Open Access Article
Open Access ArticleCell-free biology using remote-controlled digital microfluidics for individual droplet control†
Dong Liu,
Zhenghuan Yang,
Luyang Zhang,
Minglun Wei and
Yuan Lu *
*
Department of Chemical Engineering, Key Laboratory of Industrial Biocatalysis, Ministry of Education, Tsinghua University, Beijing 100084, China. E-mail: yuanlu@tsinghua.edu.cn
First published on 20th July 2020
Abstract
Cell-free biology for diverse protein expression and biodetection in vitro has developed rapidly in recent years because of its more open and controllable reaction environment. However, complex liquid handling schemes are troublesome, especially when scaling up to perform multiple different reactions simultaneously. Digital microfluidic (DMF) technology can operate a single droplet by controlling its movement, mixing, separation, and some other actions, and is a suitable scaffold for cell-free reactions with higher efficiency. In this paper, a commercial DMF board, OpenDrop, was used, and DMF technology via remote real-time control inspired by the Internet of Things (IoT) was developed for detecting glucose enzyme catalytic cell-free reactions and verifying the feasibility of programmed cell-free protein expression. A cell-free biological reaction process which can be remote-controlled visually with excellent interactivity, controllability and flexibility was achieved. As proof-of-concept research, this work proposed a new control interface for single-drop cell-free biological reactions. It is much like the “droplet operation desktop” concept, used for remote-controllable operations and distributions of cell-free biology for efficient biological screening and protein synthesis in complex reaction networks, with expanded operability and less artificial interference.
1. Introduction
Cell-free biology (CFB) has developed rapidly in recent years.1 CFB means synthesizing proteins or conducting specific enzyme-catalyzed biochemical reactions in vitro, which imitate metabolic processes in vivo.2 Cell-free biochemical reactions utilize purified enzymes for reaction synthesis or the detection of useful substances, such as carbohydrates and esters. Nucleic acids such as DNA or RNA are even used as templates to perform cell-free protein synthesis. Compared with a cellular system, a cell-free system eliminates the cell’s tightness, making the system more open, and allows more control over the component concentrations and reaction conditions.3–5 Meanwhile, CFB separates biological growth and protein expression, and is more suitable for high-throughput screening as well as engineering applications for industrial production.6 Given the above advantages, cell-free biology is widely used in the fields of biological detection, high-throughput screening of drugs and mutations, and the design and synthesis of unnatural proteins.7,8 Several groups have developed different cell-free reaction protocols to meet diverse needs and applications. Using a freeze-dried cell-free platform, rapid and portable nucleic acid detection via in situ protein synthesis reactions is expected to be applied in field, military, epidemic, and other environments.9,10 Swartz’s and Jewett’s groups have done excellent work on cell-free non-standard amino-acid incorporation into different sites using orthogonal expression systems with high fidelity.11,12 Meanwhile, as a method for protein engineering, cell-free protein synthesis technology has been scaled up to 100 liters so far, and it is likely to undergo large-scale industrialization in the near future.13However, with the widespread popularity and application of CFB as a research platform, the inconvenience of a complex liquid handling process becomes obvious, especially when scaling up to perform multiple reactions simultaneously. CFB is characterized by a wide variety of reagents, but each in a small amount. Whether a CFB kit is laboratory-made or commercial, there are usually more than five and even up to ten different reagents, each of which needs to be stored separately and then manually mixed in the experiments.14 Furthermore, multiple reactions are usually performed simultaneously to compare or contrast various conditions for optimization or screening. When the number of simultaneous reactions is scaled up, the operation simply to remove the droplets occupies 50% of the experimental time, resulting in a massive amount of wasted labor and disposable consumables, especially in the purified reconstituted system (PURE).15 F. Villarreal et al. presented an approach to produce pure translation machinery by a single culturing and purification step through exploiting microbial consortia in the PURE system. The huge innovation of the synthetic microbial consortia method helps to greatly decrease the independent reactants.16 However, it is necessary to mix the pure translation machinery with other reactants. Meanwhile, whether by manual or machine operation, the removal of a very small amount (a few microliters of liquid) by pipette may cause large errors, resulting in the poor repeatability of cell-free reactions.17
Digital microfluidic (DMF) technology is an interesting emerging technology for the direct manipulation of individual droplets. In recent years, basic physicists have often conducted research on the physical properties of droplets and fluids using DMF platforms. DMF is commonly referred to as electrowetting technology, which uses a set of planar electrodes to operate separately on individual droplets. Wheeler’s group first proposed the electrowetting method of digital microfluidics to control the shape and position of droplets on surfaces.18 In addition, a variety of principles for DMF have been developed, such as thermal gradient and acoustic or magnetic response. Magnetic-actuated programmable droplet manipulation has been achieved for basic chemical reactions.19 Compared with traditional microfluidics, where liquids are controlled in closed channels, DMF retains the advantages of microfluidics, such as low amounts of reagents and consumable consumption, enhanced heat transfer, and easy integration. Meanwhile, DMF has its own excellent advantages:20–22 (1) DMF independently controls individual droplets, eliminating the need for microdevices, such as micropipes, micropumps, microvalves, and micromixers, which can avoid the complicated design and processing of microfluidic plates; (2) a multi-threaded operation can be performed at the same time by performing the independent operation of different unit modules; (3) controlled by the electrode array, it can be reconstructed according to different experiments with the universality of different situations; (4) its open environment and relatively large volume (up to 10 microliters) makes it easy to sample later; (5) it is even possible to use solid samples without clogging.
Because of the unique and useful features, DMF technology is a promising candidate for use in biochemistry.23–27 DMF is used for chemical enzyme catalysis, immunoassays, and medical testing. These studies focus on simple reaction assays. T. B. Yehezkel et al. first reported de novo synthesis and cell-free cloning of custom DNA libraries via droplets using a DMF method. However, the manipulation of proteins, which are expressed and eventually function inside living cells, has not been explored.28 There have also been studies on proteomics, and step-by-step operations for protein separation and purification, and cell culture and adhesion on DMF as a culture pad in terms of cell applications. However, due to the limitations of a series of difficulties in cell mass expression, separation, and purification, cells have not been used to synthesize actual polymer proteins using the DMF method. Cell-free biology provides an open environment that combines DMF technology with in vitro biochemical detection and protein synthesis. It provides an excellent platform for a large amount of efficient biological screening and synthesis.29,30
Gaudenz’s group has released the OpenDrop product as an integrated DIY platform for DMF biochips; its users include academia, companies, hackerspaces, and even individuals.31 The velocity of a droplet is measured using an OpenDrop device to optimize the thickness and the types of dielectric layers to improve the droplet actuation.32 2D DMF arrays are limited to small sizes, which hinders high throughput screening applications; Ishii’s group addressed these issues by scaling electrowetting for large area droplet manipulation, enabling multiple parallel operations.33 However, the control methods are relatively basic, mainly based on preset procedures. There is no way to deal with problems that occur at any time during the operation. Furthermore, considering that many biological reactions require harsh operating environments to minimise disturbance, such as environments that require strict sterility, it will be very convenient for the experimenter if remote biological operations are enabled. Therefore, a DMF control interface in combination with remote-controlled IoT technology was further developed for use as a more smart and conveniently-operated CFB platform, which has not been done before.
In this paper, real-time remote-controlled DMF technology is developed to assist with the research of cell-free biological reactions to improve its interactivity, controllability and flexibility with higher efficiency. The feasibility of DMF with pigments was firstly verified, and the double enzyme-catalyzed detection of glucose biochemical reactions was then carried out, which served as a conceptual exploration of DMF technology for cell-free biological sample detection. A cell-free protein synthesis reaction on the DMF pad with the expression of variable-fluorescence proteins and catechol 2,3-dioxygenase (CatO2ase) was finally conducted (Fig. 1). It demonstrates the feasibility of the new mouse-like droplet operation interface with the combination of a DMF method and a cell-free biological system, followed by the key performance and mathematical analysis of movements on the DMF platform. The advantages and limitations are discussed. The droplet desktop paves the way for a smarter cell-free interface for biological detection and protein synthesis in complex reaction networks with expanded operability and less artificial interference.
2. Experimental
2.1 Pigment preparation
Aqueous dyes were purchased at the Yihang Art Supply Store, and they contained 8 colors. Of these, four colors with higher contrast were selected for experiments. 1 mL of dye was taken, diluted with 9 mL of water, and 20 μL of the solution was added to the DMF plate for operation. The control program was set on the PC according to specific droplets’ motion trajectory. For more detail, please refer to the remote control section.2.2 Glucose detection
To detect the concentration of glucose, the final concentrations of GOx, HRP, and ABTS in the reaction mixture were maintained at 0.023 mg mL−1, 0.023 mg mL−1, and 0.864 mg mL−1. The final concentrations of glucose were varied from 0.023 to 0.181 mg mL−1. 0.25 mL of 1 mg mL−1 GOx and 0.25 mL of 1 mg mL−1 HRP were added to 9.5 mL of 1 mg mL−1 ABTS, and then 20 μL of the mixture of GOx, HRP and ABTS was added to 2 μL of glucose at varied concentrations. The concentrations of glucose were varied from 0.25 to 2.0 mg mL−1. The mixture was incubated for 5 minutes at room temperature. Then water was added to the mixture until the volume was 1.5 mL.34 The absorbance of the solution after dilution was measured at 420 nm.2.3 Cell-free protein system
Cell-free reactions were carried out in a volume of 20 μL at 30 °C. The reaction system was composed of 10× salts, phosphoenolpyruvic acid (PEP), amino acid mixture, 25× NTPs mixture, Mg2+ solution, PEG8000, Crude T7 RNA polymerase, E. coli extract, and DNAs.35 10× salt was prepared by 1.75 M potassium glutamate, 27 mM potassium oxalate monohydrate, and 100 mM glutamate, and the pH was adjusted to 7.2–7.4 with ammonia during dissolution. Mg2+ solution was prepared by 1 M magnesium glutamate. Amino acid mixture was prepared by adding the amino acids in the following order (given by three-letter codes): Arg, Val, Trp, Phe, Ile, Leu, Cys, Met, Ala, Asn, Asp, Gly, Gln, His, Lys, Pro, Ser, Thr, Tyr. During preparation, it was necessary to ensure that all of the amino acid was dissolved before adding the next, and Tyr was added finally with the pH adjusted to 7.4 with ammonium hydroxide. A concentration of 883 mM PEP solution was prepared rapidly on ice and flash frozen, and 10 M KOH was added to adjust the pH to 7.4. 25× NTPs mixture was prepared by adding all the reagents one by one in the following order: 1 M putrescine, 1.5 M spermidine, 8.3 mM NAD, 30 mM ATP, 21.5 mM CTP, GTP and UTP, 6.8 mM CoA, 4.3 mg mL−1 E. coli tRNA, and 0.9 mg mL−1 folinic acid. Before adding the next reagent, the last reagent was completely dissolved. The final pH should be between 7.4 and 7.6 and the mixture was stored at −80 °C. When preparing the cell-free system, 6 μL of E. coli extract, 0.8 μL of PEP, 2 μL of 10× salt, 0.8 μL of amino acid mixture, 0.8 μL of 25× NTPs mixture, 0.2 μL of Mg2+ solution, 2.5 μL of 20% PEG8000, and 0.1 μL of crude T7 RNA polymerase, 300 ng DNA (mCherry, GFP, and YPet) was used. Water was added to the mixture until the volume of the reaction system was 20 μL. Due to the limitations of DMF control, it is best to control the droplet size at 20–60 μL, and accordingly adjust the amount of the prepared reaction system. For example, the total reaction system consists of 5 reaction units; that is, 5 × 20 μL = 100 μL.During operation on the DMF board, the cell-free reaction system was divided into 4 droplets. Droplet 1 includes E. coli extract and T7 RNA polymerase, Droplet 2 includes PEG8000, NTPs mixture, and phosphoenolpyruvic acid (PEP), Droplet 3 includes Mg2+ and other salts, and amino acid mixtures, and Droplet 4 includes plasmid aqueous solution as the trigger of the cell-free reaction. Each of the droplets is composed of separated droplets of every single component with a prepared concentration and mixed on the DMF board. Driven by the DMF plate, the first three droplets were mixed, and then the last droplet containing the plasmid was mixed, triggering the cell-free protein reaction. The mixture was incubated for 9 hours at room temperature.
2.4 Digital microfluidics board
The DMF hardware system used was the commercially available OpenDrop V3 board from Gaudi Labs,31 which was an integrated chip device based on the ATSAMD21 ARM Cortex M0 processor that goes to the high voltage output via USB. The ready-made electrode array was assembled, and air was isolated by using 5 cs silicone oil. Parafilm was cut and stretched as the hydrophobic layer film to ensure that the droplets could move freely.2.5 Remote control of DMF board
Based on the OpenDrop board with its relevant open source software on GitHub (https://github.com/GaudiLabs/OpenDrop), changes were made according to different applications and different control methods. The internal control of the single chip was conducted by Arduino, and the processing software helped to realize the interface control of the PC and the serial communication between the PC and the chip, thus achieving control of the energization of the single electrode. By rewriting the corresponding program, the USB serial port was connected to the Raspberry Pi 3B+ for control on a Linux system. Remote desktop control was achieved using the VNC protocol on the LAN. Furthermore, in order to enable the PC and mobile to perform real-time remote control under the conditions of the wide area network to realize IoT, intranet penetration of the internet was conducted through Teamviewer, connecting the Raspberry Pi with the OpenDrop pad and the control terminal to one remote network. Therefore, remote desktop access can be performed on the PC or the mobile phone for remote control. The specific control program generates the relevant json file through processing to perform control of the flow settings, which can be saved to improve its repeatability.3. Results and discussion
3.1 Principle
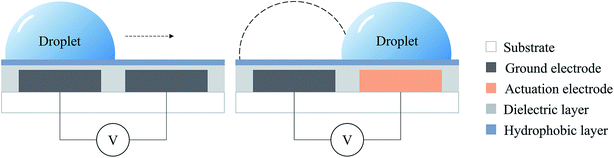
The principle of using the digital microfluidic electrowetting method to manipulate individual droplets is shown in Fig. 2. There is a set of a parallel array of electrodes on the substrate, with a dielectric layer on top of the electrodes. When energized, the electrode is at a high voltage, and the ground electrode is at a low voltage. The charges between the upper surface of the electrode and the lower surface of the droplet form a small capacitor by means of the thin dielectric layer. The hydrophobic layer strengthens the surface tension of the aqueous droplet so that it can be freely moved on the surface of the electrode. After energization, due to the interaction of the surface of the droplet and the charge between the electrodes, the droplets move from the ground electrode to the actuation electrode driven by the electrostatic force, thereby generating movement of the droplets.36,37By using the above principle, a single droplet can be controlled by continuously controlling the energization of the electrode to perform a series of cell-free reactions processed by DMF technology.
The feasibility of DMF method control was verified through experiments with different color pigments. Four pigments of different colors were dropped on the four corners of the electrode array, and the movement of the droplets was controlled through the processing interface. The motion of four aqueous solution dyes preliminarily proved the feasibility of droplet motion of the DMF aqueous solution on the hydrophobic film (Fig. 3, Movies S1 and S2†).
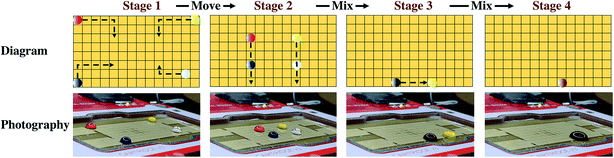 | ||
| Fig. 3 Four droplet dyes of different colors were driven freely on the DMF board. They were mixed and the color of the droplet changed. | ||
3.2 Cell-free glucose detection
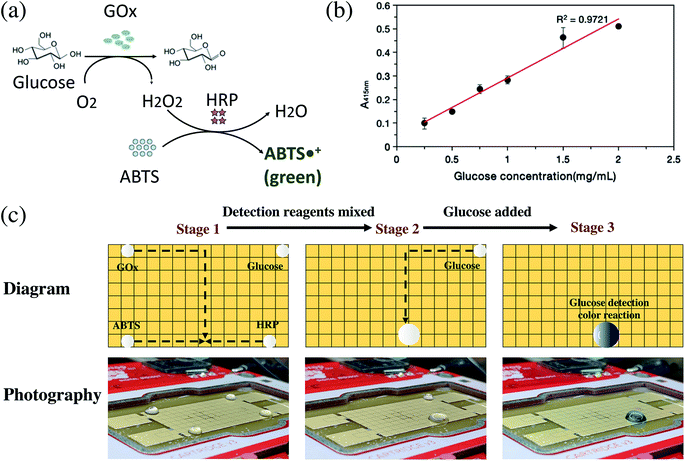
The detection of glucose in blood samples is important for major diseases such as diabetes. Using DMF technology, a conceptual reaction of double enzyme-catalyzed glucose discoloration was also performed.34Glucose produces hydrogen peroxide by the catalysis of GOx, ABTS reacts with hydrogen peroxide by the catalysis of HRP, and then the produced ABTS free radicals appear green (Fig. 4a). For cell-free glucose detection, 3 detective droplets, GOx, HRP, ABTS, and a concentrated glucose droplet were controlled on the OpenDrop board. The 3 detective droplets were mixed and shaken as a detective substrate, then the test glucose droplet was added. A green color appeared, indicating that the glucose detection reaction took place (Fig. 4b, c and Movie S3†).
The rapid DMF biodetection method has the potential to be applied to medical high-throughput detection or optimization of reaction conditions with higher efficiency. In addition to glucose detection, similar methods could be widely applied for the detection of other biochemical samples.
Because of the multi-threaded reaction, the method can speed up the optimization of the reaction conditions and perform multiple sets of experimental samples in parallel. For example, multiple parallel glucose detection reactions on the OpenDrop board were performed simultaneously. Depending on the concentration of the added glucose solution, the resulting green color is different and is measured by an ultraviolet-visible spectrophotometer to obtain the final OD value. The concentrations of GOx, HRP and ABTS were all 1 mg mL−1, and they were mixed as the glucose test reagent in the proportions GOx![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HRP
HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ABTS = 25
ABTS = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 950. Then a series of glucose solutions with concentrations of 0.25, 0.5, 0.75, 1, 1.5 and 2 mg mL−1 were prepared. The glucose detection reaction was performed on the OpenDrop DMF board (Fig. 4c), which shows a linear relationship between the fluorescence value and different glucose concentrations. The results on the DMF plate were consistent with normal operation. By setting up the program, multiple groups of reactions can be performed at the same time efficiently.
950. Then a series of glucose solutions with concentrations of 0.25, 0.5, 0.75, 1, 1.5 and 2 mg mL−1 were prepared. The glucose detection reaction was performed on the OpenDrop DMF board (Fig. 4c), which shows a linear relationship between the fluorescence value and different glucose concentrations. The results on the DMF plate were consistent with normal operation. By setting up the program, multiple groups of reactions can be performed at the same time efficiently.
3.3 Cell-free protein synthesis reaction
The cell-free protein synthesis platform can perform high-throughput screening and expression of proteins. Compared with cell systems, it is a more open and controllable reaction system. DMF technology was combined with the CFB method to express proteins. Through using DMF technology to control the individual droplets, the droplets of each component required for the cell-free reaction can be easily manipulated, such as Mg2+, phosphoenolpyruvic acid (PEP), PEG8000, nucleoside triphosphate (NTP), 19 amino acids, expression plasmid, and cell extract. The DMF method could be used to replace many manual pipetting operations, to programmatically control the mixing and shaking of the liquid, add the coloring reagent, and finally perform the cell-free reaction. Different protein expressions were achieved by changing different plasmid DNA sequences. As a conceptual detective model, catechol 2,3-dioxygenase (CatO2ase) was also expressed. The xylE gene sequence based on the pET plasmid vector was constructed. The CatO2ase protein can turn colorless catechol into brown–yellow 2-hydroxymuconic semialdehyde (HMS), which can be used as a good selection marker. It does not require special preparation of an indicator-containing medium to monitor the expression of the xylE gene. The detection system has ultra-high sensitivity, and can be quantitatively determined by simple spectrophotometry.38Cell-free reactions were carried out in a volume of 20 μL at 30 °C. The reaction system was composed of 10× salts, PEP, amino acid mixture, 25× NTPs mixture, Mg2+ solution, PEG8000, T7 RNA polymerase, Escherichia coli extract, and the expression plasmid. The cell-free reaction system was divided into 4 droplets: Droplet 1 includes Mg2+ and other salts; Droplet 2 includes E. coli extract and T7 RNA polymerase; Droplet 3 includes PEG8000 and NTPs mixture; Droplet 4 includes plasmid aqueous solution as the trigger of the cell-free reaction (Fig. 5a). The 4 droplets were mixed in turn and incubated in an anti-evaporation environment (Fig. 5b and Movie S4†).
During the reaction, the color of the reaction solution gradually changed. After the reaction at 30 °C for 3 h, the reaction solution began to appear brownish-yellow, indicating that the xylE gene was expressed, and the catechol turned into HMS under the catalysis of CatO2ase. Then the reactive solution gradually turned dark brown, which demonstrated that the reaction was carried out continuously.
The expression results of the cell-free reaction on the DMF electrode array were similar to those obtained in the 1.5 mL EP tube, demonstrating that the biochemical reaction by the DMF method has no significant effect on the reaction itself. Hence, the experimental results were sufficiently reliable.
Due to the open DMF method, tens of microliters of the reaction system were easily evaporated to dryness in an incubator at 30 °C, which may affect the experimental results. Therefore, a better method is to use a top cover. The closed DMF method can be used to better avoid the volatilization of the cell-free reaction system while ensuring the integrity of the expressed protein. Another method is to transfer the solution system to an EP tube for the reaction after mixing on the DMF board.
In the process of DMF control, the aqueous solutions of different substances had different movement speeds and sensitivities. In near-pure water or the lower concentration aqueous solution, the droplet can move very quickly, but in the relatively viscous aqueous solution, for example, the crude cell extract required for the cell-free reaction, which contains a variety of complex biological proteins including variable proteins and enzymes, results in a solution with relatively high viscosity that is relatively slow to move. It is necessary to take corresponding measures to make further improvements. The effect of fluid viscosity on motion will be further discussed in Section 3.6.
3.4 Proof-of-concept screening experiment
DMF technology allows us to quickly screen multiple proteins on the same DMF board, and proof-of-concept verification experiments were conducted. In order to more intuitively verify the protein expression of a cell-free system, different fluorescent proteins were expressed as model systems. Four different DNA plasmid templates were simultaneously reacted on the same DMF plate for rapid screening. Green fluorescent protein (sfGFP), red fluorescent protein (mCherry) and yellow fluorescent protein (YPet) were selected, with deionized water as the control group. A fast DMF experiment was performed on the OpenDrop board, and different fluorescence was observed (Fig. 5c). On the basis of significantly reducing the use of consumables and human operation, this experiment quickly screens different DNA plasmid substrates and expresses 3 different proteins at the same time in order to screen the desired type of protein.According to the same principle as protein screening, multi-parallel rapid experimental condition exploration can also be conducted. Taking mCherry as an example, the effect of Mg2+ on the cell-free protein synthesis reaction was explored. Magnesium ions play a crucial role in the process of transcription and translation, especially in transcription. Adjusting different magnesium ion concentrations will greatly affect the amount of protein expressed in the CFB system.39–41 Therefore, exploring the optimal concentration of magnesium ions by a screening method is crucial for cell-free protein synthesis. Three different concentrations of Mg2+ are prepared in advance and mixed with salts and amino acids through real-time DMF control to form Droplet 3. Each of the CFB reactants is grouped in the same way to form 4 droplets, as shown in Fig. 5a. Then all 4 droplets were mixed under real-time control. On the same board, four reactions with different Mg2+ concentrations were set up simultaneously to investigate the effect of protein expression. Therefore, in the process of optimizing experimental conditions using preliminary experiments, the ideal reaction conditions were quickly obtained, and engineering iteration and optimization were performed (Fig. 5d). Currently, the workflow still relies on some manual operations. A better way is to use DMF for the preparation of the solution gradients, which further reduces the manual operation, thereby taking advantage of DMF for CFB. Using DMF to split single droplets uniformly to quantify the volume of a split droplet is a key bottleneck in achieving gradient concentration generation of reactants. A special circuit design for HV amplifier and human intervention for calibration are needed.42 Focusing on the geometry of the electrodes will contribute to lower universality and higher machining difficulty of the complex design.43 J. Fan et al. proposed a micro-well array combining multiplexed concentration gradients that can be generated in a high-throughput manner, with which the individual gradients of 3 different kinds of solutions are also illustrated.44 This clever design provides valuable ideas for the next step of our high-throughput gradient dilution design for different solution concentrations on DMF, which is of great significance. With the combination of DMF and cell-free technology, rapid screening iterations can be performed effectively, whether for screening different types of proteins or reaction conditions.
3.5 Operation interface optimization and remote control
The OpenDrop control program from Gaudi Labs was modified. The OpenDrop source program uses a method of clicking and changing frames. This control method can control individual droplets, but it needs to be set in advance. The initial path setting is a heavy workload and not flexible enough.31 Moreover, the error rate is high, especially when there is a problem in the middle of the path. It is often impossible to move the next frame, and thus an overall motion error occurs.Our control method used a different idea to make the control of the droplet like a mouse moving on an image interface. In other words, where the finger/mouse moves on the computer touchpad/phone screen, the droplet will move to the corresponding position of the OpenDrop board, which means real-time interaction between the control panel and the host computer (Fig. 6 and Movie S5†). Inspired by the idea of the computer interface, it can provide a more practical user experience for the operator, allowing the experimenter to flexibly manipulate the droplets as desired to achieve a more ideal and fluent experimental experience. The traditional DMF control method is a pre-set time-series, and the essence of its control process is to execute a preset program. Our work puts forward the concept of the droplet–user interface, which can realize real-time interaction between the droplet and the operator through a touchpad like a mouse–desktop interface, which improves the interactivity, controllability and flexibility of the liquid handling process.
Many biochemical reactions have extremely stringent requirements for the reaction conditions, such as control over specific conditions of temperature, humidity, pressure, sterility, vacuum, and the like. This adds a lot of inconvenience for the researcher and the experiment. If the operator can operate the liquid remotely, the impact on the reaction environment can be greatly reduced, and the operation can be more convenient and efficient. For example, in the case of a large-scale outbreak of an infectious disease, high-throughput nucleic acid detection is needed in a short period of time, and cross infection should absolutely be avoided. The DMF method can not only improve the flux of sample treatment and shorten the waiting time before the detection process, but also ensure the non-infection of the operating environment with more convenient operation, which can further improve the biosafety and ensure the accuracy of the detection.45,46 This method can also be applied to many biological reaction processes, such as reactions under strict constant humidity conditions. The DMF board can be completely encapsulated in a constant humidity box with droplet operation by remote control. The reaction can be observed in real-time through a camera, so that the reaction process requires zero interference from the person involved.
By connecting the OpenDrop board to the Raspberry Pi 3B+ and using the VNC protocol or the Teamviewer intranet penetration method for remote control, a wireless connection between the manipulated droplets and the control host computer was realized, and a camera was installed on the Raspberry Pi. The conditions of the reactants can be seen in real-time and adjusted according to experimental requirements.
Similar effects can be achieved if a robot arm is used for control, but the DMF board has obvious advantages over a robot arm. On the DMF board, the reactants can react and be screened in multiple threads; the cost is extremely low, the volume is small, and the operation is more flexible. The DMF system has more portability when adapted to a variety of different reaction scenarios.
3.6 Key performance metrics and the analysis of movements
Abstractly, the entire droplet operation interface is similar to mouse–desktop coordination control. The overall response time consists of three aspects: Teamviewer remote desktop rendering time, command transmission time, and Raspberry Pi and OpenDrop processing time. The refresh rate of the remote desktop of the upgraded Teamviewer is 60 frames per s, so each frame time is about 17 ms. When transmitting through the LAN, the command transmission time is less than 1 ms under normal bandwidth. With regard to Raspberry Pi and OpenDrop processing time, through the use of a 48 MHz microprocessor and program optimization, one cycle of data input to HV voltage output can be completed within 1 ms. Therefore, the decisive step for the response time of the entire system is the remote desktop refresh rate of Teamviewer. At a unit execution rate of <20 ms, there is no impact on user experience in spite of a very slight hysteresis in real-time control via the remote desktop. Movie S5† shows the remote operation and the corresponding droplet movement.The error rate of the system depends on the balance between the mouse movement on the remote desktop and the response time. Errors will occur when the mouse is moved to different electrodes within two separate response times. In order to solve this problem, we set up a large interactive interface on the remote desktop to increase the time to move the mouse to the adjacent electrode grid. Meanwhile, the screen resolution is reduced to speed up the transmission and reduce the corresponding time, thereby reducing the error rate. On the other hand, when the droplet moves too slowly to reach the corresponding position, and the next instruction has been executed, the error rate will rise. Therefore, the operator can watch the status of the droplet movement in real time, so that feedback control can be performed on the remote desktop, which can reduce the error rate.
The accuracy of the system consists of the droplet position and droplet concentration. The most suitable droplet volume is 20–60 μL. Within the volume range, due to the continuous pulling force of the electrode, the center of the droplet can fall in the center of a single electrode uniformly. When multiple droplets are mixed and the volume of the droplet increases, one droplet occupies multiple grids. The operation of the droplet will become complicated and unstable. Under this condition, the droplet can be split into several droplets of suitable size to meet the need for appropriate operation.
In the operation of cell-free protein synthesis, the viscosity of Droplet 1 (E. coli extract and T7 RNAP) is large, which makes it difficult to move, although it can still be controlled using more time (Movie S4†). When the viscosity of the fluid increases further, DMF cannot smoothly control the movement of the droplet. Baird et al. proposed a unified model to approximate the steady-state velocity of droplets on DMF.47 According to this formulation, the total energy of the system can be expressed via the usual formula for the energy stored in a capacitor:
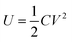 | (1) |
 | (2) |
Some studies have used OpenDrop to explore the factors that influence the speed of droplet movement. By comparing Parafilm, PTFE and ETFE dielectric layers of different thicknesses, the effect of the dielectric constant on the moving speed is verified. Meanwhile, the average droplet velocities increased rapidly as the applied voltage increased from 190 V to 330 V.
When performing the movements of different kinds of liquid on the same DMF board, the movement speed ratio of different liquids is compared using the formula given by:
 | (3) |
In order to make the different droplets move at a similar speed, the voltage can be adjusted within an appropriate range. The square relationship of V makes the adjustable range larger and can compensate for the viscosity range of most liquids in CFB reactions.
3.7 Advantages and limitations
Remote access to the CFB reaction improves the interactivity, controllability and flexibility with higher efficiency by real-time controlled DMF. By adopting the mouse-like desktop control interface, the process of operating a CFB experiment is like playing a computer game when handling the complex liquid array. In fact, the greatest advantage of this droplet desktop solution is its network effect. When the number of different types of droplets is huge and they need to be matched with each other, the droplets can be accurately distributed by the DMF method. Thereby, many heterogeneous reactions can be carried out in parallel, just like the matching of parcels and destinations in unmanned warehouses. The scenario of cell-free reactions on DMF is essentially the same as a traffic scheduling problem in logistics. Some research groups have tried to use the methodology of industrial logistics scheduling to study micro-scale motion control.48 The advanced microfluidics and micro-chemical processing can be achieved by using the same method for droplet control and integrating AI. This is also the reason why the combination of DMF and CFB reaction was chosen. CFB reactants have multiple types, and require a large parallel flux under the experimental conditions of control variables. The characteristics make CFB reactions suitable for DMF operation under a complex network, and DMF can offer remarkable advantages.The DMF method combined with cell-free synthetic biology can be used to investigate the temporal and spatial order of living organisms. Each droplet can represent an independent time and space dimension, which is an independent microenvironment. The heterogeneous and asymmetric characteristics between droplets can be used to construct complex spatiotemporal problems.49 The droplet behavior at different space–time scales simulates the interactions between different biological microenvironments in vitro and can be controlled on the time axis through a control program to study the spatiotemporal behavior at the droplet resolution.50
However, with the OpenDrop board, since the number of electrode arrays is only 128, it is still small in volume, and there is no way to perform free control of large-scale droplets. If the volume is expanded, there will be a shortage of ports and computing power. Since each electrode voltage needs to rise to 250–300 V, OpenDrop uses two HV507 chips for amplifier operation, and each HV507 has 64 OUT ports. If this amplifier method is still used, when the electrode array area needs to be enlarged, the number of pins will be greatly increased since the number of arrays is multiplicative. If the HV507 is simply connected in series to expand the control, it may cause instability at the operating point of the electronic system. An alternative method is to apply a high voltage output only to a portion of the electrodes in the electrode array. For example, a high voltage may be applied only to electrodes that have droplets present and that need to be controlled to move, thereby greatly reducing the high voltage output and the number of pins. However, this method requires a high voltage output pin that is technically more difficult to implement as the electrode moves continuously.
Other control methods have also been studied, such as using a magnetic field to control the transport of droplets, which can avoid the limitation of high voltage electricity, but the corresponding degrees of freedom and the number of controlled droplets will be reduced. With the development of magnetic control principles, it is possible to have optimized control by a magnetic field.51–53 The communication between chemical signals and physical signals can be bridged through integrated organic and inorganic interfaces for actionable decisions.54 Further DMF control of the droplets can be based on the natural chemical characteristics of different droplets to excite the electrodes in addition to visual position feedback.
4. Conclusion
As conceptual research, a remote real-time control platform of digital microfluidics for cell-free biological synthesis reaction and detection was developed. Through pigment verification, in vitro enzyme-catalyzed glucose detection and cell-free protein expression, the feasibility of the control method was demonstrated to improve the interactivity, controllability and flexibility of complex liquid handling schemes with higher efficiency. The remote access of the mouse-like droplet interface provides new insight into smart operations and distributions of cell-free biology for complex reaction networks with expanded operability and less artificial interference in laboratories and industries. Future work will be further combined with droplet splitting, input-mixing-detection closed-loop operation, high-throughput design, artificial intelligence control with complex droplet distribution networks, and further engineering iterations. The scenario of cell-free reactions on DMF is essentially the same as a traffic scheduling problem in logistics. Therefore, the smart cell-free system will also be able to learn from related industrial AI fields for cross-scale applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFA0901700) and a grant from the Institute Guo Qiang, Tsinghua University (2019GQG1016).References
- A. D. Silverman, A. S. Karim and M. C. Jewett, Nat. Rev. Genet., 2020, 21, 151, DOI:10.1038/s41576-019-0186-3.
- Y. Lu, Synthetic and Systems Biotechnology, 2017, 2, 23–27 CrossRef PubMed.
- T. Yanagisawa, M. Takahashi, T. Mukai, S. Sato, M. Wakamori, M. Shirouzu, K. Sakamoto, T. Umehara and S. Yokoyama, ChemBioChem, 2014, 15, 1830–1838 CrossRef CAS PubMed.
- D. K. Karig, S. Iyer, M. L. Simpson and M. J. Doktycz, Nucleic Acids Res., 2012, 40, 3763–3774 CrossRef CAS PubMed.
- G. Yin and J. R. Swartz, Biotechnol. Bioeng., 2004, 86, 188–195 CrossRef CAS PubMed.
- L. Thoring, S. K. Dondapati, M. Stech, D. A. Wüstenhagen and S. Kubick, Sci. Rep., 2017, 7, 11710 CrossRef PubMed.
- S. K. Joseph, D. Boehning, S. Pierson and C. V. Nicchitta, J. Biol. Chem., 1997, 272, 1579–1588 CrossRef CAS PubMed.
- A. S. Karim and M. C. Jewett, Metab. Eng., 2016, 36, 116–126 CrossRef CAS PubMed.
- K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan, N. M. Daringer, I. Bosch, D. M. Dudley, D. H. O’Connor, L. Gehrke and J. J. Collins, Cell, 2016, 165, 1255–1266 CrossRef CAS PubMed.
- M. K. Takahashi, X. Tan, A. J. Dy, D. Braff, R. T. Akana, Y. Furuta, N. Donghia, A. Ananthakrishnan and J. J. Collins, Nat. Commun., 2018, 9, 3347 CrossRef PubMed.
- C. Albayrak and J. R. Swartz, Nucleic Acids Res., 2013, 41, 5949–5963 CrossRef CAS PubMed.
- S. H. Hong, I. Ntai, A. D. Haimovich, N. L. Kelleher, F. J. Isaacs and M. C. Jewett, ACS Synth. Biol., 2014, 3, 398–409 CrossRef CAS PubMed.
- J. F. Zawada, G. Yin, A. R. Steiner, J. Yang, A. Naresh, S. M. Roy, D. S. Gold, H. G. Heinsohn and C. J. Murray, Biotechnol. Bioeng., 2011, 108, 1570–1578 CrossRef CAS.
- B. J. Des Soye, J. R. Patel, F. J. Isaacs and M. C. Jewett, Curr. Opin. Chem. Biol., 2015, 28, 83–90 CrossRef CAS PubMed.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS.
- F. Villarreal, L. E. Contreras-Llano, M. Chavez, Y. Ding, J. Fan, T. Pan and C. Tan, Nat. Chem. Biol., 2018, 14, 29–35 CrossRef CAS.
- W. Gao, E. Cho, Y. Liu and Y. Lu, Front. Pharmacol., 2019, 10, 611 CrossRef CAS.
- A. R. Wheeler, Science, 2008, 322, 539–540 CrossRef CAS.
- A. Li, H. Li, Z. Li, Z. Zhao, K. Li, M. Li and Y. Song, Sci. Adv., 2020, 6, eaay5808 CrossRef PubMed.
- R. Khnouf, D. J. Beebe and Z. H. Fan, Lab Chip, 2009, 9, 56–61 RSC.
- R. Khnouf, D. Olivero, S. Jin and Z. H. Fan, Biotechnol. Prog., 2010, 26, 1590–1596 CrossRef CAS PubMed.
- A. C. Timm, P. G. Shankles, C. M. Foster, M. J. Doktycz and S. T. Retterer, Small, 2016, 12, 690 CrossRef.
- R. P. S. de Campos, D. G. Rackus, R. Shih, C. Zhao, X. Liu and A. R. Wheeler, Anal. Chem., 2019, 91, 2506–2515 CrossRef CAS.
- B. Coelho, B. Veigas, E. Fortunato, R. Martins, H. Águas, R. Igreja and P. V. Baptista, Sensors, 2017, 17, 1495 CrossRef.
- B. Seale, C. Lam, D. G. Rackus, M. D. Chamberlain and A. R. Wheeler, Anal. Chem., 2016, 88(8), 10223–10230 CrossRef CAS PubMed.
- M. David, N. Scott, S. Raj, S. Rama, S. Vijay and P. Vamsee, Expert Rev. Mol. Diagn., 2018, 18, 701–712 CrossRef.
- J. L. He, C. An-Te, L. Jyong-Huei and F. Shih-Kang, Int. J. Mol. Sci., 2015, 16, 22319–22332 CrossRef CAS.
- T. B. Yehezkel, A. Rival, O. Raz, R. Cohen, Z. Marx, M. Camara, J.-F. Dubern, B. Koch, S. Heeb, N. Krasnogor, C. Delattre and E. Shapiro, Nucleic Acids Res., 2016, 44, e35 CrossRef PubMed.
- K. Choi, A. H. C. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef CAS.
- C. Kleanthous, P. Rassam and C. G. Baumann, Curr. Opin. Struct. Biol., 2015, 35, 109–115 CrossRef CAS.
- M. Alistar and U. Gaudenz, Bioengineering, 2017, 4, 45 CrossRef PubMed.
- S. N. I. S. Zulkepli, N. H. Hamid and V. Shukla, Biosensors, 2018, 8, 45 CrossRef.
- U. Umapathi, S. Chin, P. Shin, D. Koutentakis and H. Ishii, MRS Adv., 2018, 3, 1475–1483 CrossRef CAS.
- X. Wu, J. Ge, C. Yang, M. Hou and Z. Liu, Chem. Commun., 2015, 51, 13408–13411 RSC.
- N. Bu and Y. Lu, Methods Protoc., 2019, 2(1), 16 CrossRef.
- T. Xu, W. Shi, J. Huang, Y. Song, F. Zhang, L.-P. Xu, X. Zhang and S. Wang, ACS Nano, 2017, 11, 621–626 CrossRef CAS PubMed.
- H. Geng, J. Feng, L. M. Stabryla and S. K. Cho, Lab Chip, 2017, 17, 1060–1068 RSC.
- X. He, Acta Genet. Sin., 1990, 17(1), 46 CAS.
- A. Sargenti, S. Castiglioni, E. Olivi, F. Bianchi, A. Cazzaniga, G. Farruggia, C. Cappadone, L. Merolle, E. Malucelli, C. Ventura, J. A. M. Maier and S. Iotti, Int. J. Mol. Sci., 2018, 19, 1410 CrossRef.
- C. Zou, X. Duan and J. Wu, Appl. Microbiol. Biotechnol., 2016, 100, 7115–7123 CrossRef CAS PubMed.
- X. Ge, D. Luo and J. Xu, PLoS One, 2011, 6, e28707 CrossRef CAS.
- J. Gong and C.-J. C. Kim, Lab Chip, 2008, 8, 898–906 RSC.
- N. Y. J. B. Nikapitiya, M. M. Nahar and H. Moon, Micro and Nano Systems Letters, 2017, 5, 24 CrossRef.
- J. Fan, F. Villarreal, B. Weyers, Y. Ding, K. H. Tseng, J. Li, B. Li, C. Tan and T. Pan, Lab Chip, 2017, 17, 2198–2207 RSC.
- M. Wang, R. Cao, L. Zhang, X. Yang, J. Liu, M. Xu, Z. Shi, Z. Hu, W. Zhong and G. Xiao, Cell Res., 2020, 30, 269–271 CrossRef CAS PubMed.
- Z. Xu, L. Shi, Y. Wang, J. Zhang, L. Huang, C. Zhang, S. Liu, P. Zhao, H. Liu, L. Zhu, Y. Tai, C. Bai, T. Gao, J. Song, P. Xia, J. Dong, J. Zhao and F.-S. Wang, Lancet Respir. Med., 2020, 8, 420–422 CrossRef CAS PubMed.
- E. S. Baird and K. Mohseni, Nanoscale Microscale Thermophys. Eng., 2007, 11, 109–120 CrossRef CAS.
- T. Li, X. Chang, Z. Wu, J. Li, G. Shao, X. Deng, J. Qiu, B. Guo, G. Zhang, Q. He, L. Li and J. Wang, ACS Nano, 2017, 11, 9268–9275 CrossRef CAS.
- Q. Delacour, C. Li, M.-A. Plamont, E. Billon-Denis, I. Aujard, T. Le Saux, L. Jullien and A. Gautier, ACS Chem. Biol., 2015, 10, 1643–1647 CrossRef CAS PubMed.
- A. Levskaya, O. D. Weiner, W. A. Lim and C. A. Voigt, Nature, 2009, 461, 997–1001 CrossRef CAS.
- C. Liu, J. Ju, Y. Zheng and L. Jiang, ACS Nano, 2014, 8, 1321–1329 CrossRef CAS.
- J. Vialetto, M. Hayakawa, N. Kavokine, M. Takinoue, S. N. Varanakkottu, S. Rudiuk, M. Anyfantakis, M. Morel and D. Baigl, Angew. Chem., 2017, 129, 16792–16797 CrossRef.
- Y. Zhang and N. T. Nguyen, Lab Chip, 2017, 17(6), 994–1008 RSC.
- K. B. Justus, T. Hellebrekers, D. D. Lewis, A. Wood, C. Ingham, C. Majidi, P. R. LeDuc and C. Tan, Science Robotics, 2019, 4, eaax0765 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04588h |
| This journal is © The Royal Society of Chemistry 2020 |