Open Access Article
Open Access ArticleSupermolecule-assisted imaging of low-molecular-weight quaternary-ammonium compounds by MALDI-MS of their non-covalent complexes with cucurbit[7]uril†
Di
Chen
ab,
Jun
Han
*ac,
Juncong
Yang
a,
David
Schibli
a,
Zhenzhong
Zhang
 b and
Christoph H.
Borchers
b and
Christoph H.
Borchers
 *defg
*defg
aUniversity of Victoria – Genome British Columbia Proteomics Centre, Victoria, BC V8Z 7X8, Canada. E-mail: HanJun@proteincentre.com
bSchool of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou 450001, China
cDivision of Medical Sciences, University of Victoria, Victoria, BC V8P 5C2, Canada
dDepartment of Biochemistry and Microbiology, University of Victoria, Victoria, BC V8P 5C2, Canada
eGerald Bronfman Department of Oncology, Jewish General Hospital, McGill University, Montreal, Quebec H3T 1E2, Canada. E-mail: christoph.borchers@mcgill.ca
fSegal Cancer Proteomics Centre, Lady Davis Institute, Jewish General Hospital, McGill University, Montreal, Quebec H3T 1E2, Canada
gCenter for Computational and Data-Intensive Science and Engineering, Skolkovo Institute of Science and Technology, Moscow 121205, Russia
First published on 15th September 2020
Abstract
Cucurbit[7]uril was used to form non-covalent complexes with low-molecular-weight quaternary-ammonium compounds for their indirect analysis by MALDI-MS. By shifting the ion signals to a higher and interference-free mass region, the distributions of neurine, choline, and phosphocholine in rat brain were visualized by MALDI imaging with high selectivity and good sensitivity.
Matrix assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) is a powerful tool for the sensitive and multiplexed analysis of molecules in a tissue sample. This technique provides the localization and abundances of a molecule in situ, making it an attractive molecular histology tool in medical, pharmaceutical, and biological research.1 Despite its many advantages, some practical problems exist in the direct analysis of some low-molecular-weight (LMW) compounds because of matrix-related ion interferences in the low-mass region of MALDI-MS spectra which interfere with the selectivity of the assay.2,3
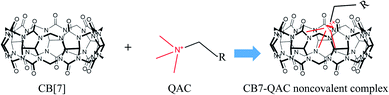
LMW quaternary-ammonium compounds (QACs) include neurine, choline, phosphocholine, as well as a few other molecules. These compounds are essential biomolecules involved in energy production, lipid metabolism, and neurotransmission.3–6 Evaluation of the distributions of these compounds in a tissue is of biological significance. Due to their low molecular weights, however, the direct detection of some of these QACs by MALDI-MS is often hampered by interferences from numerous matrix-related signals. Furthermore, the successful determination by MALDI-MS of the spatial distributions of the QAC compounds in a tissue sample is often inaccurate because the observed ion signals of these compounds can partially result from in-source fragmentation of, for example, phospholipids. Therefore, selective MALDI-MSI of such LMW QACs is challenging. In recent years, on-tissue chemical derivatization has emerged as an effective technique to enhance the in situ detection and imaging performance of some low-abundance or hard-to-ionize endogenous compounds by MALDI-MS.7–10 On-tissue chemical derivatization, however, is not applicable to most LMW QACs because these compounds do not have the functional groups in their molecular structures that are required for chemical derivatization. Therefore, new methods are needed for the molecular imaging of these compounds by MALDI-MS. To ameliorate this situation, we have developed a new approach to enhance the selectivity of MALDI-MSI for in situ imaging of the three LMW QACs using on-tissue inclusion of the analytes within a supermolecule, i.e., cucurbit[7]uril (CB[7]), to form non-covalent complexes (Fig. 1).
Cucurbit[n]urils (CB[n]) consist of n glycouril units that are bound in a ring-like arrangement via methylene bridges, and which can form stable non-covalent host–guest complexes with cations with high binding constants.11 As a soft ionization technique, MALDI has been shown to ionize non-covalent complexes under mild laser energy conditions, without dissociation during desorption or ionization.12–14 CB[6] and CB[7] have been used as mass-shifting reagents to analyze polyamines (e.g., spermidine, and spermine) in plant tissues by forming the non-covalent CB[n]–analyte complexes.15,16 By shifting the detected or expected ion signal of an analyte to a higher mass region, the interference signals originating from common MALDI matrices and other coexisting LMW compounds in a sample can be greatly reduced or eliminated, thus increasing the selectivity of the assay. In addition, non-covalent interactions between CB[n] and the LMW analytes often need no activation energy for the formation of the complex.11 In other words, the formation of non-covalent bonds is relatively easy and rapid. Because of these features, we examined the use of CB[n] as host molecules for the on-tissue formation of non-covalent supramolecular complexes with LMW QACs, for MALDI-MSI.
CB[n] supermolecules that are composed of n = 5, 6, 7, 8, or 10 repeating units of glycouril are commercially available. Due to the extremely small cavity size, CB[5] is only suitable for the encapsulation of gases, forming portal complexes with alkali, alkaline earth, and ammonium cations.11 At the other extreme, due to their large portal size and large cavity volume, CB[8] and CB[10] can interact with two identical or two different small molecules to form 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 or 1
2 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes. This feature would increase the complexity of MALDI MS spectra and would make interpreting these spectra more difficult. In contrast, CB[6] and CB[7] have moderate cavity sizes, with cavity volumes of 164 Å and 279 Å, respectively,11 and thus may have the potential for use in the MALDI-MSI of LMW QACs. CB[6], however, is essentially insoluble in either water or organic solvents, and performed poorly in our preliminary experiments. CB[7], on the other hand, possesses modest solubility in water (∼30 mM),11 and has been shown to be compatible with commonly used organic solvents such as methanol and acetonitrile,17,18 which would be important for MALDI matrix-deposition procedures using wet-chemistry techniques such as spray coating. Therefore, CB[7] was chosen for the MALDI-MSI presented in this study. To evaluate the proposed strategy for the indirect analysis of LMW QACs, 3 compounds (neurine, choline, and phosphocholine) were tested by MALDI-MS, with or without CB[7] as the non-covalent complex-forming supermolecule. For the inclusion of CB[7], dried-droplet spotting was used by, in turn, depositing 0.5 μL of a solution of one of the 3 QACs (50 μM each in 50% MeOH), 0.5 μL of a CB[7] solution (4 mM in 50% MeOH), and 0.5 μL of an α-cyano-4-hydroxycinnamic acid (CHCA) (7 mg mL−1 in 50% MeOH) MALDI matrix solution on a polished stainless-steel plate. For the CB[7]-free procedure, no CB[7] was deposited. When CB[7] was included, all 3 QACs displayed unique signals corresponding to positive ions of the formed CB[7]–QAC complex, without sodiated or potassiated adduct ions or fragment ions of the complexes being observed (Fig. S1A–C†). In Fig. S1D to F,† these 3 compounds were also able to be detected by direct MALDI-MS without the CB[7] inclusion, but the mass spectrum acquired from the spotted choline sample (Fig. S1E†) showed a weak ion signal corresponding to the m/z of neurine when the power of the laser impacting the sample for desorption/ionization was ≥21 μJ (25% of the full power), indicating in-source fragmentation of choline. Similarly, in the mass spectrum of phosphocholine, two ion signals corresponding to neurine and choline, respectively, were observed in direct MALDI-MS of phosphocholine when the same level of laser power was applied. The comparison of the MALDI-MS spectra of these 3 compounds, with and without the CB[7] inclusion, showed the high selectivity of the CB[7]-assisted strategy for the indirect MALDI-MSI of these QACs.
1 complexes. This feature would increase the complexity of MALDI MS spectra and would make interpreting these spectra more difficult. In contrast, CB[6] and CB[7] have moderate cavity sizes, with cavity volumes of 164 Å and 279 Å, respectively,11 and thus may have the potential for use in the MALDI-MSI of LMW QACs. CB[6], however, is essentially insoluble in either water or organic solvents, and performed poorly in our preliminary experiments. CB[7], on the other hand, possesses modest solubility in water (∼30 mM),11 and has been shown to be compatible with commonly used organic solvents such as methanol and acetonitrile,17,18 which would be important for MALDI matrix-deposition procedures using wet-chemistry techniques such as spray coating. Therefore, CB[7] was chosen for the MALDI-MSI presented in this study. To evaluate the proposed strategy for the indirect analysis of LMW QACs, 3 compounds (neurine, choline, and phosphocholine) were tested by MALDI-MS, with or without CB[7] as the non-covalent complex-forming supermolecule. For the inclusion of CB[7], dried-droplet spotting was used by, in turn, depositing 0.5 μL of a solution of one of the 3 QACs (50 μM each in 50% MeOH), 0.5 μL of a CB[7] solution (4 mM in 50% MeOH), and 0.5 μL of an α-cyano-4-hydroxycinnamic acid (CHCA) (7 mg mL−1 in 50% MeOH) MALDI matrix solution on a polished stainless-steel plate. For the CB[7]-free procedure, no CB[7] was deposited. When CB[7] was included, all 3 QACs displayed unique signals corresponding to positive ions of the formed CB[7]–QAC complex, without sodiated or potassiated adduct ions or fragment ions of the complexes being observed (Fig. S1A–C†). In Fig. S1D to F,† these 3 compounds were also able to be detected by direct MALDI-MS without the CB[7] inclusion, but the mass spectrum acquired from the spotted choline sample (Fig. S1E†) showed a weak ion signal corresponding to the m/z of neurine when the power of the laser impacting the sample for desorption/ionization was ≥21 μJ (25% of the full power), indicating in-source fragmentation of choline. Similarly, in the mass spectrum of phosphocholine, two ion signals corresponding to neurine and choline, respectively, were observed in direct MALDI-MS of phosphocholine when the same level of laser power was applied. The comparison of the MALDI-MS spectra of these 3 compounds, with and without the CB[7] inclusion, showed the high selectivity of the CB[7]-assisted strategy for the indirect MALDI-MSI of these QACs.
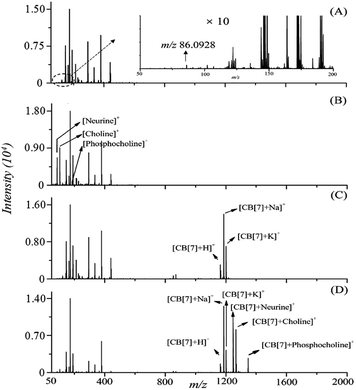
The selectivity of the CB[7]-assisted MALDI detection of the LMW QACs was further evaluated by comparing the interference background signals obtained by direct and indirect analysis of a mixture of the 3 QACs. Fig. 2 shows a comparison of the MALDI mass spectra of the matrix CHCA, 3 QACs, CB[7], and the 3 QACs–CB[7] complexes, respectively, acquired in the reflectron mode of MALDI-TOF operations. In Fig. 2A, a signal at m/z 86.0928, which may be derived from the fragmentation of CHCA, was observed, which interferes with neurine detection. In Fig. 2B, although signals for neurine, choline, and phosphocholine were observed, severe interferences from the matrix and other background ions were also seen in the lower mass region (i.e., below m/z 400), and phosphocholine was only weakly detected, compared with the signals for neurine and choline, demonstrating the inadequate selectivity of direct MALDI-MS for detection of these LMW compounds using the conventional CHCA organic matrix. In Fig. 2C, signals from the CB[7]-related ions ([CB[7] + H]+, [CB[7] + Na]+, and [CB[7] + K]+) can clearly be seen in the mass region from m/z 1160 to m/z 1210 – i.e., in the higher-mass clean-background region of the spectrum (Fig. 2C). For the sample deposited with a mixture of the 3 QACs, CB[7], and CHCA, MALDI signals corresponding to the ions of CB[7]–neurine, CB[7]–choline and CB[7]–phosphocholine were observed as ion clusters around m/z 1148, 1266, and 1346, respectively (see the measured and calculated m/z values in Table S1† for details), without matrix-related interferences (Fig. 2D). This experiment demonstrated the high selectivity of the CB[7]-assisted MALDI-MS method for the detection of these 3 LMW QACs.
The analytical sensitivity of the proposed CB[7]-assisted approach was evaluated by performing on-target CB[7] inclusion on serially diluted standard solutions containing the analytes. As a result, the limits of detection (LOD), defined as the lowest amounts of the analytes which produced MALDI signals of the CB[7]–QAC complexes with a signal-to-noise ratio of ≥3, were determined to be 0.2, 0.2, and 2 pmol for neurine, choline, and phosphocholine, respectively, indicating acceptable sensitivities for the on-spot detection using the proposed method. As a comparison, the LODs of neurine, choline, and phosphocholine by MALDI-MS without CB[7] assisting were determined to be 0.1, 0.3, and 2 pmol, respectively. The MALDI-MS analysis of QACs with and without CB[7] assistance showed similar sensitivities of detection in terms of LODs.
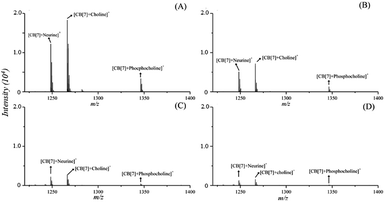
To demonstrate the capability of the proposed strategy for relative quantitation of the QACs by MALDI-MS as well as for the subsequent sensitivity study, 1 μL of standard solutions of the 3 QACs with varying concentrations were spotted on the MALDI target, followed by deposition of 1 μL of 2 mM of CB[7] solution and 2 μL of the same CHCA matrix solution using the dried-droplet approach. A comparison study without the addition of CB[7] was also performed. MALDI-MS spectra of CB[7]–QAC complexes were acquired under a set of optimized instrument operating conditions and compared, as shown in Fig. 3. The observed signal intensities of the individual QACs or CB[7]–QAC complexes showed linear responses as a function of the concentrations of the applied standard solutions, with correlation coefficients (R2) being equal to or greater than 0.977 over a concentration range of 5 pmol to 100 pmol, for each of the 3 compounds (Fig. S2†). The slopes of the calibration curves with or without CB[7] were similar, which further showed that the CB[7]-assisted approach increased the selectivity of detection of the 3 LMW QACs while maintaining the sensitivity.
Next, the performance of this method for on-tissue detection of LMW QACs was evaluated using 14 μm-thick rat brain tissue sections as representative tissue samples. Two rat brain tissue sections were deposited with the matrix of CHCA (14 mg mL−1 in 50% MeOH), with and without inclusion of CB[7] (2 mM in 50% MeOH) prior to the addition of the matrix coating, using a Bruker Daltonics ImagePrep sprayer. When no pre-coating of CB[7] on the tissue section was used, only very few weak signals were detected in the mass region of m/z 1220–1400 (Fig. 4A). In the upper part of Fig. 4B, signals corresponding to the formed complexes of CB[7]-neurine, CB[7]-choline, and CB[7]-phosphocholine can be clearly detected with the signal-to-noise ratios of ≥300 for the 3 endogenous QACs. These results indicated that the CB[7]-assisted approach delivered signals free from interferences and gave accurate results, demonstrating the high selectivity of the proposed strategy for on-tissue detection of these three compounds.
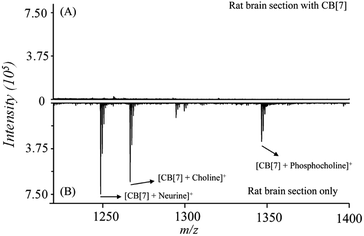 | ||
| Fig. 4 Average mass spectra obtained by conducting on-tissue inclusion. (A) A rat brain tissue section (14 μm) coated with CB[7] and CHCA, (B) rat brain section coated with CHCA only. | ||
To optimize the procedure for the MALDI imaging of these compounds in rat brain, the effects of the amount of CB[7] deposited and the percentage of organic solvent (MeOH) in the CB[7] solutions on the MALDI signal intensities were investigated. The amounts of deposited CB[7] on the tissue were controlled by varying the number of the CB[7] spray cycles. Sufficient CB[7] benefited the formation of CB[7]–QACs. However, excessive amounts of CB[7] induced suppression of the observed ion signals of the CB[7]–QACs. The most intense signals were observed with the application of 15–19 spray cycles of the supermolecule solution at a concentration of 2 mM CB[7] (Fig. S3A†). The use of 25% and 50% methanol in both CB[7] solutions resulted in about 30% higher signal intensities than with 75% methanol in the aqueous solutions (Fig. S3B†). However, the lower percentages of methanol in the solutions induced delocalization of the analytes on the tissue sections. Therefore, 75% aqueous methanol was chosen as the solvent for both the CB[7] solution and the matrix solution, and 15 cycles of spray were used for all the subsequent imaging experiments in order to avoid possible delocalization during the CB[7] inclusion and CHCA coating steps.
To simplify the procedure for tissue imaging, we explored the feasibility of performing the on-tissue CB[7] inclusion and the matrix coating in a single step, and compared the single-step procedure with the two-step procedure. In the two-step procedure, the CB[7] solution (2 mM in 75% MeOH) and the CHCA solution (14 mg mL−1 in 75% MeOH) were coated, in turn, with 15 spray cycles for each round of the coating. For the single-step operation, a mixed solution of 2 mM of CB[7] and 14 mg mL−1 of CHCA in 75% MeOH was sprayed onto the tissue sections with 15 spray cycles. The mass spectra obtained using the two different deposition procedures are shown in Fig. S4,† and both showed similar mass spectral quality for the detection of the 3 QACs, without any apparent differences in the observed signal intensities. Thus, the single-step procedure was utilized to simplify the experiment.
Under the optimized on-tissue inclusion conditions, the MALDI-MSI of neurine, choline, and phosphocholine in rat brain tissue was performed to characterize the localization and distribution patterns of the three QACs (neurine, choline, and phosphocholine) on transversely sliced tissue sections of a rat brain with a laser irradiation diameter of 200 μm. As shown in Fig. 5, the 3 QACs showed slightly different distributions in the different anatomical regions of the rat brain. Neurine and phosphocholine showed similar distribution patterns to each other, and both of these compounds were detected with higher abundances in the regions of cortex and thalamus, while choline showed the reverse distribution pattern. These observations were consistent with the observations in an earlier ammonium sulfate-assisted MALDI-MSI study.3 It should be noted that the current study could be defined as a feasibility study for the proposed CB[7]-assisted strategy. Better imaging results are expected to be obtained by using the CB[7]-assisted strategy on newer-generation instruments (including both matrix coating devices and MALDI-MS instruments).
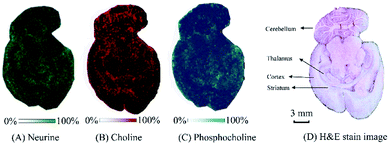 | ||
| Fig. 5 MALDI-MS ion images of three QACs obtained by using the CB[7]-assisted approach on a rat brain tissue section and the corresponding H & E staining. | ||
For the MALDI-MSI of LMW QACs, the use of ammonium sulfate-assisted MALDI-MSI (i.e., spraying ammonium sulfate together with matrix onto the tissue sections), as reported by Mitsutoshi, et al., improved the detection performance and the distributions of 5 hydrophilic QACs (carnitine, acetylcarnitine, glycerophosphocholine, choline, and phosphocholine) were visualized in rat brain tissues.3 There are also some reports on multiplexed and chemical derivatization-free MALDI-MS imaging of endogenous compounds, including a few QACs in tissue.19,20 Compared to these earlier studies on the MALDI-MSI of LMW QACs, a significant advantage of our proposed strategy is its high selectivity, resulting from shifting the LMW-QACs to an interference-free higher mass region, with no interferences derived from in-source fragmentation. To our knowledge, neurine has not been previously imaged in tissue specimens by MALDI-MS. In this work, neurine in rat brain was able to be successfully imaged, as a result of the formation of a non-covalent complex with CB[7] which improved its MALDI-MS detection.
Conclusions
In summary, a new method for MALDI-MSI of 3 LMW QACs in rat brain with high selectivity and good sensitivity has been proposed by shifting of the mass of the ion signals for these compounds to an interference-free higher mass region. By doing this, the signals of CB[7]–QAC complexes were not affected by the interfering signals derived from the MALDI matrix or the co-existing LMW compounds in the tissue. This new strategy is expected to be useful in investigations of the spatial and temporal concentration changes of these QACs in biological tissues under different disease and physiological conditions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by funding for method development from The Metabolomics Innovation Centre (TMIC), from Genome Canada, Genome Alberta, and Genome British Columbia through the Genomics Technology Platform (GTP) for operations and technology development (265MET and MC3). CHB is also grateful for support from the Leading Edge Endowment Fund (University of Victoria), and for support from the Segal McGill Chair in Molecular Oncology at McGill University, the Warren Y. Soper Charitable Trust, the Alvin Segal Family Foundation, and the Terry Fox Research Institute to the Segal Cancer Proteomics Centre at the Jewish General Hospital. The study was also supported by the MegaGrant of the Ministry of Science and Higher Education of the Russian Federation (Agreement with Skolkovo Institute of Science and Technology on December 11, 2019 No. 075-10-2019-083). We thank Dr Carol E. Parker for careful review of this manuscript.References
- M. Shariatgorji, A. Nilsson, R. J. A. Goodwin, P. Källback, N. Schintu, X. Zhang, A. R. Crossman, E. Bezard, P. Svenningsson and P. E. Andren, Neuron, 2014, 84, 697 CrossRef CAS.
- S. Guo, Y. Wang, D. Zhou and Z. Li, Anal. Chem., 2015, 87, 5860 CrossRef CAS PubMed.
- E. Sugiyama, N. Masaki, S. Matsushita and M. Setou, Anal. Chem., 2015, 87, 11176 CrossRef CAS PubMed.
- S. H. Zeisel, Ann. Nutr. Metab., 2012, 61, 254 CrossRef CAS PubMed.
- D. Tweedie, A. Brossi, D. Chen, Y. W. Ge, J. Bailey, Q. S. Yu, M. A. Kamal, K. Sambamurti, D. K. Lahiri and N. H. Greig, J. Alzheimer's Dis., 2006, 10, 9 CAS.
- A. M. A. P. Fernandes, P. H. Vendramini, R. Galaverna, N. V. Schwab, L. C. Alberici, R. Augusti, R. F. Castilho and M. N. Eberlin, J. Am. Soc. Mass Spectrom., 2016, 27, 1944 CrossRef CAS PubMed.
- Q. Wu, T. J. Comi, B. Li, S. S. Rubakhin and J. V. Sweedler, Anal. Chem., 2016, 88, 5988 CrossRef CAS.
- S. S. Wang, Y. J. Wang, J. Zhang, T. Q. Sun and Y. L. Guo, Anal. Chem., 2019, 91, 4070 CrossRef CAS.
- E. Takeo, Y. Sugiura, T. Uemura, K. Nishimoto, M. Yasuda, E. Sugiyama, S. Ohtsuki, T. Higashi, T. Nishikawa, M. Suematsu, E. Fukusaki and S. Shimma, Anal. Chem., 2019, 91, 8918 CrossRef CAS PubMed.
- Q. Cao, Y. Wang, B. Chen, F. Ma, L. Hao, G. Li, C. Ouyang and L. Li, ACS Chem. Neurosci., 2019, 10, 1222 CrossRef CAS.
- S. J. Barrow, S. Kasera, M. J. Rowland, J. del Barrio and O. A. Scherman, Chem. Rev., 2015, 115, 12320 CrossRef CAS.
- B. Baytekin, H. T. Baytekin and C. A. Schalley, Org. Biomol. Chem., 2006, 4, 2825 RSC.
- G. Bolbach, Curr. Pharm. Des., 2005, 11, 2535 CrossRef CAS.
- C. Borchers and K. B. Tomer, Biochemistry, 1999, 38, 11734 CrossRef CAS.
- J. Ding, S. Liu, H. M. Xiao, T. T. Ye, P. Zhou and Y. Q. Feng, Anal. Chim. Acta, 2017, 987, 56 CrossRef CAS.
- J. Ding, H.-M. Xiao, S. Liu, C. Wang, X. Liu and Y.-Q. Feng, Anal. Chim. Acta, 2018, 1026, 77 CrossRef CAS.
- X. Wang, J. Han, A. Chou, J. Yang, J. Pan and C. H. Borchers, Anal. Chem., 2013, 85, 7566 CrossRef CAS PubMed.
- X. Wang, J. Han, J. Pan and C. H. Borchers, Anal. Chem., 2014, 86, 638 CrossRef CAS PubMed.
- D. A. Pirman, R. F. Reich, A. Kiss, R. M. A. Heeren and R. A. Yost, Anal. Chem., 2013, 85, 1081 CrossRef CAS.
- D. A. Pirman and R. A. Yost, Anal. Chem., 2011, 83, 8575 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04604c |
| This journal is © The Royal Society of Chemistry 2020 |