A549 in comparison with EPT due to excellent bioavailability.36 Recently, NF was immobilized into poly(3-hydroxybutyrate)/polyethylene glycol–nickel oxide (PHB/PEG–NiO) nanocomposites using different NiO contents. The as-prepared loaded PHB/PEG and PHB/PEG–5% NiO nanocomposite was demonstrated to be a new potent antitumor, where the IC50 was lower than IC50 of NF accompanied with the lack of toxicity against normal cell.37
This work was aimed to study the effect of blending polyamine with PHB on the loading capacity and release profile of NF. The antimicrobial activity towards Gram-positive (Staphylococcus aureus, Streptococcus), Gram-negative bacteria (Klebsiella pneumonia, Escherichia coli, Pseudomonas aeruginosa) was studied. Moreover, the cytotoxicity of PHB/polyamine–3% NiO towards HepG-2 cell was compared with PHB and PHB/polyamine loaded drug.
2 Materials and methods
2.1. Materials
Norfloxacin (NF) (C16H18FN3O3 with a molecular weight of 319.336 g mol−1) was obtained from Epico (Egypt). Nickel acetate tetrahydrate [Ni (CH3COO)2·4H2O] was purchased from MERCK (Darmstadt). Poly(3-hydroxy butyrate) PHB (Mw ∼ 425.51 × 103 g mol−1) was kindly supplied by PHB Industrial S.A., (Brazil). Poly(propylene glycol)bis(2-amine propyl ether) (polyamine) Mw ∼ 2000 was obtained from Sigma Aldrich Chemical Co. Limited (USA) and used without further purification.2.2. Preparation of PHB/polyamine blend
0.97 g of PHB was dissolved in 10 mL chloroform and stirred for 2 h at 60 °C. Then, 0.03 g of polyamine was added, and the mixture was stirred for 12 h at 25 °C. The resulting blend was poured in a glass plate and covered at 30 °C for 24 h.2.3. Preparation of PHB/polyamine–NiO nanocomposites
NiO nanoparticles have been synthesized by the sol–gel method.38 The PHB/polyamine–NiO nanocomposites were obtained by the following steps. First, 0.96 g of PHB was dissolved in 10 mL chloroform and stirred for 2 h at 60 °C, then polyamine (0.03 g) was added and the mixture was stirred for 12 h at 25 °C. Then, 0.01 g NiO nanoparticles was added to the mixture, sonicated for 2 h and stirring was continued until a homogeneous suspension was obtained. The resulting suspension was poured in a glass plate and covered at 30 °C for 12 h to afford PHB/polyamine–1% NiO. The same experiment was done using 0.94 g of PHB, 0.03 g of NiO nanoparticles and 0.92 g of PHB, 0.05 g of NiO nanoparticles to tolerate PHB/polyamine–3% NiO and PHB/polyamine–5% NiO, respectively.2.4. Preparation of NF solution
A stock solution of NF was prepared by dissolving 0.016 g in 5 mL dimethyl sulfoxide and completed to one liter distilled water. The different concentrations of NF were prepared by successive dilutions with distilled H2O.2.5. Loading of NF into nanocarriers
0.05 g of the carrier (NiO, PHB, PHB/polyamine and PHB/polyamine–(1, 3, 5)% NiO) were stirred with 50 mL of NF (16 ppm) solution at 30 °C for 90 h. After stirring, the mixture was centrifuged, and the absorbance of a supernatant solution was measured by UV at 274 nm. The concentration of the free drug was calculated from the calibration curve of NF. The percentage of loading was calculated using the following eqn (1) | (1) |
The same experiment was done using PHB/polyamine–3% NiO nanocomposites as carriers using different concentrations of NF (8, 11, 16 ppm). All measurements are performed in triplicate and the mean value is calculated.
2.6. Preparation of different buffer media (pH 7.4 and 2)
Phosphate buffer solutions with different pH values (2 and 7.4) were prepared by mixing 50 mL of 0.1 N K2HPO4 with 50 mL of 0.1 mL KH2PO4 followed by the addition of 1 N HCl or 1 N NaOH till the pH was regulated to 2 and 7.4, respectively.2.7. In vitro drug release
The in vitro release study of NF from the carriers was performed in buffer solution (pH 2, 7.4) each in triplicate. Briefly, constant weight of (0.01 g) carrier loaded drug was introduced into 10 mL buffer. Next, the solution was stirred (200 rpm) at 37 °C and aliquots were withdrawn in specific time intervals. The amount of NF released from the carrier to the phosphate buffer solution was measured using a UV-Vis spectrophotometer by measuring the absorbance at 274 nm using a quartz cell. The concentration was calculated from the calibration curve of NF, and the release percentage was calculated according to the following eqn (2): | (2) |
2.8. In vitro drug release kinetics
The data obtained from in vitro release studies were analyzed by fitting to zero-order,45 first-order,46 Higuchi47 and Hixon–Crowell models48 kinetics equations to study the mechanism of NF release from PHB/polyamine–3% NiO nanocomposites. In addition, the release data obtained were treated according to Korsemeyer–Peppas (3)49 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cumulative percentage of NF release versus log
cumulative percentage of NF release versus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) time) and the release exponent value (n) is calculated in different pH values.
time) and the release exponent value (n) is calculated in different pH values.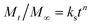 | (3) |
2.9. Characterization of the carriers
Fourier Transform Infra-Red (FTIR) analysis (Shimatzu FTIR-8101 A) was done for NiO nanoparticles, PHB, polyamine, PHB/polyamine and PHB/polyamine–NiO nanocomposites within the range (4000–400 cm−1) using potassium bromide disc.X-ray diffraction analysis (XRD) was achieved for NiO nanoparticles, PHB, PHB/polyamine and PHB/polyamine–NiO nanocomposites to determine their crystallinity. The XRD instrument (Philips PW 1710) was equipped with Cu-Kα radiation (λ = 1.54060 Å), voltage (40 kV) and current (30 mA). The samples were analyzed in a scanning speed of 0.02° min−1.
Thermogravimetric analysis (TGA) was done to evaluate the thermal stability of nanocomposites and their thermal behavior during the heating process. 7 mg of each sample was located separately in the TG instrument (TGA-50) pan to record the weight residue with increasing temperature at a heating rate of 10°C min−1 under a nitrogen atmosphere with a gas flow of 10 mL min−1.
The size distribution of the nanoparticles was determined by dynamic light scattering (DLS; Zetasizer Nano ZS; Malvern Instruments, Malvern, UK). The morphology and particle size of the samples were determined using scanning electron microscopy SU8000 at 2.0 kV and transmission electron microscopy (TEM) with a JEOL (Japan) JEM 2100 high-resolution transmission electron microscope operating at 200 kV. In brief, the nanoparticles were suspended in deionized water and then the sample was sonicated using a sonicator bath until the sample formed a homogeneous suspension. Then, a drop of aqueous nanoparticles suspension was placed onto a carbon-coated copper grid and dried in air to obtain TEM images.
The concentration of the drug (NF) was determined spectrophotometrically by using a UV-visible spectrophotometer (UV-1800 double beam).
2.10. Antibacterial activity
The antibacterial activity of free carriers and NF@carriers was tested against Gram-positive bacteria (Staphylococcus aureus, Streptococcus pyogenes) and Gram-negative bacteria (Klebsiella pneumonia, Escherichia coli, Pseudomonas aeruginosa)50 by well diffusion method. A specified quantity of the nutrient sterile agar at 40–50 °C was poured into a Petri dish to give a depth of 3–4 mm and allowed to solidify. Suspension of the microorganism was prepared to contain approximately 5 × 10−5 CFU per mL. The culture plates were seeded with tested organisms and allowed to solidify thereafter punched with a sterile cork borer (5.0 mm diameter) to cut uniform wells. The open wells were filled with 0.01 g of the tested compounds and incubated at 37 °C for 24 h. The experiment was repeated three times and the zones of inhibition were then measured and the average value was calculated.2.11. Cytotoxicity and antitumor evaluation
Cell proliferation was assessed by MTT assay51,52 in HepG-2 and WISH cell lines. 100 μL of cell suspension at a density of 1 × 104 cells per mL was seeded into 96-well tissue culture test plates and incubated for 48 h at 37 °C under 5% CO2. The samples with different concentrations (100, 50, 25, 12.5, 6.25, and 1.56 μg mL−1) were added and incubated for 24 h. Then, the medium with the samples was removed and MTT solution (20 μL of 5 mg mL−1) was added to the cultures and incubated for 4 h. The medium with MTT solution was discarded and formazan crystals were solubilized using 100 μL DMSO. The absorbance was measured at 570 nm using a plate reader (EXL 800, USA).3 Results and discussion
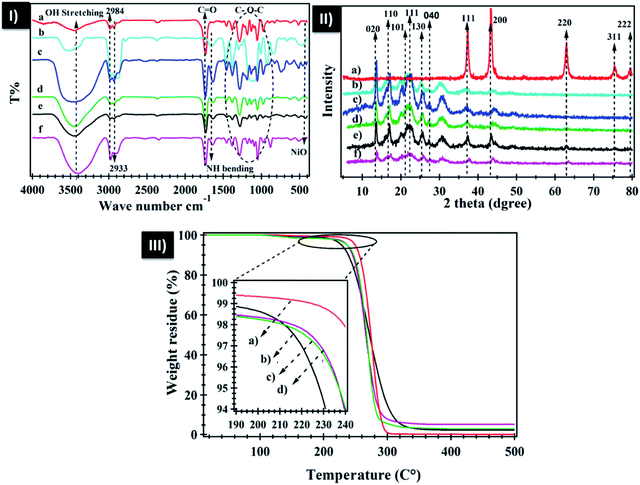
The FTIR spectra of neat PHB, polyamine, PHB/polyamine blend, PHB/polyamine–NiO with different contents of NiO nanoparticles are represented in Fig. 1-I. The FTIR spectrum of neat PHB shows a prominent band at 1731 cm−1 due to the stretching of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester.53 The band at 1458 cm−1 belongs to CH3 asymmetric bending while H–C–O in plan bending appears at 1384 cm−1.54,55 The C–O–C stretching vibration emerges within the range of 1286–1054 cm−1 while the C–H stretching bands lies in 2984, 2933 cm−1. FTIR spectrum of polyamine shows a band at 740 cm−1 that was attributed to primary and secondary amine bending,56 a band at 1653 cm−1 that is assigned to NH bending vibration. This band was shifted to 1636 cm−1 for PHB/polyamine blend and 1639, 1636 and 1635 cm−1 for PHB/polyamine–1% NiO, PHB/polyamine–3% NiO and PHB/polyamine–5% NiO, respectively. The C
O ester.53 The band at 1458 cm−1 belongs to CH3 asymmetric bending while H–C–O in plan bending appears at 1384 cm−1.54,55 The C–O–C stretching vibration emerges within the range of 1286–1054 cm−1 while the C–H stretching bands lies in 2984, 2933 cm−1. FTIR spectrum of polyamine shows a band at 740 cm−1 that was attributed to primary and secondary amine bending,56 a band at 1653 cm−1 that is assigned to NH bending vibration. This band was shifted to 1636 cm−1 for PHB/polyamine blend and 1639, 1636 and 1635 cm−1 for PHB/polyamine–1% NiO, PHB/polyamine–3% NiO and PHB/polyamine–5% NiO, respectively. The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester band in PHB was slightly shifted to 1734 cm−1 in PHB/polyamine blend and 1733 cm−1 in PHB/polyamine–NiO nanocomposites. The band at 740 cm−1 that is attributed to primary and secondary amine bending56 was shifted to 742, 707, 709 and 735 cm−1 for PHB/polyamine blend, PHB/polyamine–1% NiO, PHB/polyamine–3% NiO and PHB/polyamine–5% NiO, respectively confirming the physical interaction. A strong band at 422 cm−1 assigned to Ni–O stretching has appeared in the nanocomposites.
O ester band in PHB was slightly shifted to 1734 cm−1 in PHB/polyamine blend and 1733 cm−1 in PHB/polyamine–NiO nanocomposites. The band at 740 cm−1 that is attributed to primary and secondary amine bending56 was shifted to 742, 707, 709 and 735 cm−1 for PHB/polyamine blend, PHB/polyamine–1% NiO, PHB/polyamine–3% NiO and PHB/polyamine–5% NiO, respectively confirming the physical interaction. A strong band at 422 cm−1 assigned to Ni–O stretching has appeared in the nanocomposites.
The X-ray diffraction pattern of NiO nanoparticles, PHB, PHB/polyamine and PHB/polyamine–NiO nanocomposites using different contents of NiO nanoparticles are displayed in Fig. 1-II. The X-ray diffraction pattern of NiO nanoparticles, reveals sharp peaks at 2θ of 37.53°, 43.60°, 63.09°, 75.67° and 79.58° related to (111), (200), (220), (311) and (222) crystal planes, respectively, according to the Joint committee on powder diffraction standards (JCPDS no 004-0835). The XRD reveals peaks (crystal planes) of neat PHB at 13.1°(020), 16.9°(110), 19.6°, 21.3°(101), 22.5°(111), 25.6°(130), and 27.1°(040) and provides evidence of the crystallinity of the polymer.57 The XRD pattern of PHB/polyamine reveals the existence of crystalline PHB and confirms that the crystallinity of PHB did not affected by the inclusion of polyamine.
XRD patterns of PHB/polyamine–NiO nanocomposites show that the characteristic peaks of PHB are also observed along with the three sharp peaks at 2θ of 37.53°, 43.60° and 63.09° characteristic to NiO nanoparticles. This confirms the incorporation of NiO into the polymer matrixes. The crystallite size was calculated using Scherrer formula d = 0.89λ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ where d = average particle size, β is full width at half maxima (FWHM), θ is Bragg's angle and λ is the wavelength of Cu Kα radiations. The crystallite size of the NiO nanoparticles and PHB was found to be 14 and 7.6 nm, respectively.
θ where d = average particle size, β is full width at half maxima (FWHM), θ is Bragg's angle and λ is the wavelength of Cu Kα radiations. The crystallite size of the NiO nanoparticles and PHB was found to be 14 and 7.6 nm, respectively.
The thermogravimetric analysis (TGA) of PHB, PHB/polyamine–NiO nanocomposite are shown in Fig. 1-III. The TGA thermogram of PHB displays one step of weight loss at 237 °C. It is seen that the weight loss of nanocomposites are located at 210, 219, 221 °C depending on NiO nanoparticles contents followed by stationary thermal stability without degradation at 500 °C. This behavior confirms that the addition of polyamine reduces the thermal stability of the PHB. The weight residue (2.3, 3.1, 5.1%) in PHB/polyamine–1% NiO, PHB/polyamine–3% NiO, and PHB/polyamine–5% NiO are approximately close to the amount added in the feeding process confirming the good dispersion of the nanoparticles in the matrix.
The average particle size along with its polydispersity index (PdI) and the zeta potential (ZP) of the NiO nanoparticles and PHB/polyamine–3% NiO nanocomposite were analyzed by dynamic light scattering (DLS) to confirm the size and stability of nanoparticles in cell culture media. The measurement of the hydrodynamic size of NiO by dynamic light scattering shows particles with a mean diameter of 1701 ± 121 nm. The PDI value of NiO is 0.352 ± 0.04. However, PHB/polyamine–3% NiO nanocomposite shows stable non-aggregated particles with a mean diameter of 403.6 ± 8.5 nm and the PDI value of 0.42 ± 0.07 confirming a good stability in water.
Zeta potential of NiO, PHB/polyamine–3% NiO nanocomposite are −17.6 ± 0.26 and −20.2 ± 0.58 mV, respectively. These values corroborate the stability of synthesized particles in water.
SEM images of PHB/polyamine–NiO nanocomposites using different contents of NiO nanoparticles were recorded in Fig. 2A and B. Obviously, at low magnification, the surface morphology shows the absence of any aggregation of NiO nanoparticles and the particles are dispersed homogeneously through the polymer matrix. This means that the presence of polyamine greatly enhanced the dispersion of nanoparticles. Energy dispersive X-ray (EDX) measurements indicate that the white areas on the SEM micrographs (Fig. 2C) are rich in Ni and accordingly represent the NiO nanoparticles. The atomic percentages of Ni and oxygen are 7.33% and 31.08%, respectively. The high percentage of oxygen in comparison with Ni is attributable to the existence of oxygen in the polymer blend. The carbon signal was due to the existence of the polymer matrix.
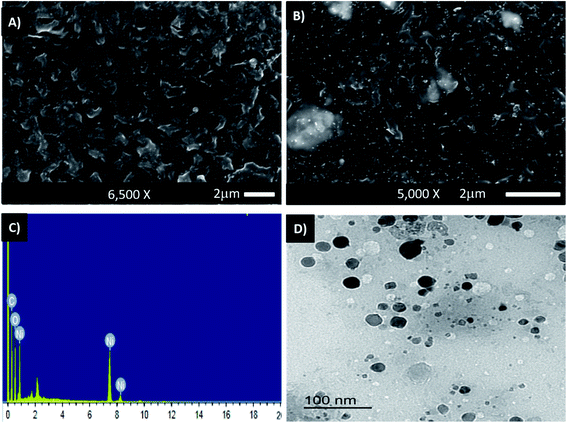 | ||
| Fig. 2 SEM images of PHB/polyamine–1% NiO (A), PHB/polyamine–5% NiO (B) and EDX analysis plot of PHB/polyamine–3% NiO (C); TEM of PHB/polyamine–3% NiO (D). | ||
The TEM of PHB/polyamine–3% NiO nanocomposite is represented in Fig. 2D. The particles are non-agglomerated with a range size of 10–20 nm. The size of the particles can't be correlated with a hydrodynamic diameter measured from DLS experiment.15
The amount of NF uptake onto NiO nanoparticles, PHB, PHB/polyamine and nanocomposites are demonstrated in Fig. 3. The combination of NiO nanoparticles and PHB/polyamine blend slightly enhanced the loading affinity of drugs on the adsorbents. The most efficient drug loading was found on PHB/polyamine–5% NiO (68%) followed by PHB/polyamine–3% NiO (63%) then PHB/polyamine/1% NiO (57%), PHB/polyamine (56.6%), PHB (56%) and NiO nanoparticles (41%). Higher capacity is one of the basic needs for the drug delivery system. The high loading capacity may be due to the presence of interaction between NiO nanoparticles in the nanocomposite and hydroxyl groups present in the NF.
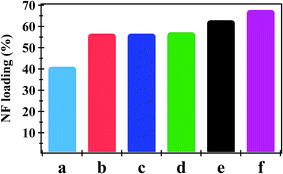 | ||
| Fig. 3 NF Loading (%) onto (a) NiO, (b) PHB, (c) PHB/polyamine, (d) PHB/polyamine–1% NiO, (e) PHB/polyamine–3% NiO and (f) PHB/polyamine–5% NiO using 16 ppm NF. | ||
FTIR spectrum of NF and NF loaded NiO, PHB, PHB/polyamine, PHB/polyamine–3% NiO, PHB/polyamine–5% NiO are shown in Fig. S1.† The spectrum of NF@NiO in Fig. S1† shows that the peaks characteristic to the C–N stretching aromatic amine in NF at 1394 was shifted to 1381 cm−1 in NF@NiO nanoparticles. The peak characteristic of the stretching of Ni–O bonds at 422 cm−1 was shifted to 462 cm−1 in NF@NiO and 439, 459 cm−1 in NF@PHB/polyamine–3% NiO and NF@PHB/polyamine–5% NiO, respectively. The spectrum of NF@PHB shows that the peak characteristic to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group at 1731 was disappeared and the peak characteristic to NH bending at 1625 cm−1 in NF was shifted to 1638, 1637, 1635 cm−1 in NF@PHB, NF@PHB/polyamine, NF@PHB/polyamine–NiO nanocomposites, respectively. The shifting of the bands can be assigned to the interaction between NF and NiO nanoparticles.
O group at 1731 was disappeared and the peak characteristic to NH bending at 1625 cm−1 in NF was shifted to 1638, 1637, 1635 cm−1 in NF@PHB, NF@PHB/polyamine, NF@PHB/polyamine–NiO nanocomposites, respectively. The shifting of the bands can be assigned to the interaction between NF and NiO nanoparticles.
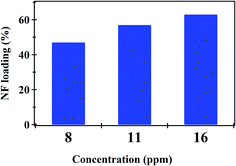
Fig. 4 shows the effect of NF concentration on the loading% on to PHB/polyamine–3% NiO. It was observed that with increasing NF concentration from 8 to 16 ppm, loading efficiency started to increase from 47 to 62%. The most efficient drug loading concentration was obtained at 16 ppm with 62% efficiency. It was reported that the encapsulation efficiency of Ellipticine (EPT) in PHBV nanoparticles depends on molecular weight of copolymer which could entrap more EPT within its structure with higher molecular weight.36 The adsorption kinetics and adsorption isotherms of PHB/polyamine–3% NiO nanocomposite are studied as shown in Fig. 5 and R2 are recorded in Table 1. The regression coefficient (R2) value for the intra-particle diffusion model is higher than those for the pseudo-first-order model and pseudo-second-order model. As seen, the Freundlich and Temkin isotherms show poor agreement with empirical data with low correlation coefficients. However, the high fitted model is the Langmuir isotherm model which shows well agreement with the empirical data with the highest regression coefficient.
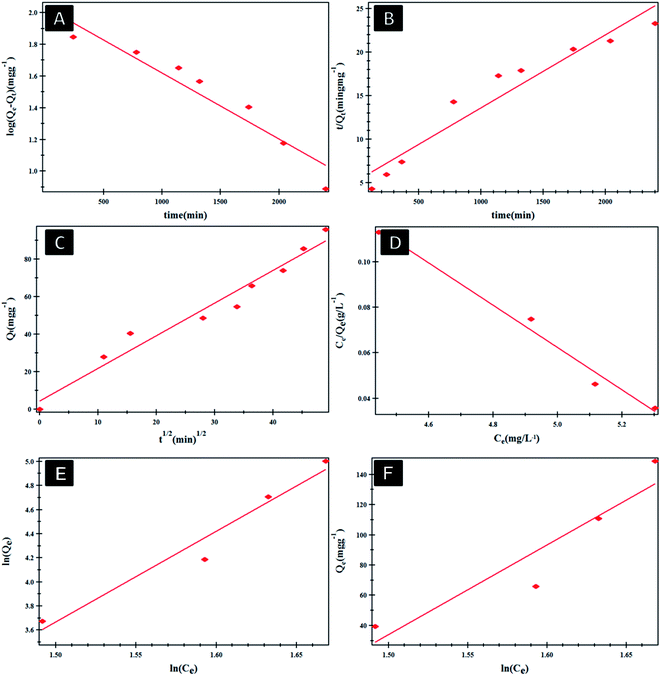 | ||
| Fig. 5 Pseudo-first order (A), pseudo-second order (B) and intra-particle diffusion model (C) Langmuir (D), Freundlich (E) and Temkin (F) isotherms of NF@PHB/polyamine–3% NiO nanocomposites. | ||
| Models | Model coefficient | R 2 |
|---|---|---|
| Pseudo-first order | Q e = 108.543 mg g−1 | 0.946218 |
| K 1 = 9.587 × 10−4 min−1 | ||
| Pseudo-second order | Q e = 119.28 mg g−1 | 0.933423 |
| K 2 = 2.11 × 10−5 g mg−1 min−1 | ||
| Intraparticle diffusion | K 3 = 4.417 mg g−1 min−1/2 | 0.961602 |
| I = 1.738 mg g−1 | ||
| Langmuir model | b = 0.17625 L mg−1 | 0.992499 |
K
![[small script l]](https://www.rsc.org/images/entities/char_e146.gif) = 1.8993 L mg−1 = 1.8993 L mg−1 |
||
| Freundlich model | 1/n = 4.249n = 0.23534 | 0.957927 |
| K f = 12.996 L mg−1 | ||
| Temkin model | B = 856.39 J mol−1 | 0.875362 |
| K t = 4.23168 L mg−1 |
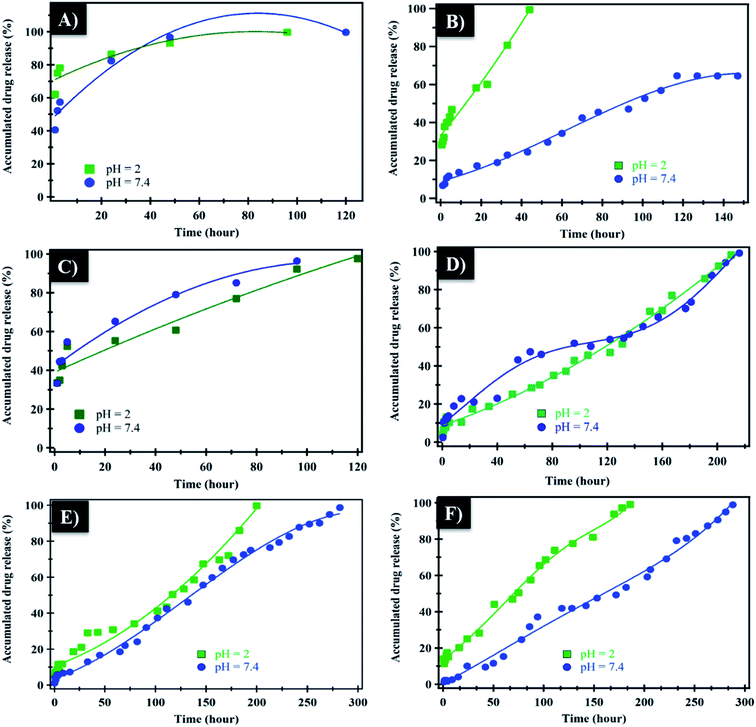
The in vitro controlled release of loaded NF from NiO nanoparticles, PHB, PHB/polyamine and PHB/polyamine–NiO nanocomposites were studied by immersing these films in buffer solution under different pH conditions (2, 7.4). In the case of NiO nanoparticles (Fig. 6A), the rate of NF release at pH 2 was 100% after 96 h. However, at pH 7.4 the release of NF from NiO nanoparticles was continued to reach 100% after 120 h. The release of caffeic acid phenethyl ester (CAPE) from NiO nanoparticles conjugated with guanidine was rapid (80%) during the 10 h.14 In the case of PHB (Fig. 6B) the rate of NF releases at pH 2 was 100% after 40 h. However, the release of NF from PHB was continued to reach 64% after 117 h at pH 7.4. For PHB/polyamine (Fig. 6C), the rate of NF release at pH 2 was 100% after 120 h. However, the release of NF from PHB/polyamine reached 100% after 96 h at pH 7.4. The rate of release was slower, and a linear release profile was observed in PHB/polyamine–1% NiO, PHB/polyamine–3% NiO, PHB/polyamine–5% NiO and reached 100% after 210, 200, 186 h at pH = 2, respectively. The release of NF from PHB/polyamine–(1, 3, 5)% NiO was continued to reach 100% after 216, 282, and 288 h at pH 7.4. The highest ratio of NiO contents to the matrix (highest drug loading), leads to a slower rate of drug release. In acidic medium, protonation for lone pairs of heteroatoms (N) in the drug will reduce the formation of H-bonding between the carrier and drug facilitating the release of the drug from the carrier. It was reported that In vitro release of progesterone (pro) from the PHB membrane was affected by the original progesterone loaded in the membrane, the higher loading, the slower the drug release. This is due to the presence of a high amount of drug dissolution on the membrane and the drug is hindered by the poor solubility of the drug in the medium. Incorporation of nifedipine into polyacrylate showed a decrease in the release rate with increasing drug concentration due to the formation of crystalline drug in microspheres.58 Maximum release rate was obtained in the case of 1/0.125 poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) (PHBHHX)/etoposide initial ratio while the lowest value was obtained with 1/0.5 ratio.59 Increased initial burst was caused by the drug closer to the surface of the nanoparticles while the drug in the core of the nanoparticles is responsible for the prolonged drug release from the nanoparticle.60 A similar trend of higher encapsulation efficiency of tetracycline neutralized (TCN) loaded microspheres with higher 3-hydroxyvalerate (3HV) comonomer unit in the composition was observed by Sendil et al.61 Amphiphilic core–shell nanoparticles PHA–mPEG (PHA-monomethoxy poly(ethylene glycol)) prepared using emulsification–solvent evaporation method showed slow and sustained release kinetics of the nanoparticles which was attributed to the hydrophobic interactions between the hydrophobic drugs and hydrophobic polymer core.62 This result indicated that the PHA–mPEG nanoparticles effectively extended the release of thymoquinone (TQ) and can be used for the controlled delivery of hydrophobic drugs.62 One possible explanation was the slower in vitro degradation of PHAs over the poly(lactic-co-glycolic acid) (PLGA) polymer systems that might be due to the molecular weight differences. The molecular weight of the PHAs used was higher than other aliphatic polyesters, such as PLGA or PLA, which are often used to prepare drug-loaded NPs with faster release rates.63
The mechanism which controls the drug release kinetic process was studied by various kinetics equations, including zero-order, first-order, Higuchi, Hixson–Crowell and Korsmeyer–Peppas models which are formulated below.64 For each model, the slope regression coefficient (R2), release exponent value (n), and rate constant (K) are graphically determined and used to indicate the mechanisms – as well as the kinetics release of the drug (Table 2 and Fig. 7). The NF release from NiO nanoparticles was found to follow first-order kinetics rather than other models at pH = 2 and 7.4. From Korsmeyer–Peppas equation at pH = 2, 7.4 media, the release mechanism follows the Fickian diffusion mechanism (Table 2 and Fig. 7). However, the NF release from PHB/polyamine–3% NiO was found to follow zero-order kinetics at pH = 2and 7.4. From Korsmeyer–Peppas equation at pH = 2, 7.4 media, the release mechanism follows the non Fickian diffusion mechanism (Table 2 and Fig. 8).
| pH | Zero-order | First-order | Higuchi | Hixson–Crowell | Korsemeyer–Peppas | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| K 1 | R 2 | K 2 | R 2 | K 3 | R 2 | K 4 | R 2 | K 5 | R 2 | n | |
| Controlled release kinetics of NF from NiO at different pH media | |||||||||||
| 2 | 0.315 | 0.771 | −0.023 | 0.993 | 3.55 | 0.875 | −0.025 | 0.966 | −0.171 | 0.92 | 0.088 |
| 7.4 | 0.440 | 0.679 | −0.016 | 0.977 | 5.88 | 0.869 | -0.026 | 0.88 | −0.352 | 0.97 | 0.184 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||||
| Controlled release kinetics of NF from PHB/polyamine–3% NiO nanocomposites at different pH media | |||||||||||
| 2 | 0.45 | 0.973 | −0.011 | 0.693 | 6.94 | 0.91 | −0.018 | 0.82 | 1.28 | 0.938 | 0.51 |
| 7.4 | 0.35 | 0.992 | −0.004 | 0.82 | 6.01 | 0.93 | 0.01 | 0.941 | 1.67 | 0.953 | 0.646 |
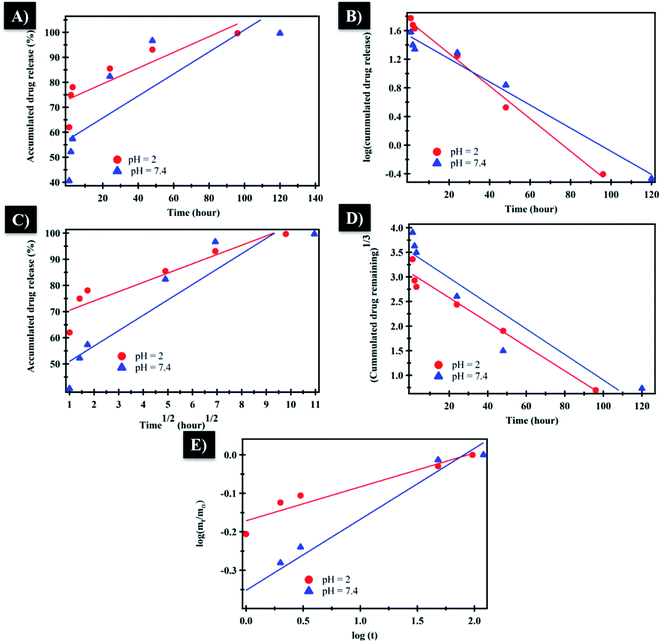 | ||
| Fig. 7 (A) Zero-order, (B) first-order, (C) Higuchi, (D) Hixson–Crowell and (E) Korsmeyer–Peppas kinetic equations of NF from NiO at pH = 2 and 7.4. | ||
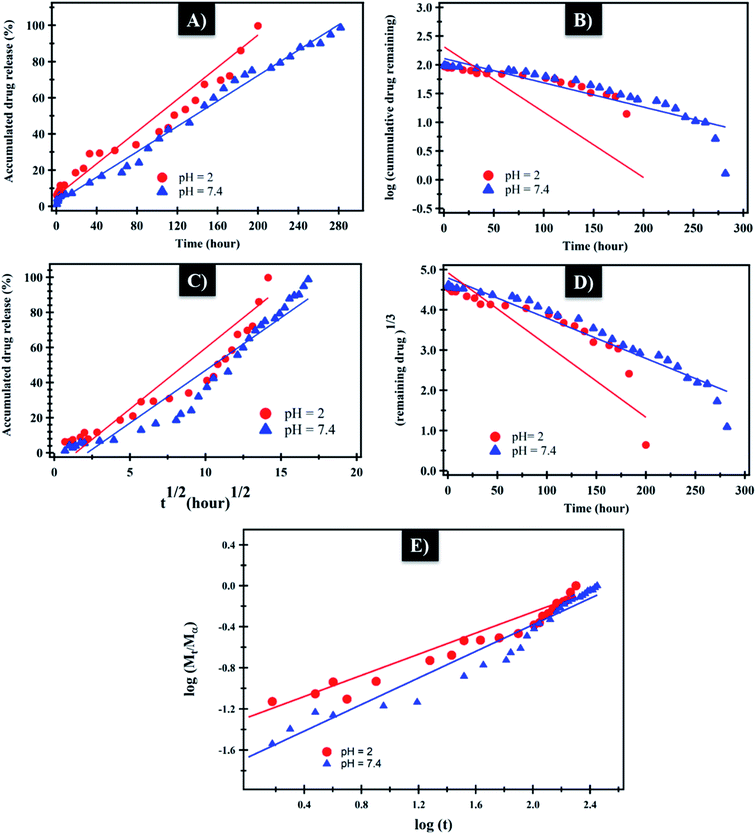 | ||
| Fig. 8 (A) Zero-order, (B) first-order, (C) Higuchi, (D) Hixson–Crowell and (E) Korsmeyer–Peppas kinetic equations of NF from PHB/polyamine–3% NiO at pH = 2 and 7.4. | ||
All samples were tested against five humans' pathogens: Escherichia coli, Staphylococcus aureus, Streptococcus pyogenes, Klebsiella pneumonia, Pseudomonas aeruginosa. Unloaded NiO nanoparticles and PHB/polyamine have no effect against Gram +ve bacteria.65 It was reported that NiO-NPs were not demonstrated bactericidal effect on six different bacterial strains tested, Bacillus subtilis, Staphylococcus aureus, and Enterococcus faecalis (Gram-positive) and Proteus vulgaris, Salmonella typhimurium, and Shigella sonnei (Gram-negative) implying that the NiO-NPs may not perturb the human normal gut microbiome.11 NF loaded on NiO nanoparticles showed significant activity against Klebsiella pneumonia (Gram-negative), Pseudomonas aeruginosa (Gram-negative) and Streptococcus pyogenes (Gram-positive) as shown in Table 3. In addition, NF loaded on PHB, PHB/polyamine and PHB/polyamine–NiO showed significant activity in comparison with the unloaded drug carriers. Notably, an antibacterial activity was observed for loaded PHB/polyamine–(1, 3, 5)% NiO against Escherichia coli, Staphylococcus aureus. Inclusion of NiO onto PHB/polyamine showed higher efficacy against Streptococcus pyogenes and Pseudomonas aeruginosa than the free NF. It worth to mention that only a low percentage of the drug (not exceed 25%) was released during the period of test (24 h).
| Code | S. aureus | E. coli | S. pyogenes | P. aeruginosa | K. pneumonia |
|---|---|---|---|---|---|
| NF | 35 ± 4 | 18 ± 2 | 18.5 ± 0.5 | 21 ± 2.0 | 24 ± 1 |
| NiO | −ve | −ve | −ve | −ve | −ve |
| PHB | −ve | −ve | −ve | 18 ± 1.0 | 11 |
| PHB/polyamine | −ve | −ve | −ve | −ve | −ve |
| PHB/polyamine–1% NiO | −ve | −ve | −ve | 16.5 ± 2.5 | −ve |
| PHB/polyamine–3% NiO | −ve | −ve | 29 ± 2.0 | 21.5 ± 1.5 | −ve |
| PHB/polyamine–5% NiO | −ve | −ve | 31 | 27.5 ± 1.5 | −ve |
| NF@NiO | −ve | −ve | 15 | 20 | 14 |
| NF@PHB | 23 ± 3 | −ve | 25 | 20 | 22.5 ± 2.5 |
| NF@PHB/polyamine | −ve | 24 | 22.5 ± 2.5 | 14.5 ± 2.5 | −ve |
| NF@PHB/polyamine–1% NiO | 22 ± 5 | 14.5 ± 2.5 | 23 ± 3.0 | 12 ± 2.0 | −ve |
| NF@PHB/polyamine–3% NiO | 20 ± 4 | 19 ± 2 | 29 ± 1.02 | 16.5 ± 0.5 | 19 ± 2 |
| NF@PHB/polyamine–5% NiO | 21 ± 1 | 18 ± 3 | 28 ± 2.02 | 18.5 ± 3.5 | 14 ± 1 |
The cytotoxic effect of NF loaded PHB, PHB/polyamine, PHB/polyamine–(1, 3, 5%) NiO nanocomposites against HepG-2 tumor cell was evaluated. The cell viability (%) was determined at different concentrations (Fig. 9A). The compound concentrations which produce a reduction in cellular viability (IC50) was calculated. IC50 values were calculated to be 67.22, 56.3, 43.1, 29.67 and 32.7 μg mL−1 for NF@PHB, NF@PHB/polyamine, and NF@PHB/polyamine–NiO% nanocomposites using 1, 3, 5% NiO, respectively. The most efficient IC50 was found for NF@PHB/polyamine–3% NiO (29.67 μg mL−1). It was reported that treatment of HT-29 cells with NiO-NPs showed the downregulation of anti-apoptotic Bcl2 and Bcl-xL proteins. Induction the marker of apoptosis of PARP, crucial protein of apoptosis that involve controlling programmed cell death and DNA repair, was observed when the HT-29 cells were treated with NiO-NPs.11 Lipid bases prodrugs of ciprofloxacin and norfloxacin provided promising on anthelmintic activity against Indian earthworm (Pheretima posthuma), due to improved lipophilicity of these compounds which exhibited good activity against human lung cancer cell line A-549 when compounding with the parent drug. This is due to lipophilicity and the better penetration of prodrugs to cellular membrane compounds (N-Mannich) with parent drug.66 In another report, the cytotoxicity of drug-loaded NPs particularly made with PHA polymers was delayed, compared with the free drug. Death of HeLa cells treated with free doxorubicin were observed at 24 h. The comparable cytotoxicity of doxorubicin encapsulated in the PHBV with HV content 6.5 wt% (P(HB-6.5HV)) particles was found at 72 h.67 Likewise, a two-day delay was observed for the cytotoxic effect of paclitaxel-containing 12 wt% (P(HB-12HV)) NPs (1 M paclitaxel) in Ishikawa cells for drug release.68 The delayed PDT effect of PHA NP system might offer an advantage for using the NPs in vivo, since they might prevent too early release and subsequent degradation/clearance of the anti-cancer drug before reaching the tumor sites. This prevents drug interaction with normal tissues and reducing systemic side effects.63
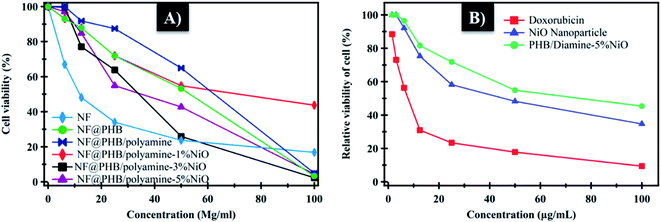 | ||
| Fig. 9 (A) Relative viability of cells at different concentration; (B) cytotoxicity of NiO and PHB/polyamine–3% NiO on normal cell (WISH) expressed as 50% inhibitory concentration (μg m−1). | ||
NiO nanoparticles and PHB/polyamine–3% NiO nanocomposite were also evaluated against the normal human cell line (WISH) (Fig. 9B). No effect on cell proliferation and IC50 was detected to be 44.95 and 70 μg mL−1 for NiO nanoparticles and PHB/polyamine–3% NiO nanocomposite, respectively indicating a selectivity of action towards tumor cells coupled with a lack of cytotoxicity towards normal cells. However, the effect of DOX was significantly higher than NiO nanoparticles and nanocomposite against the normal cell.
4 Conclusion
The present work reports the loading of NF onto NiO nanoparticles, PHB, PHB/polyamine blend and PHB/polyamine–NiO nanocomposites using different contents of NiO nanoparticles. The in vitro release of loaded NF was described in different pH media and the highest ratio of NiO nanoparticles contents to the matrix, leads to a slower rate of release of the drug. The release kinetics was studied and NF release of PHB/polyamine–NiO was found to follow zero-order kinetics in pH = 2 and 7.4. Moreover, the antimicrobial and anticancer efficiency were tested for the loaded nanocomposites and compared with free NF. Inclusion of NiO onto PHB/polyamine showed higher efficacy against Streptococcus pyogenes and Pseudomonas aeruginosa than the free NF. PHB/polyamine–3% NiO showed high efficacy against the HeG-2 cell line (IC50 = 29.67) with poor cytotoxic activity against normal cells (IC50 = 70) confirming the selectivity of the drug carrier and targeting the tumor cells.Conflicts of interest
There are no conflicts to declare.References
- M. Zakariah, S. Khan, A. A. Chaudhary, C. Rolfo, M. M. Ben Ismail and Y. A. Alotaibi, Molecules, 2018, 23, 994–1006 CrossRef PubMed.
- S. Khan, M. Zakariah, C. Rolfo, L. Robrecht and S. Palaniappan, Oncotarget, 2017, 8, 30830–30843 CrossRef PubMed.
- F. Caputo, C. Santini, C. Bardasi, K. Cerma, A. Casadei-Gardini, A. Spallanzani, K. Andrikou, S. Cascinu and F. Gelsomino, Int. J. Mol. Sci., 2019, 20, 5369 CrossRef CAS PubMed.
- K. Lokesh, G. Kavitha, E. Manikandan, G. K. Mani, K. Kaviyarasu, J. B. B. Rayappan, R. Ladchumananandasivam, J. S. Aanand, M. Jayachandran and M. Maaza, IEEE Sens. J., 2016, 16, 2477–2483 CAS.
- E. Manikandan, J. Kennedy, G. Kavitha, K. Kaviyarasu, M. Maaza, B. Panigrahi and U. K. Mudali, J. Alloys Compd., 2015, 647, 141–145 CrossRef CAS.
- K. Kaviyarasu and P. A. Devarajan, Der Pharma Chem., 2001, 43(3), 248–254 Search PubMed.
- K. Kaviyarasu, A. Raja and P. A. Devarajan, Spectrochim. Acta, Part A, 2013, 114, 586–591 CrossRef CAS PubMed.
- F. Fang, J. Kennedy, D. Carder, J. Futter, P. Murmu and A. Markwitz, J. Nanosci. Nanotechnol., 2010, 10, 8239–8243 CrossRef CAS.
- A. K. Gupta and M. Gupta, Biomaterials, 2005, 26, 1565–1573 CrossRef CAS.
- S. Khan, A. A. Ansari, A. Malik, A. A. Chaudhary, J. B. Syed and A. A. Khan, J. Trace Elem. Med. Biol., 2019, 52, 12–17 CrossRef CAS.
- S. Khan, A. A. Ansari, A. A. Khan, M. Abdulla, O. Al-Obaid and R. Ahmad, Colloids Surf., B, 2017, 153, 320–326 CrossRef CAS.
- Q. Saquib, A. A. Al-Khedhairy, M. A. Siddiqui, F. M. Abou-Tarboush, A. Azam and J. Musarrat, Toxicol. In Vitro, 2012, 26, 351–361 CrossRef CAS.
- S. Khan, A. A. Ansari, A. A. Khan, R. Ahmad, O. Al-Obaid and W. Al-Kattan, JBIC, J. Biol. Inorg. Chem., 2015, 20, 1319–1326 CrossRef CAS PubMed.
- V. K. Gupta, A. Fakhri, S. Agarwal, E. Ahmadi and P. A. Nejad, J. Photochem. Photobiol., B, 2017, 174, 235–242 CrossRef CAS.
- J. Adhikary, P. Chakraborty, B. Das, A. Datta, S. K. Dash, S. Roy, J.-W. Chen and T. Chattopadhyay, RSC Adv., 2015, 5, 35917–35928 RSC.
- A. K. Gupta and M. Gupta, Biomaterials, 2005, 26, 3995–4021 CrossRef CAS PubMed.
- S. H. Wang, X. Shi, M. Van Antwerp, Z. Cao, S. D. Swanson, X. Bi and J. R. Baker, Adv. Funct. Mater., 2007, 17, 3043–3050 CrossRef CAS.
- M. A. Abdelwahab, A. A. El-Barbary, K. S. El-Said, S. A. El Naggar and H. M. ElKholy, Int. J. Biol. Macromol., 2019, 122, 793–805 CrossRef CAS.
- M. A. Abdelwahab, A. A. El-Barbary, K. S. El-Said, M. Betiha, H. M. Elkholy, E. Chiellini and M. A. El-Magd, J. Appl. Polym. Sci., 2019, 136, 46924 CrossRef.
- P. Sasikumar and P. Ayyasamy, Int. J. Curr. Microbiol. Appl. Sci., 2015, 4, 311–317 CAS.
- A. C. Kassab, K. Xu, E. Denkbas, Y. Dou, S. Zhao and E. Piskin, J. Biomater. Sci., Polym. Ed., 1997, 8, 947–961 CrossRef CAS.
- H. Vardhan, P. Mittal, S. K. R. Adena and B. Mishra, Eur. J. Pharm. Sci., 2017, 99, 85–94 CrossRef CAS.
- K. Soda, Med. Hypotheses, 2010, 75, 299–301 CrossRef CAS.
- N. C. de la Peña, J. A. Sosa-Melgarejo, R. R. Ramos and J. D. Méndez, Arch. Med. Res., 2000, 31, 546–550 CrossRef.
- Y. Ramot, S. Tiede, T. Bíró, M. H. A. Bakar, K. Sugawara, M. P. Philpott, W. Harrison, M. Pietilä and R. Paus, PLoS One, 2011, 6, e22564 CrossRef CAS PubMed.
- C. LoÈser, Br. J. Nutr., 2000, 84, 55–58 CrossRef.
- Y.-L. Chen, K.-C. Fang, J.-Y. Sheu, S.-L. Hsu and C.-C. Tzeng, J. Med. Chem., 2001, 44, 2374–2377 CrossRef CAS.
- A.-M. Alaa, Y. A. Asiri and M. H. Al-Agamy, Eur. J. Med. Chem., 2011, 46, 5487–5497 CrossRef.
- R. Mather, L. M. Karenchak, E. G. Romanowski and R. P. Kowalski, Am. J. Ophthalmol., 2002, 133, 463–466 CrossRef CAS.
- H. Halkin, Rev. Infect. Dis., 1988, 10, S258–S261 CrossRef CAS PubMed.
- S. Norrby, Eur. J. Clin. Microbiol. Infect. Dis., 1991, 10, 378–383 CrossRef CAS.
- M. Sobczak, K. Nurzyńska and W. Kolodziejski, Molecules, 2010, 15, 842–856 CrossRef CAS PubMed.
- M. E. Katsarou, E. K. Efthimiadou, G. Psomas, A. Karaliota and D. Vourloumis, J. Med. Chem., 2008, 51, 470–478 CrossRef CAS PubMed.
- A. A.-M. Abdel-Aziz, A. S. El-Azab, A. M. Alanazi, Y. A. Asiri, I. A. Al-Suwaidan, A. R. Maarouf, R. R. Ayyad and T. Z. Shawer, J. Enzyme Inhib. Med. Chem., 2016, 31, 796–809 CrossRef CAS PubMed.
- V. Dave, R. B. Yadav, K. Kushwaha, S. Yadav, S. Sharma and U. Agrawal, Bioact. Mater., 2017, 2, 269–280 Search PubMed.
- F. Masood, P. Chen, T. Yasin, N. Fatima, F. Hasan and A. Hameed, Mater. Sci. Eng., C, 2013, 33, 1054–1060 CrossRef CAS.
- M. Abdelwahab, N. Salahuddin, M. Gaber and M. Mousa, Int. J. Biol. Macromol., 2018, 114, 717–727 CrossRef CAS PubMed.
- S. Thota and J. Kumar, J. Phys. Chem. Solids, 2007, 68, 1951–1964 CrossRef CAS.
- S. Lagergren, K. Sven. Vetenskapsakad. Handl., 1898, 24, 1–39 Search PubMed.
- Y.-S. Ho and G. McKay, Process Saf. Environ. Prot., 1998, 76, 183–191 CrossRef CAS.
- W. J. Weber and J. C. Morris, J. Sanit. Eng. Div., Am. Soc. Civ. Eng., 1963, 89, 31–60 CrossRef.
- G. Mckay, H. Blair and J. Gardner, J. Appl. Polym. Sci., 1982, 27, 3043–3057 CrossRef CAS.
- H. Freundlich, Z. Physiol. Chem., 1907, 57, 385–470 CrossRef CAS.
- M. Temkin and V. Pyzhev, Acta Physicochim. URSS, 1940, 12, 217–222 Search PubMed.
- C. G. Varelas, D. G. Dixon and C. A. Steiner, J. Controlled Release, 1995, 34, 185–192 CrossRef CAS.
- S. Kitazawa, I. Johno, y. Ito, S. Teramura and J. Okada, J. Pharm. Pharmacol., 1975, 27, 765–770 CrossRef CAS.
- S. J. Desai, P. Singh, A. P. Simonelli and W. I. Higuchi, J. Pharm. Sci., 1966, 55, 1224–1229 CrossRef CAS.
- A. Hixson and J. Crowell, Ind. Eng. Chem., 1931, 23, 923–931 CrossRef CAS.
- R. W. Korsmeyer, R. Gurny, E. Doelker, P. Buri and N. A. Peppas, Int. J. Pharm., 1983, 15, 25–35 CrossRef CAS.
- P. Doley and D. K. Jha, Ann. Plant Sci., 2016, 4, 1243–1247 Search PubMed.
- M. Metwally, M. Gouda, A. N. Harmal and A. Khalil, Eur. J. Med. Chem., 2012, 56, 254–262 CrossRef CAS.
- F. Denizot and R. Lang, J. Immunol. Methods, 1986, 89, 271–277 CrossRef CAS.
- N. Grassie, E. Murray and P. Holmes, Polym. Degrad. Stab., 1984, 6, 127–134 CrossRef CAS.
- A. M. Díez-Pascual and A. L. Díez-Vicente, Int. J. Mol. Sci., 2014, 15, 10950–10973 CrossRef PubMed.
- S. Yalcin, G. Unsoy, P. Mutlu, R. Khodadust and U. Gunduz, Am. J. Phytomed. Clin. Ther., 2014, 21, 453–461 Search PubMed.
- T.-S. Hu, P.-K. Hong, D. Saikia, H.-M. Kao and M.-C. Chen, Ionics, 2014, 20, 1561–1571 CrossRef CAS.
- G. Uzun and D. Aydemir, Bull. Mater. Sci., 2017, 40, 383–393 CrossRef CAS.
- S. Benita, A. Barkai and Y. Pathak, J. Controlled Release, 1990, 12, 213–222 CrossRef CAS.
- X. Y. Lu, Y. Zhang and L. Wang, J. Appl. Polym. Sci., 2010, 116, 2944–2950 CrossRef CAS.
- A. Budhian, S. J. Siegel and K. I. Winey, Int. J. Pharm., 2008, 151–159 CrossRef CAS.
- D. Sendil, I. Gürsel, D. L. Wise and V. Hasırcı, J. Controlled Release, 1999, 59, 207–217 CrossRef CAS PubMed.
- M. Shah, M. I. Naseer, M. H. Choi, M. O. Kim and S. C. Yoon, Int. J. Pharm., 2010, 400, 165–175 CrossRef CAS PubMed.
- S. Pramual, A. Assavanig, M. Bergkvist, C. A. Batt, P. Sunintaboon, K. Lirdprapamongkol, J. Svasti and N. Niamsiri, J. Mater. Sci.: Mater. Med., 2016, 27, 40 CrossRef.
- H. K. Shaikh, R. Kshirsagar and S. Patil, World J. Pharm. Pharm. Sci., 2015, 4, 324–338 CAS.
- M. R. Abdulbaqi, N. K. Maraie and A. H. Dawood, Int. J. Pharm. Pharm. Sci., 2016, 8, 322–333 CAS.
- M. Piplani, H. Rajak and P. C. Sharma, J. Adv. Res., 2017, 8, 463–470 CrossRef CAS.
- A. Murueva, E. Shishatskaya, A. Kuzmina, T. Volova and A. Sinskey, J. Mater. Sci.: Mater. Med., 2013, 24, 1905–1915 CrossRef CAS PubMed.
- L. Shang, S. C. Yim, H. G. Park and H. N. Chang, Biotechnol. Prog., 2004, 20, 140–144 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04784h |
| This journal is © The Royal Society of Chemistry 2020 |