Open Access Article
Open Access ArticleControlled assembly of filamentous viruses into hierarchical nano- to microstructures at liquid/liquid interfaces†
Michihiro Tanakaa,
Toshiki Sawada *ab,
Xiang Li
*ab,
Xiang Li c and
Takeshi Serizawa
c and
Takeshi Serizawa *a
*a
aDepartment of Chemical Science and Engineering, School of Materials and Chemical Technology, Tokyo Institute of Technology, 2-12-1 Ookayama, Meguro-ku, Tokyo 152-8550, Japan. E-mail: sawada@mac.titech.ac.jp; serizawa@mac.titech.ac.jp
bPrecursory Research for Embryonic Science and Technology, Japan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi-shi, Saitama 332-0012, Japan
cInstitute for Solid State Physics, The University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa, Chiba 227-8581, Japan
First published on 14th July 2020
Abstract
Recently, viruses have been regarded as useful molecular assemblies for materials applications rather than as disease-causing agents. The orderly assembled structures of the viruses are highly related to the resultant properties and functions of the assemblies; however, methods to control the assembly are still limited. Here, we demonstrated the assembly of filamentous viruses into hierarchical nano- to microstructures at liquid/liquid interfaces through emulsification in a controlled manner. The viruses form fibrous nanostructures of several micrometers length, which are much longer than the original virus. Subsequently, the fibers self-assemble into well-packed ordered microstructures. Furthermore, the resultant hierarchically assembled structures showed long-term stability and potential applicability through the desired functionalization.
Introduction
The development of dynamic functions of molecular assemblies using biomolecular components is an important key term for progress in various materials fields.1–3 Researchers have recently begun to view viruses as more than disease-causing agents and as molecular assemblies useful for various applications.4,5 Such virus-based materials have many beneficial properties owing to their highly ordered and controlled structures.6 Although animal viruses need to be handled with great skill in appropriate facilities following required safety protocols, plant and bacterial viruses (bacteriophages) are not pathogenic to animals and therefore more easily utilized in material components. Rod-shaped or filamentous viruses such as tobacco mosaic virus and M13 bacteriophage (M13 phage, Fig. 1a) are widely utilized as self-assembling components.7 Importantly, genetic engineering for M13 phage to display foreign peptides or proteins for the evolutional screening process has been well established;8 therefore, the resultant M13 phages with desired functions are utilized as versatile building blocks for virus-based functional soft materials for application in electronic devices, sensors, and biomedical scaffolds as well as conventional antibacterial reagents and drug delivery vehicles.9–21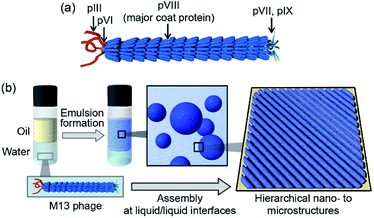 | ||
| Fig. 1 (a) Schematic representation of the assembled structure of M13 phage. (b) Schematic representation of the assembly of M13 phages into ordered microstructures at liquid/liquid interfaces. | ||
Viruses have protein shells that assemble several copies of the same protein into assemblies (called capsids) with precisely controlled three-dimensional structures. Their genetic code in the form of DNA or RNA is located in the interior of the coat protein-assemblies, and electrostatic interactions between the genetic material and surrounding proteins are essential to form these ordered structures. The M13 phage is a regularly assembled filamentous structure with a high aspect ratio (4.5 nm width and 900 nm length) and is constructed by the self-assembly of approximately 2700 copies of the helical major coat protein (pVIII) and 3–7 copies of minor coat proteins at the termini around the DNA.22 The framework of M13 phage is competent for liquid crystal mesogens to form various phases owing to their high aspect ratio, dipole properties, and charge densities, with the use of liquid crystal formation-based ordered assemblies that are indispensable for constructing functional virus-based soft materials.23–25 We aimed to develop a novel strategy to assemble the M13 phage into alternative structures, allowing development of novel class of virus-based materials.
Herein, we demonstrate the assembly of M13 phage into single-layered and well-packed aligned microstructures composed of fibrous structures different from the original phage. The liquid/liquid interface formed during emulsion formation was suitably utilized for the construction of hierarchical nano- to microstructures (Fig. 1b). Mixing of M13 phage aqueous solutions with various organic solvents resulted in the formation of stable oil-in-water (O/W) emulsions. Immediately after emulsification, M13 phages formed unique fibrous nanostructures of several micrometers in length, longer than that of the original M13 phage, but similar in height to that of the original M13 phage. The fibers further self-assembled into uniformly aligned and well-packed microstructures, resulting in extremely stable emulsions. Furthermore, the biomineralization capability of the original M13 phage coat proteins towards converting tetrachloroaurate ions (AuCl4−) into gold nanoparticles (AuNPs) was observed even after assembly, resulting in highly aligned AuNPs on the assembled M13 phages. We show that controlled assembly of filamentous viruses into well-ordered microstructures at liquid/liquid interfaces will widen the applicability of this novel class of functional soft materials.
Experimental
Materials
Fluorescein isothiocyanate (FITC) was purchased from Sigma-Aldrich. SYBR Green II was purchased from Takara Biotechnology Co., Ltd. All other reagents were purchased from Nacalai Tesque. Ultrapure water with a resistivity greater than 18.2 MΩ cm was obtained from a Milli-Q Advantage A-10 (Merck Millipore) and used throughout all experiments.Preparation of M13 phages
M13 phages were expressed using the Ph.D. Peptide Display Cloning System (New England Biolabs, Inc.). Phagemid DNA was transformed into competent Escherichia coli ER2738 cells through heat-shock. The M13 phages expressed were amplified using E. coli and purified by precipitation and re-dispersion procedures using a mixed solution composed of 20 w/v% poly(ethyleneglycol) and 2.5 M NaCl.Modification of M13 phages with FITC
FITC solution was prepared at 5 mg mL−1 in 1 M Na2CO3/1 M NaHCO3, pH 9.0 (conjugation buffer). M13 phage solution (2.5 mg mL−1, 200 μL) was incubated in the dark on a rotator for 1 h at ambient temperature with 10 μL of FITC solution. Following incubation, FITC-conjugated M13 phages were purified three times using PEG and NaCl as described above. Additional rounds of PEG precipitation minimized the residual free fluorophore.Preparation of M13 phage-based emulsions
400 μL of aqueous M13 phage solution (1.5, 15, 150, or 1500 nM) was mixed with 400 μL of organic solvents such as toluene, hexane, cyclohexane, and chloroform, followed by shaking the mixture by hand for 30 s. The prepared emulsions were allowed to incubate at 25 °C.Characterization of emulsions
The emulsions were diluted 5-fold with water, mounted on a concave glass slide (Toshin Riko Co., Ltd.) and examined using an optical microscope (ECLIPSE LV100ND, Nikon) equipped with a digital camera (DS-Fi2, Nikon). Size measurements of the emulsions were performed using ImageJ software (version 1.52, developed at the National Institutes of Health) with at least 90 droplet measurements per sample. The stability of the emulsions against creaming was evaluated by comparing the volume fraction of the stable emulsion. The volume fraction was calculated by dividing the height of the emulsion phase by the total height of the initial emulsion phase.Fluorescence microscopy observation
The emulsions prepared using the FITC-conjugated M13 phage or SYBR Green II-stained (purchased SYBR Green II solution was diluted to 1/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) emulsions prepared using the original M13 phage were observed under a fluorescence microscope (ECLIPSE LV100ND, Nikon). Fluorescence images were acquired using a digital camera (DS-Fi2, Nikon), and the appropriate filter set for fluorophores was used.
000) emulsions prepared using the original M13 phage were observed under a fluorescence microscope (ECLIPSE LV100ND, Nikon). Fluorescence images were acquired using a digital camera (DS-Fi2, Nikon), and the appropriate filter set for fluorophores was used.
Atomic force microscopy (AFM) observation
The morphology of the assembled structures at the emulsion surfaces was imaged by AFM (SPM-9600, Shimadzu) in tapping mode in air at ambient temperature. The emulsions were dispersed in distilled water to remove the freed M13 phage, and 2 μL aliquots of the diluted dispersion were deposited onto a freshly cleaved mica substrate and dried for at least 6 h at ambient temperature in a desiccator with silica gel.Small-angle X-ray scattering (SAXS) measurement
SAXS measurements of the emulsions prepared with 150 nM of M13 phage were performed at BL10C in the Photon Factory, High Energy Accelerator Research Organization, KEK (Tsukuba, Japan). The emulsion was sealed in a glass capillary with an inner diameter of 2 mm. The wavelength of the incident X-ray was 0.1 nm, and the beam diameter at the sample position was approximately 0.25 × 0.50 mm. The exposure time per sample was 30 s. The sample-to-detector distance was 2.54 m. The scattering profile was collected using a 2D hybrid pixel detector (PILATUS3 2M, DECTRIS Ltd., Switzerland). The reduction was performed using a custom-made reduction package Red2D (https://github.com/hurxl/Red2D). The scattering intensity was corrected for the time, transmittance, cell scattering, sample thickness, and then normalized to the absolute scale using glassy carbon (NIST SRM 3600) as a reference. Curve fitting was performed using a core–shell model with a polydisperse core and a constant shell thickness (PolyCoreForm model function) with a data analysis package of the National Institute of Standards and Technology (NIST).26 The average core size, scattering length density (SLD) of the core, shell, and solvent were fixed at 80 μm, 8 × 10−6 Å−2, 1.2 × 10−5 Å−2, and 9.5 × 10−6 Å−2, respectively. The SLD of the core and solvent were estimated using the NIST SLD calculator. A typical SLD of proteins was used for the shell.27Attenuated total reflection/Fourier transform infrared (ATR/FT-IR) absorption spectroscopy
To record the ATR/FT-IR spectra of the emulsions, aliquots of 20 μL of the diluted emulsions were deposited on a gold substrate several times and dried under vacuum conditions. The IR absorption spectra of the samples were measured using a JASCO FT/IR-4100 spectrometer with a cumulative number of 100 and a resolution of 2.0 cm−1 under ambient conditions.Results and discussion
M13 phage-based emulsions were successfully produced using various organic solvents in the oil phase. Briefly, M13 phage aqueous solution (M13 phage concentration 1.5, 15, 150, and 1500 nM) and organic solvents (toluene, hexane, cyclohexane, and chloroform) were mixed at a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
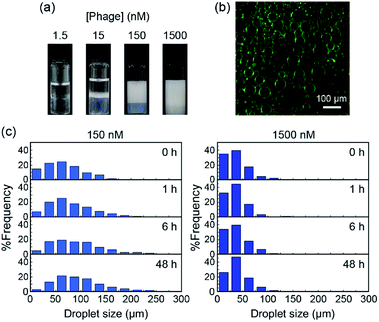
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the mixtures were shaken vigorously for 30 s (Fig. S1a†). The emulsion prepared using toluene was the most stable and the size of droplets showed a narrow size distribution as compared to those prepared using other organic solvents (Fig. S1b and c†). Toluene was therefore selected as the oil phase for further experiments. Emulsions were prepared using different concentrations of M13 phage to determine the influence of the concentration on emulsion formation and their properties. It was previously reported that concentration of emulsifiers was inversely proportional to droplet size, surface coverage density, and emulsion stability.28 As Fig. 2a shows, emulsion volumes clearly increased with increasing concentration of M13 phage, demonstrating the contribution of M13 phage to emulsification. To track M13 phage coat proteins in the formed emulsions, FITC was conjugated to the amino groups of the M13 phage as previously reported.29 The resultant FITC-conjugated M13 phage was applied to the emulsion formation at a concentration of 150 nM, and fluorescence microscopy was performed. In the fluorescence image, it was evident that coat proteins of M13 phage were mostly located at the emulsion interfaces (Fig. 2b). The results indicated that the coat proteins (surrounded with DNA, see below) accumulated at the interface to stabilize emulsion droplets.
1, and the mixtures were shaken vigorously for 30 s (Fig. S1a†). The emulsion prepared using toluene was the most stable and the size of droplets showed a narrow size distribution as compared to those prepared using other organic solvents (Fig. S1b and c†). Toluene was therefore selected as the oil phase for further experiments. Emulsions were prepared using different concentrations of M13 phage to determine the influence of the concentration on emulsion formation and their properties. It was previously reported that concentration of emulsifiers was inversely proportional to droplet size, surface coverage density, and emulsion stability.28 As Fig. 2a shows, emulsion volumes clearly increased with increasing concentration of M13 phage, demonstrating the contribution of M13 phage to emulsification. To track M13 phage coat proteins in the formed emulsions, FITC was conjugated to the amino groups of the M13 phage as previously reported.29 The resultant FITC-conjugated M13 phage was applied to the emulsion formation at a concentration of 150 nM, and fluorescence microscopy was performed. In the fluorescence image, it was evident that coat proteins of M13 phage were mostly located at the emulsion interfaces (Fig. 2b). The results indicated that the coat proteins (surrounded with DNA, see below) accumulated at the interface to stabilize emulsion droplets.
We measured at least 90 different emulsion droplets stabilized by 15, 150, and 1500 nM of M13 phage for statistical analyses of the distribution. More uniform and smaller size emulsion droplets were observed with increasing concentration (Fig. S2†), further supporting the contribution of the M13 phage towards stabilization of the interface. In fact, the creaming behavior of the emulsion clearly demonstrated that the stability of the emulsions increased with increasing concentration of M13 phage (Fig. S3†). Furthermore, the time-dependent size distribution of emulsion droplets prepared with 150 and 1500 nM was characterized in detail (Fig. 2c and S4†). The size of the emulsion droplets prepared with 150 nM M13 phage increased with increasing incubation time after emulsification at constant phage concentration. It is well known that emulsion droplets with insufficient surface coverage of emulsifiers generally lead to coalescence and/or flocculation of the droplets to decrease the surface free energy.30 Therefore, M13 phages at the emulsion interface were considered to assemble in order to cover the interface, resulting in high stability (see below) during the creaming process when 150 nM of M13 phage aqueous solution was used for emulsification. On the other hand, utilization of the M13 phage at 1500 nM, which is higher than the concentration required for liquid crystal formation, would lead to more stable assemblies being formed immediately after emulsification.
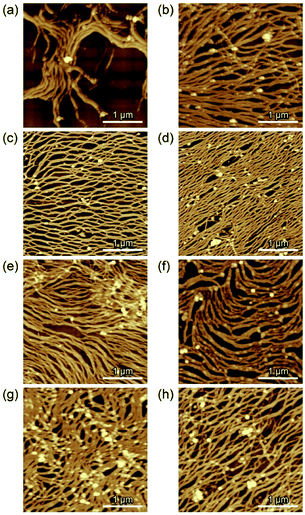
AFM was performed to characterize the assembled nano- to microstructures of M13 phages at the emulsion interfaces during the creaming associated with the coalescence process (Fig. 3). Immediately after emulsification, fibrous nanostructures of several micrometers, which is longer than that of the original M13 phage (approximately 1 μm), were observed at the interface of the emulsion prepared using 150 nM of the M13 phage (Fig. 3a). As the incubation time was increased, the fibrous nanostructures proceeded to self-assemble into aligned and dense microstructures without overlapping, resulting in a decrease in the surface free energy (Fig. 3b–d). After sufficient incubation to form a stable emulsion (48 h), microstructures composed of highly aligned nanofibers were observed (Fig. 3d). These hierarchical nano- to microstructures were observed on a scale of several tens of micrometers (Fig. S5a†). Importantly, the height of the non-overlapping nanofibers in all the images was approximately 4–5 nm (Fig. S5b†), which was comparable to the width of the original M13 phage, indicating that the nanofibers formed assemblies with the same thickness as the original virus structure.
When the M13 phage is used at 1500 nM, immediately after emulsification, more densely assembled structures composed of similar long nanofibers were observed (Fig. 3e). A negligible change was observed in the assembled structures during the incubation (Fig. 3f–h), reflecting the unchanged droplet size. This is possibly due to that preparation of sufficient amounts of the assembled fibrous structures to cover the interface immediately with the mixing, resulting in “kinetically trapped” less-ordered structures at high concentration (1500 nM), whereas less amounts of the assembled nanofibers enabled formation of ordered microstructures at “thermodynamic equilibrium” with structural relaxation during incubation at low concentration (150 nM). These results strongly indicate that the liquid/liquid interface formed under suitable emulsification conditions offered assembly of M13 phage into hierarchical nano- to microstructures in a controlled manner.
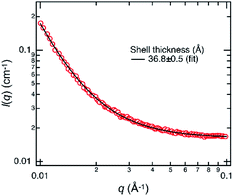
SAXS measurement of the emulsion prepared with 150 nM of M13 phage was performed to clarify that a single-layered microstructure was formed during the emulsification and subsequent creaming process, not during the sample drying process for AFM. The observed profile can be fitted well with a theoretical equation of core–shell form with a polydisperse core and a constant shell thickness (Fig. 4). The estimated shell thickness was 3.68 ± 0.05 nm, which is similar to the height of assemblies determined by AFM. These results strongly indicate that a single-layered microstructure was formed at the emulsion interface in situ. Although AFM was performed for dried assemblies, it was suggested that the observed morphologies possibly represented those at the liquid/liquid interface.
In order to investigate the structural denaturing of M13 phage at the interface, DNA staining experiments using SYBR Green II with the capability to specifically bind to single-stranded nucleic acids were performed (Fig. S6†). Immediately after emulsification, obvious fluorescence was observed from the surface of the emulsion droplets prepared with 150 nM, as compared to those prepared with 1500 nM, indicating that the original capsid structure at 150 nM was denatured to allow access by the small molecule. The fluorescence intensity for the emulsion with 1500 nM was comparable to that for the original M13 phage as a control experiment, demonstrating less denaturing of the capsid structure at 1500 nM. Liquid crystal formation at a high concentration (1500 nM) seems to have suppressed the denaturation. Furthermore, ATR/FT-IR spectroscopy measurements indicated that the secondary structure of the assemblies of M13 phage prepared with 150 nM underwent a partial structural transition from the original α-helix to β-sheet structures as compared to that with 1500 nM (Fig. S7†), further supporting a greater amount of denaturing for the capsid of M13 phage at 150 nM.
The stability of the emulsions was evaluated to better understand the structural features of the assembled microstructures. Although the emulsion prepared using 1500 nM of M13 phage demulsified after incubation for 4 months, the one prepared with 150 nM was still stable after 1 year (Fig. S8a†). This indicates that assembly of M13 phages into ordered hierarchical nano- to microstructures through an incubation process under suitable concentration conditions (e.g., 150 nM) is essential for the formation of stable structures rather than less-ordered assembly under higher concentration conditions. In fact, the nano- to microstructures at the emulsion were still observed after incubation for 1 year (Fig. S8b†). These results strongly indicate that the assembled microstructures composed of well-packed nanofibers (that is, assembled M13 phages) show high structural stability even though the droplet size was over several tens of micrometers.
Biomineralization of AuCl4− on the assembled M13 phage was performed. Previously, it was found that the major coat protein of M13 phage showed sorption and reduction capabilities of AuCl4− into AuNPs in situ under mild conditions (aqueous buffer solution (pH 6.8, 37 °C)).31 An aqueous solution of AuCl4− was incubated with the assembled M13 phage, and the surface morphology was observed by AFM after removal of excess amounts of AuCl4− and AuNPs (Fig. 5). As a result, densely aligned nano-objects with a height of approximately 15 nm, considered to be AuNPs, were observed. The morphology of the aligned AuNPs was similar to that of the assemblies, indicating selective sorption and subsequent reduction of AuCl4− for the preparation of AuNPs on the assemblies. The results demonstrated that the originally expressed function of M13 phage was maintained after assembly at the interface although the assemblies showed different assembled structures with slightly denatured states. Thus, similar to the desired functionalization of coat proteins through genetic engineering, various other functions can be introduced to the ordered assembled structures.
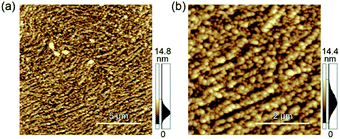 | ||
| Fig. 5 Morphological characterization of the AuNPs on the microstructures by AFM. Observed area are (a) 15 μm × 15 μm and (b) 5 μm × 5 μm, respectively. | ||
Conclusions
In conclusion, the assembly of filamentous viruses into ordered hierarchical nano- to microstructures was demonstrated using liquid/liquid interfaces. Emulsion formation using the filamentous virus resulted in the formation of nanofibers with a non-native form. During the creaming process of the emulsion, the fibrous nanostructures further self-assembled into single-layered and well-packed microstructures to decrease the surface free energy under suitable concentration conditions. The resultant hierarchical structures showed long-term stability compared to the less-ordered structures and potential applicability with desired functionalization. Our results showed that the assembly of filamentous viruses in a controlled manner at the liquid/liquid interfaces will provide new insights for the potential application of structurally ordered protein-based stable assemblies.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Japan Science and Technology Agency (JST) through the Precursory Research for Embryonic Science and Technology (PRESTO) Grant Number JPMJPR17I4, The Moritani Scholarship Foundation, the Iketani Science and Technology Foundation, and The Ogasawara Foundation for the Promotion of Science & Technology for T. Sawada. The SAXS experiments were performed the approval of the Photon Factory Program Advisory Committee (Proposal No. 2019G055).Notes and references
- T. Sawada, H. Mihara and T. Serizawa, Chem. Rec., 2013, 13, 172–186 CrossRef CAS PubMed.
- L. Zhao, Q. Zou and X. Yan, Bull. Chem. Soc. Jpn., 2019, 92, 70–79 CrossRef CAS.
- K. Fukunaga, H. Tsutsumi and H. Mihara, Bull. Chem. Soc. Jpn., 2019, 92, 391–399 CrossRef CAS.
- C. M. Soto and B. R. Ratna, Curr. Opin. Biotechnol., 2010, 21, 426–438 CrossRef CAS PubMed.
- K. S. Sunderland, M. Yang and C. Mao, Angew. Chem., Int. Ed., 2017, 56, 1964–1992 CrossRef CAS PubMed.
- Z. Liu, J. Qiao, Z. Niu and Q. Wang, Chem. Soc. Rev., 2012, 41, 6178–6194 RSC.
- C. E. Flynn, S. W. Lee, B. R. Peelle and A. M. Belcher, Acta Mater., 2003, 51, 5867–5880 CrossRef CAS.
- G. P. Smith and V. A. Petrenko, Chem. Rev., 1997, 97, 391–410 CrossRef CAS PubMed.
- W. J. Chung, J. W. Oh, K. Kwak, B. Y. Lee, J. Meyer, E. Wang, A. Hexemer and S. W. Lee, Nature, 2011, 478, 364–368 CrossRef CAS PubMed.
- T. Sawada and T. Serizawa, J. Mater. Chem. B, 2013, 1, 149–152 RSC.
- S. H. Yang, W. J. Chung, S. McFarland and S. W. Lee, Chem. Rec., 2013, 13, 43–59 CrossRef CAS PubMed.
- T. Sawada, S. Kang, J. Watanabe, H. Mihara and T. Serizawa, ACS Macro Lett., 2014, 3, 341–345 CrossRef CAS.
- T. Sawada, H. Chen, N. Shirakawa, S. Kang, J. Watanabe and T. Serizawa, Polym. J., 2014, 46, 511–515 CrossRef CAS.
- T. Sawada, H. Otsuka, H. Yui and T. Serizawa, Polym. Bull., 2015, 72, 1487–1496 CrossRef CAS.
- K. Mohan and G. A. Weiss, ACS Chem. Biol., 2016, 11, 1167–1179 CrossRef CAS PubMed.
- T. Sawada, M. Yanagimachi and T. Serizawa, Mater. Chem. Front., 2017, 1, 146–151 RSC.
- T. Sawada, Polym. J., 2017, 49, 639–647 CrossRef CAS.
- T. Sawada and T. Serizawa, Bull. Chem. Soc. Jpn., 2018, 91, 455–466 CrossRef CAS.
- T. Sawada, Y. Murata, H. Marubayashi, S. Nojima, J. Morikawa and T. Serizawa, Sci. Rep., 2018, 8, 5412 CrossRef PubMed.
- T. Sawada, H. Inomata and T. Serizawa, J. Membr. Sci., 2020, 595, 117595 CrossRef CAS.
- T. Sawada, T. Tsuruoka, N. Ueda, H. Marubayashi, S. Nojima, J. Morikawa and T. Serizawa, Polym. J., 2020, 52, 803–811 CrossRef CAS.
- C. Mao, A. Liu and B. Cao, Angew. Chem., Int. Ed., 2009, 48, 6790–6810 CrossRef CAS PubMed.
- Z. Dogic and S. Fraden, Curr. Opin. Colloid Interface Sci., 2006, 11, 47–55 CrossRef CAS.
- Z. Zhang and E. Grelet, Soft Matter, 2013, 9, 1015–1024 RSC.
- Z. Dogic, P. Sharma and M. J. Zakhary, Annu. Rev. Condens. Matter Phys., 2014, 5, 137–157 CrossRef CAS.
- S. R. Kline, J. Appl. Crystallogr., 2005, 39, 895–900 CrossRef.
- F. Zhang, F. Roosen-Runge, M. W. A. Skoda, R. M. J. Jacobs, M. Wolf, P. Callow, H. Frielinghaus, V. Pipich, S. Prévost and F. Schreiber, Phys. Chem. Chem. Phys., 2012, 14, 2483–2493 RSC.
- D. J. McClements and C. E. Gumus, Adv. Colloid Interface Sci., 2016, 234, 3–26 CrossRef CAS PubMed.
- D. L. Jaye, C. M. Geigerman, R. E. Fuller, A. Akyildiz and C. A. Parkos, J. Immunol. Methods, 2004, 295, 119–127 CrossRef CAS PubMed.
- M. Shields, R. Ellis and B. R. Saunders, Colloids Surf., A, 2001, 178, 265–276 CrossRef CAS.
- M. I. Setyawati, J. Xie and D. T. Leong, ACS Appl. Mater. Interfaces, 2014, 6, 910–917 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Preparation and characterization of emulsions with various organic solvents or different concentrations of M13 phage, creaming behaviour of the emulsions, an expanded AFM image, height characterization of the assembled structures, DNA staining experiments, IR measurements and characterization of the emulsions after 1 year-incubation. See DOI: 10.1039/d0ra04529b |
| This journal is © The Royal Society of Chemistry 2020 |