Open Access Article
Open Access ArticleA facile assay for rapid detection of COVID-19 antibodies†
Caixia Liu‡
a,
Baiping Mao‡a,
Vanessa Martinezb,
Xiaojian Chena,
Yanan Lia,
Lingyun Hea,
Sian Chena,
Xiaoling Guoa,
Xian Shena,
Xiandan Baoc,
Haifa Shend,
Stefania Lennaef,
Pinyi Qiang,
Lingzhi Wu*c and
Chao Li *a
*a
aThe Second Affiliated Hospital, Yuying Children's Hospital of Wenzhou Medical University, 109 Xueyuan West Road, Wenzhou, 325027, PR China. E-mail: dishboy@163.com
bHouston Methodist Research Institute, University of St. Thomas, 3800 Montrose Blvd, Houston, TX 77006, USA
cWenzhou Kont Biotechnology Co., Ltd, 128 Luoxi Street, Oubei, Wenzhou, 325101, PR China. E-mail: wlz20121126@163.com
dHouston Methodist Research Institute, Weill Cornell Medical College, 6670 Bertner Avenue, Houston, TX 77030, USA
eCenter for Musculoskeletal Regeneration, Houston Methodist Research Institute, 6670 Bertner Avenue, Houston, TX 77030, USA
fOrthopedics and Sports Medicine, Houston Methodist Hospital, 6670 Bertner Avenue, Houston, TX 77030, USA
gPhoner (Wenzhou) Pharmaceutical Biotechnology Research Institute, Room 401, South Building, Danan Road, Wenzhou, 325028, PR China
First published on 28th July 2020
Abstract
The outbreak of new coronavirus disease (COVID-19) has quickly spread all over the world. Real time reverse transcriptase polymerase chain reaction (rRT-PCR) for nucleic acid detection has become the standard method for clinical diagnosis of COVID-19 infection. But these rRT-PCR tests have many inherent limitations, and carry a high false negative rate. It is an urgent to develop a method to accurately identify the vast infected patients and asymptomatic viral carriers from the population. In this article, we present the principle and procedure of developing a colloidal gold immunochromatographic assay (GICA) for rapid detection of COVID-19-specific antibodies. The detection kit can be used to detect immunoglobulin M (IgM) and IgG of COVID-19 in human blood samples within 15 minutes, and to identify different stages of viral infection. Test results can be digitalized using an office scanner and a FiJi software with appropriate confidence interval (CI) setting. Based on analysis from 375 samples, we calculated that overall sensitivity and specificity of the assay were 95.85% and 97.47%, respectively. Compared with rRT-PCR, this assay has many advantages including convenience and rapid detection. The detection kit can be widely used in hospitals, clinics and laboratories for rapid screening of both symptomatic and asymptomatic COVID-19 carriers in large scale.
1. Introduction
In December 2019, there were many cases of pneumonia with unknown etiology in Wuhan, China.1 The patients' clinical manifestations were very similar to SARS-CoV in 2003 (ref. 2) and MERS-CoV in 2012.3 The later genomic sequencing confirmed that the pathogen is a new type of coronavirus. The International Committee on Taxonomy of Viruses (ICTV) classified it as “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2), and it was officially named as COVID-19 by the World Health Organization (WHO) and the International Society for Infectious Diseases (ISID).4 At present, the COVID-19 infection has been listed as the Public Health Emergency of International Concern (PHEIC).5 Up to date, COVID-19 has spread all over the world, with more than 3.76 million diagnoses and 264![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths (Google data on May 7). In this context, most countries have laid different regulation policies and strict quarantine measures to suppress the infection transmission. However, the rapid spread and nonselective infection of COVID-19 is often fatal for patients with severe underlying diseases.6 Facing this sudden strike, the serious medical resource shortage and insufficient Personal Protective Equipment (PPE) have become the insurmountable difficulties for CoIVD-19 infection control and the health care systems are in the edge of collapse in many countries.
000 deaths (Google data on May 7). In this context, most countries have laid different regulation policies and strict quarantine measures to suppress the infection transmission. However, the rapid spread and nonselective infection of COVID-19 is often fatal for patients with severe underlying diseases.6 Facing this sudden strike, the serious medical resource shortage and insufficient Personal Protective Equipment (PPE) have become the insurmountable difficulties for CoIVD-19 infection control and the health care systems are in the edge of collapse in many countries.
Facts have proven that earlier detection, report, isolation, diagnosis and treatment are useful strategies in COVID-19 patient treatment.7 As we all know, real time reverse transcriptase polymerase chain reaction (rRT-PCR) for nucleic acid detection, CT imaging and parameter changes in blood have been important indies for clinical diagnosis of COVID-19, and high-throughput sequencing of viral nucleic acids can be used as the “gold standard” for COVID-19 diagnosis.8 Various countries and companies have developed a variety of rRT-PCR test kits, some of them have been certified by the China CDC or the FDA of USA. However, these technologies including genomic sequencing and rRT-PCR have some inherent limitations, including long time testing, expensive equipment, the need of certified laboratories and the well trained staff, etc.9 Moreover, the rRT-PCR for COVID-19 is often interfered by many uncontrolled factors, such as sample quality, RNA extraction, resulting in high false negative rate (the positive rate of rRT-PCR for SARS-CoV was 50–79%).10 These reasons make the use of rRT-PCR unsuitable for a quick screening of the infected patients from healthy people on the spot, and in the large scale population. The most concerning problem is the incapability of the current global nucleic acid detection to meet the demands of large scale population screening. Therefore, it is urgent to expand the type and number of test kits to close the gap made by the deficiency of rRT-PCR.
In this background, an accurate, rapid and sensitive test method is needed to quickly identify the large number of COVID-19 infected patients and asymptomatic carriers to prevent the widespread of the COVID-19, and ensure that the infected patients obtain the effective treatment on time. Since COVID-19 and SARS-CoV belong to the same family, the transmission route and infection symptoms are very similar.11 Therefore, we speculate that the first defensive line of immune globulin M (IgM) against COVID-19 appears at 1–6 days after infection.12 The high affinity IgG, which is essential for long-term protection and immune memory, appears at 8 days after infection.13 In addition, IgM can early resist the invasion of COVID-19 to multiple organs in the human body, and the appearance of IgM indicates recent exposure to COVID-19. The appearance of IgG indicates the patient has been exposed to the virus for a period of time, and the body has developed strong immunity to against COVID-19. Therefore, the rapid detection of IgM and IgG can provide information of timeline and stage of infection, and supply the references for COVID-19 large scale screening, diagnosis and treatment, as well as references of appropriate protection for healthcare staff.
The “Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (7th edition)”8 issued by the National Health Commission (NHC) of China on March 4, 2020 has recommended that any suspicious cases with positive IgM and or IgG antibodies can be identified as COVID-19 confirmed cases.14 However, there are many kinds of IgM/IgG detection products in the market, and the sensitivity and specificity data are insufficient. Currently, still are missing detailed information and referable publications about how to determine positive or negative, and how to improve the specificity and accuracy of IgM/IgG combined detection.
As the AuNPs have size-dependent diffusion rate, the smaller particle size and narrow size distribution are preferable for almost all applications. What's more, the smaller particle size has the higher specific surface area, which facilitates the more protein conjugation, as well as the improved detection sensitivity. Based on these contexts, we developed a colloidal gold immunochromatography kit with smaller AuNPs for the specific detection of IgM and IgG antibodies of COVID-19 in human blood sample within 15 minutes. In order to evaluate its sensitivity, specificity, accuracy and clinical effectiveness, we tested the kit at the designated hospital for patients with COVID-19 infection. The results showed that the rapid test kit has high sensitivity and specificity for the IgM/IgG detection of COVID-19. Combined with our simple data analysis method, it can be used in hospitals, clinics and laboratories, which make it a powerful force against the global threat of COVID-19.
2. Material and methods
2.1 The materials for producing the COVID-19 IgM/IgG test kit
The COVID-19 nucleocapsid recombinant antigen ncov-ps-Ag8, anti-human IgG (#8) and anti-human IgM were purchased from Fapon Biological Company, Shenzhen, China. The antigen design is based on the published sequence of COVID-19, and several different antigen designs have been tested and optimized, and Ag8 was finally selected for the product manufacture. Bovine serum albumin (BSA) was purchased from sigma. Rabbit IgG (D110502-0010) and goat anti-rabbit IgG (D111018-0001) antibodies were purchased from Sangon Biotech, Shanghai. Nitrocellulose (NC) membrane was provided by Merck Millipore. Ahlstrom Advanced Filtration and PVC substrate were purchased from Joey Bio, Shanghai. Gold chloride trihydrate (G141105), sodium citrate (S116131), potassium carbonate (P112404) and polyethylene glycol (PEG, MW 20,000, P103730) were purchased from Aladdin, Shanghai, China.2.2 Preparation of the 40 nm Au nanoparticles (AuNPs)
2 ml of 0.02% (w/w) gold chloride trihydrate aqueous solution was added to 98 ml of deionized water and mixed evenly. After heating to boiling, 2 ml of 2% (w/w) sodium citrate aqueous solution was added to the heating system quickly and kept boiling for 20 min. After cooling to room temperature (RT), colloidal AuNPs with a hydrated diameter about 40 nm were obtained.2.3 Preparation and purification of AuNPs conjugations
The AuNP–Ag8 conjugations: 140 μl 0.2 M K2CO3 solution was added to 12 ml AuNPs solution (OD = 1.6) to adjust the pH to 8.5. Then, 120 μg of ncov-ps-Ag8 in 1 ml PBS was added to AuNPs solution, incubated and stirred at RT for 30 min. After that, 0.4 ml of 200 mg ml−1 BSA aqueous solution was added to the system to block the excessive antigen binding epitopes on AuNPs surface, and incubated at RT for another 15 min. After that, 0.4 ml of 200 mg ml−1 PEG aqueous solution was added to the system and incubated at RT for extra 30 min. The mixture was then centrifuged at 4 °C and 3000 rpm for 10 min, the supernatant was collected and recentrifuged at 9000 rpm for another 30 min, and the precipitate was collected. The precipitate was resuspended in 12 ml of 1 mg ml−1 BSA solution and recentrifuged at 4 °C and 9000 rpm for 30 min. The final precipitate was collected and resuspended in PBS buffer. The identical procedure was used to prepare and purify the AuNP–rabbit IgG conjugation. Finally, the AuNP–Ag8 and AuNP–rabbit-IgG were mixed evenly at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v), and sprayed in 15 μl cm−1 to the conjugation pad (details of the optimization process in ESI, S1†).
1 (v/v), and sprayed in 15 μl cm−1 to the conjugation pad (details of the optimization process in ESI, S1†).
2.4 Preparation of COVID-19 IgM/IgG rapid test strip
The test strip is composed with five parts, including PVC substrate, sample pad, conjugate pad, absorption pad and NC membrane. The following procedures were followed to prepare the test strip. Firstly, the NC membrane were connected to the PVC substrate, and the anti-human IgM, anti-human IgG and anti-rabbit IgG were sprayed to the areas of M, G and C position of the NC membrane respectively. Secondly, the mixture of AuNP–Ag8 and AuNP–rabbit-IgG conjugate were sprayed to Ahlstrom Advanced Filtration to prepare the conjugation pad. The sample pad was pretreated with BSA (1%, w/v) and Triton X-100 (2%, w/v). After preparation and assembling of all components, the assembled card was loaded into the CNC precision cutting machine, and the strip was cut with a width of 4 mm. At last, the strip was loaded into the plastic holder to finish the test strip preparation (details of the optimization process in ESI, S1†).2.5 Patient and sample collection
This study is a retrospective observation that included the patients admitted to COVID-19 designated hospital from Feb. 6 to March 10, 2020. According to the national guidance,8 the specimen collection including whole blood and pharyngeal swab, as well as serology and RT-PCR tests, all were carried out in the biosafety level (BSL-) 2 laboratory, and the BSL-3 PPE was used throughout all the process. All of the testing materials were treated as infectious medical waste. After examination, the waste was processed by high-temperature and steam sterilization, and then treated as medical waste as routine. The 217 patients were all confirmed cases of COVID-19 by rRT-PCR (Table S1 and ESI S2†). 158 serums of non-COVID-19 patients (three times rRT-PCR test negative) were used as the negative control (Table S2†). Approximately 3 ml of whole blood of the patient were collected, and the serums were separated and for the IgM/IgG tests immediately. 10 μl of serum was mixed with 60 μl of sample buffer, and pipetted into the sample well. The sample flowed laterally along the NC membrane and reacted with the antibodies pre-coated on the NC membrane. After 10–15 min, the colored lines appeared at M, G and C positions.2.6 Results display
The test kit includes a vial of sample buffer (1.5 ml) and 20 individually packaged test strips. The strip has 3 lines, a distal control line (C), and the proximal IgM line (M) and IgG line (G). If the sample contained anti-COVID-19 IgM and/or anti-COVID-19 IgG, these antibodies would specifically bound to AuNP–Ag8 that pre-coated on the conjugation pad, and forming the M, G and C lines.2.7 Data collection and analysis
After the visual observation of the test results, the test strips were placed on the scanning panel of a Kyocera printer (Ecosys FS-1020MFP) with a scanning resolution set at 600dpi. After scanning and a tilt correction with the Photoshop software, the images were saved as an 8 bit file in .jpg format independently. The FiJi software (http://fiji.sc/) was used to obtain the color density and intensity of M, G and C lines.Take the total date of the three lines of the sample as 100%, calculate the percentage of M and G peaks, and set the cutoff value at 95% confidence interval (CI). In this study, IgM values larger than 3% and IgG values larger than 6% were set as the cutoff values for IgM and IgG respectively. Based on the cutoff value, to determine the negative or positive of the test results, the specificity and sensitivity of the rapid test kit were calculated according to the following formula:
| Specificity (%) = 100 × [true negative/(true negative + false positive)] |
| Sensitivity (%) = 100 × [true positive/(true positive + false negative)] |
3. Results
3.1 Design and accomplish the IgM/IgG rapid test kit of COVID-19
The IgG/IgM rapid test kit for COVID-19 applies a colloidal gold immunochromatography assay (GICA) to determine COVID-19-specific IgM and IgG antibodies in human serum or plasma samples. The mechanism of this test is based on the hydration and capillary transport of solution, and the substances contained in the solution interact with the reagent pre-coated on the strip. In this study, we prepared the AuNPs with 40 nm of the hydrodynamic size and the maximum absorption at 532 nm (Fig. S1†). For the optical properties of spherical AuNPs are highly dependent on the diameter, the test strips prepared by this AuNPs have flesh-red detection bonds (40 nm), which is slightly different from the previously reported GICA strips with color from burgundy to purple of the detection bonds (80–100 nm).The mixture of sample and buffer solution migrates on the NC membrane by capillary action. If the IgM and or IgG of anti-COVID-19 present in the sample, it would be conjugated to AuNP–Ag8 of COVID-19 that was pro-coated on the conjugation pad, and formed the antigen–antibody complex (Ag–Ab, both IgM and IgG). The complex will be captured by the anti-human IgM and or anti-human IgG antibodies that were sprayed on the NC membrane in advance, and forming the M and or G line in the corresponding area (Fig. 1A and B). It also means the anti-COVID-19 IgM or anti-COVID-19 IgG or both antibodies were present in the sample.
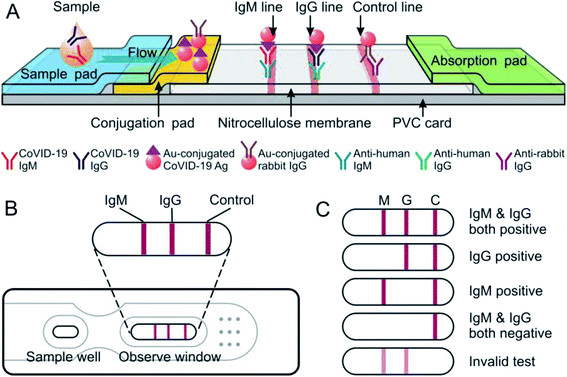 | ||
| Fig. 1 Schematic view of the COVID-19 IgM/IgG rapid test strip. (A) Preparation and principle of COVID-19 IgM/IgG rapid test strip. (B) Assembled products. (C) Scheme of test results. | ||
The excessive sample solution will continue to move along the NC membrane and will reach the position of C line, where the AuNP–rabbit-IgG complex will be captured by pre-coated anti-rabbit IgG, thereby forming the C line, and will also indicate that the sample has fully migrated throughout the test strip and confirm the test validity (Fig. 1C). If the control line did not appear, it meant the test was invalid and the test should be repeated with another test strip.
Fig. 2 showed the different test results from different patients, representing several different types of results. The results showed that neither IgM nor IgG were detected in No. 9 sample, while both IgM and IgG were detected in No. 20 sample. In No. 27 sample, high concentration of IgG and low concentration of IgM were detected, and instead in No. 30 sample, only low concentration of IgG was detected.
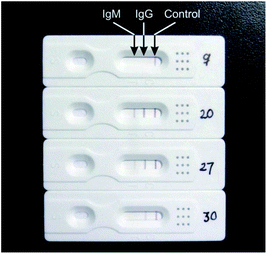 | ||
| Fig. 2 Representative photos of test results. No. 9, IgM and IgG: negative. No. 20, IgM and IgG: positive. No. 27, IgM: weak positive, IgG: positive. No. 30, IgG: weak positive. | ||
3.2 The data analysis, the sensitivity and specificity evaluations of IgM/IgG rapid test kit of COVID-19
After digitizing the test results with a scanner, the FiJi software was used to analyze the digitized images as follows: file–open, use the 1st rectangle tool to select the strip image of the first sample, and use the ctrl + 1 key to define sample 1; move the selection to the images of next test strip (copy automatically), and use ctrl + 2 to define the sample 2; the subsequent stripes are all numbered sequentially by ctrl + 2 until all the test strips are correctly numbered (Fig. 3A). Analyze–gels–plot lanes, to draw intensity peak of all strips and the bonds; use the 5th Straight tool to close the peak of the line (optional assist with magnifying glass and scrolling tools); use the 8th tracing tool to click on the peaks that were closed, the data under the peak appeared, copy the data to the Excel table for later analysis (Fig. 3A).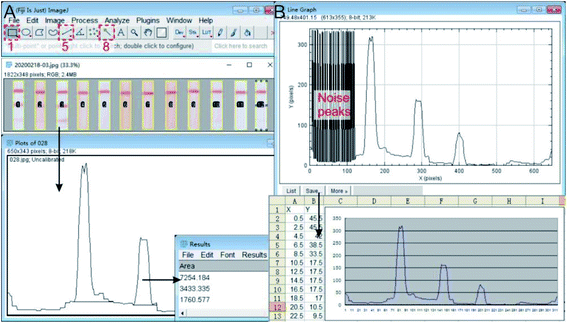 | ||
| Fig. 3 Analysis of the digitalized results. (A) Datamation the density and intensity of peak in digitalized result with FiJi software; (B) import the intensity curve into Excel to redraw the graphic. | ||
Redraw the peak curve: analyze–gels–re-plot lanes, use the tracing tool to click to the peak line. Try a few times until the peak line was selected (yellow); analyze–tools–analyze line graph, to digitize the curve again; save the data in .csv format, and delete the noise peaks of analysis errors in Excel. The corresponding peak curve can be drawn in any digital graphics software (Origin, Graphpad Prism, etc. Fig. 3B).
For each test strip, the total amount of the Ag8 conjugated AuNPs on the conjugation pad is identical in process of the strip manufacture. Under the premise of the same volume of sample and buffer were added to the sample well, the total value of the three bands should be a basic fixed value for every test. Therefore, the peak density and intensity of the IgM and IgG bands can objectively reflect the relative concentration of the IgG and IgM in the sample. Thus, the quantitative detection can be achieved. In data analysis, the total peak values of the three bands were set to 100%, and the percentage of IgM and IgG were calculated. Within 95% CI, the cutoff values of IgM and IgG were set as 3% and 6%, respectively. Total of 375 specimens were tested, including 217 clinical confirmed COVID-19 infection patients (rRT-PCR test positive) and 158 non-COVID-19 infection patients (three times rRT-PCR test negative). Table 1 summarizes the test results of the patient's serums (details in Table S1†). The data analysis showed that 208 of 217 COVID-19 samples were positive, with sensitivity of 95.85%. For 40 non-COVID-19 samples, 1 sample was IgG weak positive with specificity of 97.47%. It was also found that 51.61% (112 of 217) positive samples had both IgM and IgG antibodies (Table 1). Generally, in COVID-19 confirmed cases, the overall positive rate of IgM was 52.53%. We speculated that these patients had passed the peak period of IgM when they were admitted to the hospital, therefore, the IgM levels had decreased below the cutoff value, but the overall positive rate of IgG was still in high level (95.85%).
| Clinical positive samples | Clinical negative samples | |
|---|---|---|
| Sample quantity | 217 | 158 |
| IgG & IgM positive | 112 | 0 |
| IgG positive | 208 | 3 |
| IgM positive | 2 | 1 |
| Sensitivity (%) | 95.85 | |
| Specificity (%) | 97.47 |
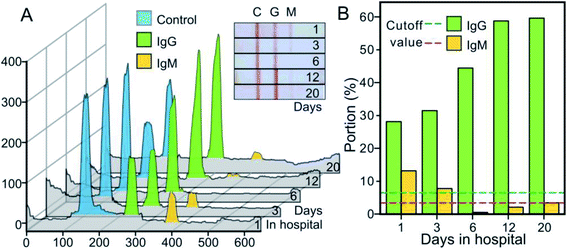
For the transmission control of infectious diseases and the treatment of patients, it is important to understand the time point of infection from clinical specimens. For a COVID-19 patient with moderate clinical symptoms, we performed a series of IgM/IgG rapid tests from the day of admission (the second day after rRT-PCR test positive) and the following three weeks treatment. The test results were digitized for quantitative analysis, as shown in Fig. 4. We noticed that the IgM was at a high level (13.15%) on the day of admission, while the IgG level was relatively low (28.16%). After treatment, the IgM level decreased rapidly, and the value was below the Cutoff by the 6th day after admission. However, the IgG level increased rapidly (31.50%) on the 6th day of admission, and remained above 45% in the three consecutive tests. During the clinical rehabilitation and until the patient was discharged from our hospital, the IgG level remained at a relatively high level (60.09%, 20th day). Due to limited time, we did not conduct more tests of the IgM/IgG levels of patients after discharge.
3.3 Interference
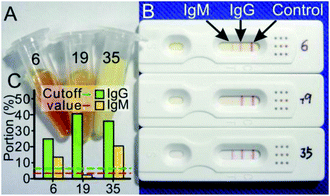
A total of 5 cases with hyperbilirubinemia and 9 cases with hyperlipidemia of confirmed cases were included in this study (the serum concentrations of the bilirubin, cholesterol and triglyceride were higher than the normal range, details in Table S3†), of which the highest values of the samples and the visual and quantitative results were shown in Fig. 5. The results manifested that these interfering substances did not affect the IgM/IgG rapid test results.4. Discussions
We have developed a rapid IgM/IgG antibody detection kit for COVID-19. Sensitivity and specificity of this kit have been evaluated. Combined with our data analysis method, this kit can meet the need for rapid COVID-19 detection in any institution with medium configuration.In this manuscript, the detailed working documents for the preparation of the kit are also provided to facilitate any countries and institutions to develop and manufacture the products earlier to meet the emergency needs of the current global pandemic of COVID-19.
Because the COVID-19 recombinant antigen was used in this kit, the test result is fairly specific for COVID-19. At the same time, within the titer ranges of positive samples of the COVID-19 IgM/IgG antibodies, the test results of our products did not show a hook effect. The sensitivity and specificity of this kit can reach 95.85% and 97.47%, respectively. However, false negative results still can be found (Table 1). We speculate the reasons for the false negative results are as follows: (1) low antibody concentration. When the IgM and IgG levels are below the limit (cutoff value) of this kit, the test result will be assessed as negative. (2) The variation of individual immune response of COVID-19.
According to the experience of SARS-CoV treatment, the IgM antibody is the first defence and can be detected in the serum after 1 to 6 days of infection.13 After 8 days, the most abundant antibody IgG can be detected.13 Based on the results of the patient with moderate clinical symptoms, we speculate that by the 6 days of admission, the IgM level had decreased below the cutoff value (Fig. 4). Since the IgM antibody will decrease and disappear after 2 weeks of infection, the single IgM test will lead to more false negative results and is likely not effective enough. Because the false negative results will bring more opportunities to infect other people, we suggest that the multiple and combined IgM/IgG test should be carry out for the patients and suspected cases, and the quantitative analysis should be conduct simultaneously. It means a lot for infection analysis, avoid false negative results and better control of the transmission.
Because COVID-19 infections originate from lungs rather than the upper respiratory tract,1 at the early stage of infection, the rRT-PCR examination with nasopharyngeal swab or sputum may not correctly detect the nucleic acid of COVID-19, and this may be one of the reasons of the higher false negative rate of rRT-PCR. The rapid COVID-19 IgM/IgG test kit owns multiple advantages: it can be performed at the bedside, in any clinic or laboratory, airport or railway station.15 Compared with complicated sample collection of nasopharyngeal swab or sputum for rRT-PCR, it uses serum or plasma as a test sample, and blood samples collection is much easier than nasopharyngeal swab.16 Therefore, it can greatly reduce the risk of cross infection for medical staff. In addition, it is rapid, simple and fast to operate, does not require expensive equipment or certified laboratories or specific training, making the large scale screening more convenient.17
Another potential application of this kit is to screen the asymptomatic COVID-19 carriers. It is reported that asymptomatic carriers can transmit COVID-19 virus constantly,18 and this makes the current COVID-19 outbreak control more complicated because of the lack of convenient test kit or method to discriminate the asymptomatic carriers from large crowds.19 This IgG/IgM rapid test kit can also be applied to large scale screening of patients with asymptomatic infections. Of course, this rapid test cannot replace rRT-PCR in confirmation of COVID-19 infection, but it can be used as an alternative supplement for rRT-PCR. Together with other test results, we speculate that combining the IgM/IgG rapid test with rRT-PCR, it can provide more accurate information for the diagnosis of COVID-19 infection.
Currently, due to the rapid outbreak of COVID-19 and personal behaviour regulation, we are unable to carry out the larger range of research activities to cover all genetic mutation-related antigens, and not yet conducted sufficient tests to verify the cross-reaction with antibodies of other virus infections (such as influenza virus). However, our results have verified that the most common interfering substances such as bilirubin and lipid (triglycerides, cholesterol) will not interfere with the test.
Another significance of this study is the detailed and objective method for non-naked eye assessment. Due to personal experience and subjectivity, it will inevitably cause the false judgments of the test results, especially for the results with low antibody concentrations. We use an office scanner to digitalize the test results, and FiJi software to quantify the test bands. Together with the appropriate confidence interval (CI), it effectively avoids the possible mistakes of naked eye judgment. Of course, if the portable device can be developed to assist the real-time data analysis, it ensured that more patients or suspected cases can be benefit from this kit.
According to current knowledge and information, IVD companies in several countries are developing or have developed similar products, including only IgM and IgM/IgG combination tests. For various reasons, we are unable to obtain more details about these techniques and performances, and there are not enough publications for reference. We believe that with the development of epidemics and better products for clinical use, more publications about COVID-19 examination will be available, which will be very helpful for our subsequent comparative research.
In addition, some limitations must be pointed out in this study. Firstly, for time limited, the performance and efficiency of this product was only verified in serum and plasma, the fingertip blood or venous whole blood have not been fully evaluated yet. Secondly, only a moderate number of samples were tested in this study. Because the number of COVID-19 patients has decreased significantly and scattered in China, larger-scale tests cannot be performed in a short time. At last, the interference validation of potential interfering substances, only a few number cases of bilirubin, cholesterol and triglyceride were verified, the other substances, such as rheumatoid factor (RF) and antinuclear antibody (ANA) on the product are on-going.
5. Conclusions
We have used the colloidal gold immunochromatography technology to develop a kit for rapid COVID-19 IgM/IgG detection. It only takes 10–15 minutes to generate results that are visible to the naked eye, without special requirements for equipment and personnel. With the simple and easy digitalization method, the test results can be semi-quantified and analysed to determine whether the patient has been infected with COVID-19 recently. The results indicated that this kit is sensitive and specific to COVID-19, and showed huge potential for rapid and large scale screening. At the present stage, it cannot replace rRT-PCR for nucleic acid examination and diagnosis, but it can be used as an effective supplementary test in clinical applications.Ethical statement
(1) The Chinese novel coronavirus pneumonia diagnosis and treatment plan (trial version 7) issued by National Health Commission of the People's Republic of China in March 04, 2020.(2) All experiments were followed the work flow for prevention and detection of COVID-19 of our Hospital (issued in Jan 25, 2020).
(3) In Dec 5, 2017, our laboratory had been approved for PCR examination of clinical sample by the Zhejiang Center for Clinical Laboratory (No. ZJ3086). In pandemic, we were also the designated test center for COVID-19 detection (nucleic acid and antibody) approved by the Health Commission of Zhejiang Province.
(4) This study has been supported by Ethics Committee of this Hospital and the Scientific Research Center (No. 2020-KY-0039). The informed consent had been signed for all samples collection and examination, and now stored in this Center.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the National Natural Scientific Foundation of China (No. 81771636, 81701426), and the Major Project of Wenzhou Science & Technology Bureau (No. ZS2017012).References
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu and X. Gu, Lancet, 2020, 395, 497–506 CrossRef CAS.
- T. G. Ksiazek, D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer and W. Lim, N. Engl. J. Med., 2003, 348, 1953–1966 CrossRef CAS PubMed.
- A. M. Zaki, S. V. Boheemen, T. M. Bestebroer, A. D. Osterhaus and R. A. Fouchier, N. Engl. J. Med., 2012, 367, 1814–1820 CrossRef CAS PubMed.
- A. E. Gorbalenya, S. C. Baker, R. S. Baric, R. J. de Groot, C. Drosten, A. A. Gulyaeva, B. L. Haagmans, C. Lauber, A. M. Leontovich, B. W. Neuman, D. Penzar, S. Perlman, L. L. M. Poon, D. V. Samborskiy, I. A. Sidorov, I. Sola and J. Ziebuhr, Nat. Microbiol., 2020, 5, 536–544 CrossRef PubMed.
- WHO Timeline COVID-19, This statement is updated on an ongoing basis, in response to evolving events and common media queries, https://www.who.int/news-room/detail/08-04-2020-who-timeline---covid-19, accessed May, 7. 2020.
- Centers for Disease Control and Prevention, People who are at higher risk for severe illness, Centers for Disease Control and Prevention, https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-at-higher-risk.html, accessed May, 7. 2020 Search PubMed.
- W. Guan, Z. Ni, Y. Hu, W. Liang, C. Ou, J. He, L. Liu, H. Shan, C. Lei and D. S. Hui, N. Engl. J. Med., 2020, 382, 1708–1720 CrossRef CAS PubMed.
- National Health Commission of the People's Republic of China, The Chinese novel coronavirus pneumonia diagnosis and treatment plan (trial version 7), National Health Commission of the People's Republic of China, http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml, accessed May, 7. 2020 Search PubMed.
- Roundups for journalists, Expert comments on different types of test for COVID-19, https://www.sciencemediacentre.org/expert-comments-on-different-types-of-test-for-covid-19/, accessed May, 7. 2020 Search PubMed.
- W. Yam, K. Chan, L. Poon, Y. Guan, K. Yuen, W. Seto and J. Peiris, J. Clin. Microbiol., 2003, 41, 4521 CrossRef CAS PubMed.
- Medical News Today, How do SARS and MERS compare with COVID-19?, Medical News Today, https://www.medicalnewstoday.com/articles/how-do-sars-and-mers-compare-with-covid-19, accessed May, 7. 2020 Search PubMed.
- FDA, Cellex emergency use authorization, FDA, https://www.fda.gov/media/136622/download, accessed May, 7. 2020 Search PubMed.
- H. Lee, B. Lee, S. Seok, B. MinWon, L. HuiYoung, K. DongJae, N. YiRang, N. KyoungJin, P. SungHoon, K. D. Noton, K. Hiroaki, N. Mina, H. SukJin and P. JaeHak, J. Vet. Sci., 2010, 11, 165–167 CrossRef PubMed.
- National Health Commission and National Administration of Traditional Chinese Medicine, Chin. Med. J., 2020, 133, E027 Search PubMed.
- A. J. Singer, J. Williams, M. Taylor, D. L. Blanc and H. C. T. Jr, Am. J. Emerg. Med., 2015, 33, 776–780 CrossRef PubMed.
- J. Xiang, M. Yan, H. Li, T. Liu, C. Lin, S. Huang and C. Shen, medRxiv, 2020 DOI:10.1101/2020.02.27.20028787.
- H. Gao, Y. Li, Z. Xu, Y. Wang, H. Wang, J. Cao, D. Yuan, L. Li, Y. Xu, Z. Zhang, Y. Huang, J. Lu, Y. Liu and E. Dai, Chin. Med. J., 2020, 133, E024 Search PubMed.
- L. Zou, F. Ruan, M. Huang, L. Liang, H. Huang, Z. Hong, J. Yu, M. Kang, Y. Song, J. Xia, Q. Guo, T. Song, J. He, H.-L. Yen, M. Peiris and J. Wu, N. Engl. J. Med., 2020, 382, 1177–1179 CrossRef PubMed.
- C. Rothe, M. Schunk, P. Sothmann, G. Bretzel, G. Froeschl, C. Wallrauch, T. Zimmer, V. Thiel, C. Janke, W. Guggemos, M. Seilmaier, C. Drosten, P. Vollmar, K. Zwirglmaier, S. Zange, R. Wolfel and M. Hoelscher, N. Engl. J. Med., 2020, 382, 970–971 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04107f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |