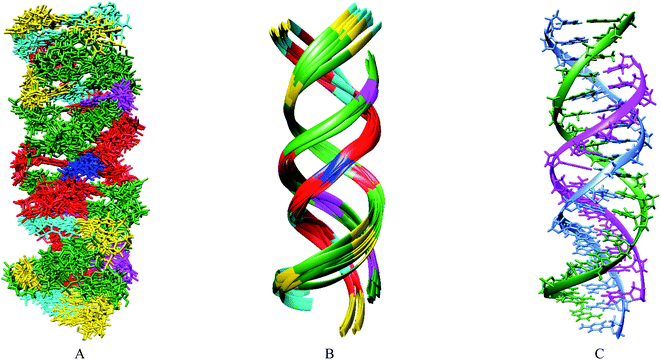Open Access Article
Open Access ArticleParallel motif triplex formation via a new, bi-directional hydrogen bonding pattern incorporating a synthetic cyanuryl nucleoside into the sense chain†
Akihiko Hatano *a,
Kei Shimazakia,
Maina Otsub and
Gota Kawaib
*a,
Kei Shimazakia,
Maina Otsub and
Gota Kawaib
aDepartment of Chemistry, Shibaura Institute of Technology, 307 Fukasaku, Minuma-ku, Saitama-City, Saitama 337-8570, Japan. E-mail: a-hatano@sic.shibaura-it.ac.jp
bDepartment of Life and Environmental Sciences, Faculty of Engineering, Chiba Institute of Technology, 2-17-1 Tsudanuma, Narashino, Chiba 275-0016, Japan
First published on 12th June 2020
Abstract
This research presents the first example of the formation of a triplex via hydrogen bonding of a synthetic cyanuryl nucleoside (Cn) composed of a 6-membered ring compound (triazine-2,4,6-trione) which functions as a pyrimidine base. The Cn was able to form the triad via hydrogen bonding in two directions with two adenines, one in the antisense and one in the parallel chain. The thermal stability of the duplex between the antisense and sense chains was evaluated via its UV melting temperature. The melting curves of triplexes possessing the cyanuryl nucleoside (sense chain) and two adenines (antisense and parallel chains) were biphasic. To prove the formation of hydrogen bonding between the cyanuryl nucleoside and the adenine base toward the major groove, structural analysis via NMR was undertaken. A cyanuryl nucleoside containing three 15N in triazine-2,4,6-trione was synthesized first, and then the 15N cyanuryl nucleoside was incorporated into target sequences. The triplex containing the A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A triad was analyzed via 1H NMR, coupled and decoupled with 15N. This triad has two imino proton signals derived from the cyanuryl nucleoside, split according to the 15N coupling condition, at low field. These results are evidence of the formation of hydrogen bonds between the Cn and adenosine. The solution structure of the triplex was analyzed via NOE information from the imino proton. The cyanuryl nucleoside-containing triplex forms a right-handed helical structure similar to natural triplex DNA, albeit DNA having an enhanced pyrimidine analog in the sense chain capable of bidirectional hydrogen bonding with high sequence selectivity.
A triad was analyzed via 1H NMR, coupled and decoupled with 15N. This triad has two imino proton signals derived from the cyanuryl nucleoside, split according to the 15N coupling condition, at low field. These results are evidence of the formation of hydrogen bonds between the Cn and adenosine. The solution structure of the triplex was analyzed via NOE information from the imino proton. The cyanuryl nucleoside-containing triplex forms a right-handed helical structure similar to natural triplex DNA, albeit DNA having an enhanced pyrimidine analog in the sense chain capable of bidirectional hydrogen bonding with high sequence selectivity.
Introduction
Triplex DNA can be used as a sequence reading agent,1 with potential use as an anti-gene agent2 and a tool in molecular biology.3–6 The characterization of triplex DNA differs according to the orientation and the base composition of the third strand.7 A pyrimidine-rich third strand binds to the duplex purine strand in parallel (T·A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T and C+·G
T and C+·G![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C),8–11 while a purine base-rich third strand assumes an antiparallel orientation (G·G
C),8–11 while a purine base-rich third strand assumes an antiparallel orientation (G·G![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, A·A
C, A·A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T and T·A
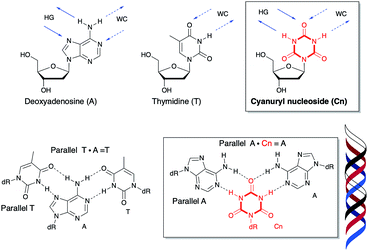
T and T·A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T). The presence of a purine-rich sequence in the sense chain has a large impact on the stability of triplex formation. Purine bases in the sense chain can interact with the corresponding base of the third strand forming a Hoogsteen base pair in the triplex.12–15 Pyrimidine bases have only a single heteroatom which can participate in hydrogen bonding toward the major groove. No natural nucleic acids can recognize the pyrimidine part of the sense chain (G·C and A·T) with high sequence selectivity and affinity. If a six-membered ring compound in the sense chain could interact with the two bases of the antisense chain and the third strand toward the major groove via bidirectional hydrogen bonding, it would be the first such example.
T). The presence of a purine-rich sequence in the sense chain has a large impact on the stability of triplex formation. Purine bases in the sense chain can interact with the corresponding base of the third strand forming a Hoogsteen base pair in the triplex.12–15 Pyrimidine bases have only a single heteroatom which can participate in hydrogen bonding toward the major groove. No natural nucleic acids can recognize the pyrimidine part of the sense chain (G·C and A·T) with high sequence selectivity and affinity. If a six-membered ring compound in the sense chain could interact with the two bases of the antisense chain and the third strand toward the major groove via bidirectional hydrogen bonding, it would be the first such example.
Recently, a variety of nucleosides have been designed for inversion site recognition via multi- and expanded hydrogen bonding.16–25 There are no natural bases which recognize the sense chain containing a pyrimidine-rich sequence via stable hydrogen bonding toward the major groove. Ts'o's group previously modeled and tested deoxypseudocytidine and deoxypseudouridine in the sense chain for their third strand binding ability.26,27 Ganesh's group reported that 5-aminouracil in the sense chain was capable of recognizing T (pyrimidine), G (purine), and 2-aminopurine (unnatural purine) in the antiparallel chain.28–30 Cyanuric acid as triazine-2,4,6-trione has the six-membered ring triazine compound constructed of three ketones and three NH groups (Fig. 1). This compound is structurally similar to the hydrogen bonding portion of thymine,31–33 and thus, has the potential to hydrogen bond with an adenine base two ways. We hypothesize that introducing a cyanuryl nucleoside into a DNA sequence could allow the sequence to function as a sense chain possessing the functional groups necessary for two directional interaction. This sense chain possessing six-membered rings could then form a triplex with the adenine of an antisense chain and the adenine of a third strand (Fig. 1).
In this study, the triplex formation ability of a sense chain containing a cyanuryl nucleoside was evaluated. In addition, the tertiary structure of the triplex was calculated using the NOE in 1H NMR by incorporating a 15N into the base moiety.
Results and discussion
Thermal denaturation analysis of the duplex
The ability of the cyanuryl nucleoside (Cn) to hydrogen bond with the adenine base was evaluated via the duplex melting behavior. Table 1 shows the melting point of the duplex with the cyanuryl nucleoside introduced at various positions in the sequences, and Fig. 2 shows the Tm profiles of the duplexes. The normal complementary duplex of dA15:dT15 melted at 46 °C (Entry 1). The duplex containing one cyanuryl nucleoside at the center position of the dT15 sequence had a Tm of 40 °C (Entry 1, dT7CnT7). Following the introduction of two cyanuryl nucleosides at the center position of dT15 (Entry 2, dT6Cn2T7), the Tm was lowered drastically to 27 °C. However, T rich sequences containing one or two 2′-deoxycytosines at the center position (a mismatch sequence) melted at 25, 14 °C, respectively (Entry 1, dT7C1T7, Entry 2, dT6C2T7). The ability to hydrogen bond of an A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cn base pair was lower than the ability of an A
Cn base pair was lower than the ability of an A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T complementary base pair; however, the ability to hydrogen bond of an A
T complementary base pair; however, the ability to hydrogen bond of an A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cn base pair was more effective than that of an A
Cn base pair was more effective than that of an A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mismatch pair. In the case of an A
C mismatch pair. In the case of an A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mismatch pair, the Tm varied depending on the position of the mismatch base. For instance, the Tm of the mismatch oligonucleotide at the center position above (Entry 1) was 25 °C, while the Tm of the A:C mismatch oligonucleotide at the penultimate base position was 35 °C (Entry 5) under the same conditions. On the other hand, the oligonucleotide obtained by introducing the cyanuryl nucleoside into the 5′ end of the sequence had a ΔTm of −7 °C (Entry 5), and the oligonucleotide introducing a cyanuryl nucleoside at each end had a ΔTm of −4 °C (Entry 3). Although the stability of the base pair between adenine and triazine-2,4,6-trione (A
C mismatch pair, the Tm varied depending on the position of the mismatch base. For instance, the Tm of the mismatch oligonucleotide at the center position above (Entry 1) was 25 °C, while the Tm of the A:C mismatch oligonucleotide at the penultimate base position was 35 °C (Entry 5) under the same conditions. On the other hand, the oligonucleotide obtained by introducing the cyanuryl nucleoside into the 5′ end of the sequence had a ΔTm of −7 °C (Entry 5), and the oligonucleotide introducing a cyanuryl nucleoside at each end had a ΔTm of −4 °C (Entry 3). Although the stability of the base pair between adenine and triazine-2,4,6-trione (A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cn) was lower than the stability of the matching pair between adenine and thymine (A
Cn) was lower than the stability of the matching pair between adenine and thymine (A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T), the duplex containing the base pair derived of adenine and triazine-2,4,6-trione (A
T), the duplex containing the base pair derived of adenine and triazine-2,4,6-trione (A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cn) was more stable than the duplex containing the A
Cn) was more stable than the duplex containing the A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mismatch pair (Entry 1–6). Thus, we thought that triazine-2,4,6-trione as a base might form hydrogen bonds with adenine.
C mismatch pair (Entry 1–6). Thus, we thought that triazine-2,4,6-trione as a base might form hydrogen bonds with adenine.
| Entry | Sequence | Tm/°C | ||
|---|---|---|---|---|
| X | ||||
| T | Cn | C | ||
| 1 | 5′-TTT TTT TXT TTT TTT | 46 | 40 | 25 |
| 2 | 5′-TTT TTT XXT TTT TTT | 27 | 14 | |
| 3 | 5′-TXT TTT TTT TTT TXT | 42 | 30 | |
| 4 | 5′-TXX TTT TTT TTT TTT | 38 | 33 | |
| 5 | 5′-TXT TTT TTT TTT TTT | 39 | 35 | |
| 6 | 5′-TXT TTT TXT TTT TXT | 20 | 15 | |
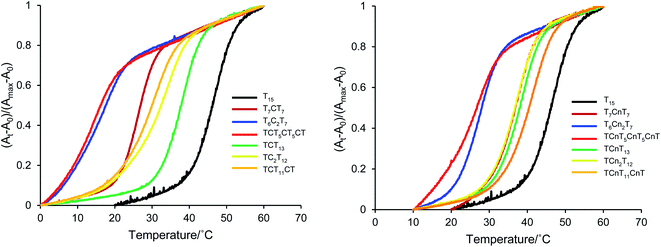 | ||
| Fig. 2 Melting profile of duplexes between A15 and T rich 15 mer oligonucleotides incorporating unnatural cyanuryl nucleosides or natural deoxycytidines. | ||
Thermal denaturation analysis of the triplex
The predicted formation of triads consisting of Cn in the central position of the oligonucleotide sequence are shown in Fig. 1. Parallel triplex formation was studied with respect to melting temperature, as shown in Table 2 and Fig. 3. First, the melting temperature profiles of triplexes were obtained at pH 5.8 and pH 6.8. The triplex did not form at pH 6.8, because the nitrogen atom at the 3 position of cytidine in the sequence is not protonated at this pH. At pH 5.8, stable triplex formation was observed for the control triplex with a T·A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T triad (Tm1 45 °C, black line, Entry 8). The triplex derived from the Cn·A
T triad (Tm1 45 °C, black line, Entry 8). The triplex derived from the Cn·A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T triad melted at 38 °C (brown line, Entry 9). Thus, the stability of hydrogen bonding upon incorporation of Cn into the parallel sequence was enhanced relative to the thymine base. However, the melting point of this Cn·A
T triad melted at 38 °C (brown line, Entry 9). Thus, the stability of hydrogen bonding upon incorporation of Cn into the parallel sequence was enhanced relative to the thymine base. However, the melting point of this Cn·A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T triad was almost same as that of the triplex possessing the A·Cn
T triad was almost same as that of the triplex possessing the A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A triad (42 °C, red line, Entry 11), and higher than Tm of the A·T
A triad (42 °C, red line, Entry 11), and higher than Tm of the A·T![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A triplex (27 °C, green line, Entry 10). That is, the parallel triplex having a modified base in the central position melts at a temperature 15 °C higher than the third strand mismatch triad. In the pyrimidine analogue triazine-2,4,6-trione, N–H substitutes for C–H at the 5 position of uracil, and also substitutes C
A triplex (27 °C, green line, Entry 10). That is, the parallel triplex having a modified base in the central position melts at a temperature 15 °C higher than the third strand mismatch triad. In the pyrimidine analogue triazine-2,4,6-trione, N–H substitutes for C–H at the 5 position of uracil, and also substitutes C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O for C–H at the 6 position of uracil. Thus, the cyanuryl nucleoside can form Hoogsteen-like hydrogen bonds with an adenine of the third parallel chain facing the major groove; that is, the cyanuryl base can serve as the middle base of a triplex triad. Although the melting temperature profile of the A·Cn
O for C–H at the 6 position of uracil. Thus, the cyanuryl nucleoside can form Hoogsteen-like hydrogen bonds with an adenine of the third parallel chain facing the major groove; that is, the cyanuryl base can serve as the middle base of a triplex triad. Although the melting temperature profile of the A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T triplex shows one sigmoidal curve (blue line, Fig. 3, Entry 12), this triplex curve is different from the Cn
T triplex shows one sigmoidal curve (blue line, Fig. 3, Entry 12), this triplex curve is different from the Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T duplex curve. We believe that the observed A·Cn
T duplex curve. We believe that the observed A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T curve represents two dissociation processes, for both the duplex and triplex, simultaneously. The Tm1 of triplex A·Cn
T curve represents two dissociation processes, for both the duplex and triplex, simultaneously. The Tm1 of triplex A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T (42.0 °C, Entry 11) is almost the same as for A·Cn
T (42.0 °C, Entry 11) is almost the same as for A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A (42.0 °C, Entry 12). This phenomenon can be explained as recognition of the middle base and the third strand base for Hoogsteen-type hydrogen bonding by A·Cn.
A (42.0 °C, Entry 12). This phenomenon can be explained as recognition of the middle base and the third strand base for Hoogsteen-type hydrogen bonding by A·Cn.
| Entry | Oligodeoxynucleotides and inserted nucleoside | Tm/°C | |||||
|---|---|---|---|---|---|---|---|
| Parallel: Z | · | Sense: X | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) |
Antisense: Y | Tm1 (triplex) | Tm2 (duplex) | |
| 8 | T | · | A | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) |
T | 45 | 63 |
| 9 | Cn | · | A | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) |
T | 38 | 62 |
| 10 | A | · | T | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) |
A | 27 | 62 |
| 11 | A | · | Cn | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) |
T | 42 | 53 |
| 12 | A | · | Cn | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) |
A | 42 | 60 |
| Sequence | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parallel | 5′ | T | C | T | T | C | T | T | Z | T | T | T | T | C | T | T | ||||
| Sense | 5′ | C | C | A | G | A | A | G | A | A | X | A | A | A | A | G | A | A | G | C |
| Antisense | 3′ | G | G | T | C | T | T | C | T | T | Y | T | T | T | T | C | T | T | C | G |
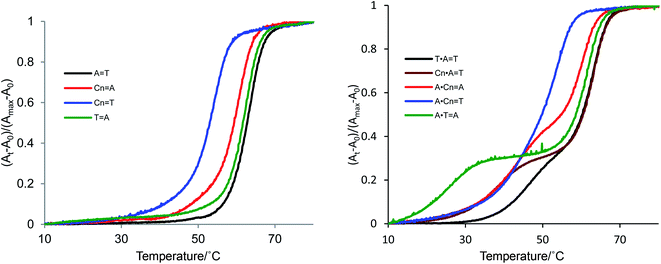 | ||
| Fig. 3 Melting profiles of duplexes and triplexes incorporating synthetic cyanuryl nucleosides (Cn) in the center of the sequences in Table 2. Conditions of duplex formation: UV absorption detected at 260 nm; concentrations of oligodeoxynucleotides were 1.0 μM in 10 mM phosphate buffer (pH 6.8) and 200 mM NaCl. Conditions of triplex formation: UV absorption detected at 260 nm; concentrations of oligodeoxynucleotides were 0.8 μM per chain of triplex and 1.0 M of NaCl in 10 mM Mcllvaine buffer (phosphate–citric acid, pH 5.8). | ||
The Tm values of the duplexes containing T![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A and A
A and A![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T in the central positions of the sequences were fundamentally identical (63 and 62 °C, respectively). The Tm of duplex Cn
T in the central positions of the sequences were fundamentally identical (63 and 62 °C, respectively). The Tm of duplex Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T was 53 °C, less than the Tm of duplex Cn
T was 53 °C, less than the Tm of duplex Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A (60 °C). The Cn
A (60 °C). The Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T duplex possesses the mismatch pair, lowering the melting point.
T duplex possesses the mismatch pair, lowering the melting point.
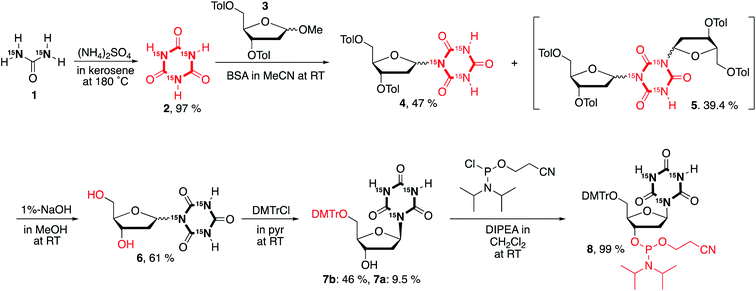
Synthesis of a cyanuryl nucleoside composed of three 15N atoms in the triazine-2,4,6-trione
An attempt was made to synthesize the cyanuryl nucleoside according to ref. 31; however, the coupling reaction of triazine-2,4,6-trione (cyanuric acid) composed of three 15N atoms and the protected ribosyl moiety varied somewhat from the literature. Thus, the procedure was modified as follows. Triazine-2,4,6-trione incorporating three 15N isotopes was produced by condensation of urea containing two 15N in kerosene and (NH4)2SO4 by heating at high temperature.34,35 The trimerization of urea takes place at 180 °C in kerosene, and produces 15NH3 gas, giving triazine-2,4,6-trione incorporating three 15N stable isotopes in good yield (97%) as a white-brown precipitate. This 15N-containing compound shows a strong singlet in the 15N-NMR at 130.4 ppm (see ESI†), indicating successful incorporation of three identical nitrogen isotopes into the triazine-2,4,6-trione. In addition, the 13C-NMR signal of triazine-2,4,6-trione is split into an overlapping doublet of doublets (a triplet at 150.5 ppm) by the effectively magnetically equivalent neighboring two 15N isotopes. Mass spectrometry also confirmed the introduction of three stable 15N isotopes into the synthetic cyanuric acid (2, EIMS: 132). A coupling reaction was also performed between O-silylated 15N-containing triazine-2,4,6-trione and 3,5-ditoluoyl-1-α/β-methoxy-2-deoxy-D-ribose (3) in the presence of a Lewis acid catalyst (TMSOTf) according to ref. 31. Interestingly, the behavior of this coupling reaction differs from that of the 14N-containing heterocyclic triazine-2,4,6-trione. For the triazine-2,4,6-trione incorporating three stable 15N isotopes, coupling of the silylated 15N cyanuric acid and the protected 1-methoxyribose was favored over the reaction giving the target cyanuryl nucleoside, producing an unexpected compound composed of one triazine-2,4,6-trione and two 3,5-ditoluoyl-2-deoxy-D-riboses (5) in 39.4% yield. It is believed that the nucleophilicity of 15N in the silylated triazine-2,4,6-trione is higher than that of the 14N. When the concentration of 15N-containing triazine-2,4,6-trione was doubled, the yield of target cyanuryl nucleoside 6 was 47%, and the yield of compound 5 was 39%, slightly less than when using the initial concentration. Ditoluoyl nucleoside 4 was hydrolyzed with 1% NaOH in MeOH to produce a mixture of α/β cyanuryl nucleosides 6. The α/β isomers 7 varied in mobility on TLC, leading to isolation of isomers 7a (α) and 7b (β) by column chromatography. NOESY analysis showed that the primary isomer is the β form (7b), because this compound was observed to have a correlation between the anomeric 1′ and 4′ protons. The 3′ OH of compound 7b was phosphitylated to give 2-cyanoethyl phosphoramidite 8 in 99% yield. Having obtained compound 8, oligodeoxynucleotides containing the 15N cyanuryl β-nucleoside were readily prepared via standard automated DNA synthetic methods (Scheme 1).Structural analysis via NOE NMR spectroscopy
To study the formation of hydrogen bonds and the structures of triplexes of oligodeoxynucleotides having the cyanuryl nucleoside triad in the center position, NMR spectra were obtained, and the structural calculation was performed.Fig. 4 shows the imino proton region of the 600 MHz 1H-NMR spectra of the triple helix with and without 15N decoupling. Imino proton signals were assigned except for G1 of the antisense chain and T3 of the parallel chain. The imino proton signals for the three cytosine residues of the parallel chain were clearly observed (9.3–10.0 ppm), indicating that these cytosine residues are protonated. Notably, intra-residual NOEs between the imino proton (H3) and amino proton (H41/H42) were also observed (data not shown). The two signals at 12.5 (Cn10, H5) and 13.4 (Cn10, H3) ppm are split by coupling between 1H and 15N. These two signals are shifted to lower magnetic field and are coupled to a 15N; therefore, these two signals were assigned to the two protons binding to an 15N atom in the cyanuryl nucleoside. In addition, these protons clearly show the formation of hydrogen bonds between the cyanuric acid moiety and adenine, resulting in triplexes of antisense and parallel chains with the sense chain containing a cyanuryl nucleoside.
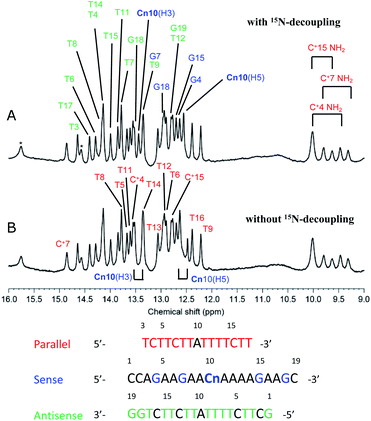 | ||
Fig. 4 600 MHz 1H-NMR spectra of the A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A triple helix (Table 2, Entry 11). Imino and amino proton regions are shown with (A) and without (B) 15N-decoupling. Signal assignments are indicated by magenta, cyan, and green for parallel, sense, and antisense, respectively. A triple helix (Table 2, Entry 11). Imino and amino proton regions are shown with (A) and without (B) 15N-decoupling. Signal assignments are indicated by magenta, cyan, and green for parallel, sense, and antisense, respectively. | ||
Structural analysis via NMR
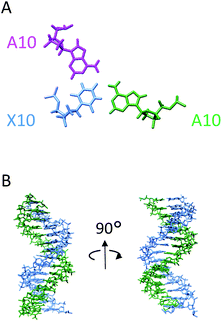
Based primarily on the formation of the base triple and the NOEs of the imino and amino protons, tertiary structures were calculated (Table 3). As shown in Fig. 5, the parallel chain located at the major groove of the duplex which is composed of the antisense chain and the sense chain. The triplex possessing A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A in the center position of the sequence has a small distortion at the end (Fig. 5B). These results indicate a right-handed triple helix that is globally similar to that of a normal DNA triplex. The X (
A in the center position of the sequence has a small distortion at the end (Fig. 5B). These results indicate a right-handed triple helix that is globally similar to that of a normal DNA triplex. The X (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cn) residue successfully formed the triplex as shown in Fig. 6A. The triad of two adenine and triazine-2,4,6-trione is located on the same plane in the triplex. The cyanuryl nucleoside base moiety is composed of a 6 membered ring, like a pyrimidine base, and can hydrogen bond in two directions 120° apart from each other. Thus, the cyanuryl base analog can form the triad between two adenines of the antisense and parallel chains via hydrogen bonding (Fig. 6A and 1). A normal DNA triplex has a triad constructed by Watson–Crick-type hydrogen bonding between the sense and antisense chains, and Hoogsteen-type hydrogen bonding for the parallel chain on the major groove side. The antisense and sense strands adopt a conformation similar to that in B-DNA during MD simulations (Fig. 6). All base pairs were maintained during the 10 ns MD simulation.
Cn) residue successfully formed the triplex as shown in Fig. 6A. The triad of two adenine and triazine-2,4,6-trione is located on the same plane in the triplex. The cyanuryl nucleoside base moiety is composed of a 6 membered ring, like a pyrimidine base, and can hydrogen bond in two directions 120° apart from each other. Thus, the cyanuryl base analog can form the triad between two adenines of the antisense and parallel chains via hydrogen bonding (Fig. 6A and 1). A normal DNA triplex has a triad constructed by Watson–Crick-type hydrogen bonding between the sense and antisense chains, and Hoogsteen-type hydrogen bonding for the parallel chain on the major groove side. The antisense and sense strands adopt a conformation similar to that in B-DNA during MD simulations (Fig. 6). All base pairs were maintained during the 10 ns MD simulation.
| a Average RMSD values between an average structure and the 10 converged structures were calculated. The converged structures did not contain experimental distance violations of >0.5 Å or dihedral violations of >5°. | |
|---|---|
| Number of experimental restraints | |
| Distance restraints | 125 |
| Sequential | 36 |
| Inter strands | 19 |
| Hydrogen bonding | 70 |
| Dihedral restraints | 150 |
| Planarity for base pairs | 33 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Number of experimental restraints | |
| Distance restraints | 125 |
| Sequential | 36 |
| Inter strands | 19 |
| Hydrogen bonding | 70 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| RMSD around the ideal values | |
| Bonds (Å) | 0.0026 ± 0.0001 |
| Angle (°) | 0.425 ± 0.005 |
Conclusion
In summary, a cyanuryl nucleoside has been developed which functions as the center base for triplex DNA. This synthetic nucleoside possesses two N–H hydrogen bonding donors and three C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O hydrogen bonding acceptors approximately 120° apart in a triazine-2,4,6-trione moiety. The cyanuryl nucleoside forms base triads via hydrogen bonding, some composed of Watson–Crick base pairs as the dominant pattern, and others utilizing a new pattern involving inversion at the major groove. It was hypothesized that the two imido sites of the triazine-2,4,6-trione cyanuryl nucleoside could recognize two adenine bases from two different directions (in the antisense and parallel chains). The melting points of duplexes and triplexes incorporating the cyanuryl nucleoside into the sequence were studied. For A·Cn
O hydrogen bonding acceptors approximately 120° apart in a triazine-2,4,6-trione moiety. The cyanuryl nucleoside forms base triads via hydrogen bonding, some composed of Watson–Crick base pairs as the dominant pattern, and others utilizing a new pattern involving inversion at the major groove. It was hypothesized that the two imido sites of the triazine-2,4,6-trione cyanuryl nucleoside could recognize two adenine bases from two different directions (in the antisense and parallel chains). The melting points of duplexes and triplexes incorporating the cyanuryl nucleoside into the sequence were studied. For A·Cn![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) A in the center of each sequence, biphasic melting was observed (Tm1 = 42 °C, Tm2 = 60 °C). The Tm results are indicative of triad formation via hydrogen bonding among triazine-2,4,6-trione and two adenine bases. Imino proton NMR analysis of the formation of triads containing the 15N-labeled cyanuryl nucleoside revealed the formation of hydrogen bonds between an adenine of the antisense chain and a Cn of the complementary sense chain, as well as between Cn (sense chain) and adenine (parallel chain). Imino and amino proton NOE information was used to calculate the triplex structure, showing that the parallel chain interacted in the major groove in the duplex, ultimately forming the triplex shown in Fig. 5. This study shows that a six-membered ring compound can function in the center sense chain of a triplex formed via bi-directional hydrogen bonding. This result may prove useful in the design of other novel synthetic nucleosides.
A in the center of each sequence, biphasic melting was observed (Tm1 = 42 °C, Tm2 = 60 °C). The Tm results are indicative of triad formation via hydrogen bonding among triazine-2,4,6-trione and two adenine bases. Imino proton NMR analysis of the formation of triads containing the 15N-labeled cyanuryl nucleoside revealed the formation of hydrogen bonds between an adenine of the antisense chain and a Cn of the complementary sense chain, as well as between Cn (sense chain) and adenine (parallel chain). Imino and amino proton NOE information was used to calculate the triplex structure, showing that the parallel chain interacted in the major groove in the duplex, ultimately forming the triplex shown in Fig. 5. This study shows that a six-membered ring compound can function in the center sense chain of a triplex formed via bi-directional hydrogen bonding. This result may prove useful in the design of other novel synthetic nucleosides.
Experimental section
General
All solvents and reagents were of reagent-grade quality and used without further purification. The TLC analysis was carried out on silica gel 60 F254 1.05554 (Merck). Column chromatography was performed using Wakogel C-300 (silica gel, Wako) or Silica gel 60 N (Kanto Chemical Co.). The NMR spectra obtained during the synthesis of the cyanuryl nucleoside were recorded on a JEOL ECS 400 (400.0 MHz for 1H; 100.4 MHz for 13C) spectrometer. The spectra were referenced to TMS in CDCl3 or CD3OD (internal standard). Complete assignment of all NMR signals was performed using a combination of 1H, 13C, COSY, HMQC, DEPT135 and NOESY experiments. The chemical shifts (δ) are reported in ppm; multiplicity is indicated by s (singlet), d (doublet), t (triplet), q (quartet), dd (doublet of doublets), ddd (doublet of doublet of doublets), m (multiplet), and br (broad). The coupling constants, J, are reported in Hz. Mass spectra were measured using ESI on a JEOL AccuTOF via the direct analysis in real time (DART) method.Cyanuryl nucleoside31
The cyanuryl nucleoside was synthesized according to the literature method.31 1H, 13C-NMR, and UV data matched the literature.15N3 cyanuric acid 2
15N2 urea (1.0 g, 17 mmol, 15NH2CO15NH2) and (NH4)2SO4 (100 mg, as catalyst) were added to 2 mL of kerosene at 180 °C and stirred for 7 h. The reaction mixture was cooled to room temperature at which point precipitate was visible at the bottom of the flask. The precipitate was filtered and dried in vacuo to afford compound 2 (15N3 cyanuric acid) as a pale yellow solid (0.657 g, 93%).1H NMR (DMSO-d6): δ 11.3 (br). 13C NMR (DMSO-d6): δ 150.5 (J = 17.7 Hz, t). 15N NMR (DMSO-d6): δ 130.5. EIMS m/z 135 [M+].
Compound 4 (α/β mixture)
1H NMR (CDCl3): δ 9.60 (2H, br), 7.87 (4H, m), 7.15 (4H, m), 6.57 (1H, m), 5.70 (0.6H, J = 5.6, 12.8 Hz, dd), 5.43 (0.4H, J = 7.6, 12 Hz, dd), 4.40–4.98 (3H, m), 3.18 (1H, br), 2.13–3.34 (1H, m), 2.92 (1H, br), 2.76 (1H, br), 2.31 (6H, m). ESIMS m/z 507 [M + Na]+. ESI-HRMS calcd for C24H36N3NaO8 [M + Na]+: 507.1294; found 507.1382.Compound 5 (dimer)
This compound has three diastereomeric mixtures (dR-α-Cy-α-dR, dR-α-Cy-β-dR, and dR-β-Cy-β-dR) varying in the coupling mode of the glycosyl bond between the triazine-2,4,6-trione and the two 3,5-ditoluoyl-1-α/β-methoxy-2-deoxy-D-riboses (3). Therefore, both 1H and 13C NMR resulted in multiple, broad signals.1H NMR (CDCl3): δ 7.93 (8H, br), 7.19 (8H, m), 6.62 (2H, br), 5.74 (1H, m), 5.45 (1H, m), 4.97 (1H, m), 4.34–4.68 (4H, m), 3.18 (1H, br), 2.98 (1H, br), 2.80 (1H, br), 2.36 (12H, m). ESIMS m/z 859 [M + Na]+. ESI-HRMS calcd for C24H2315N3NaO8 [M + Na]+: 859.8161; found 859.8289.
Compound 6 (α/β mixture)
1H NMR (CD3OD): δ 6.53 (1H, m), 6.45 (1H, m), 4.46 (1H, br), 4.24 (2H, m), 3.54–3.82 (5H, m), 4.97 (1H, m), 4.34–4.68 (4H, m), 2.82 (1H, m), 2.59 (2H, m), 2.11 (1H, m). 13C NMR (DMSO-d6): δ 150.8 (m), 150.1 (m), 88.8, 87.7, 84.1 (m), 72.7, 72.6, 63.9, 63.0, 38.2, 38.0. 15N NMR (CD3OD): δ 147.4, 144.7, 128.4. ESIMS m/z 271 [M + Na]+. ESI-HRMS calcd for C8H1115N3NaO6 [M + Na]+: 271.0457; found 271.0440.Compound 7b (β-form)
1H NMR (CDCl3): δ 7.46 (2H, m), 7.32 (4H, m), 7.24 (2H, m), 7.17 (1H, m), 6.81 (4H, m), 6.53 (1H, J = 4.4, 8.8, 11.0 Hz, ddd), 4.42 (1H, m), 3.94 (1H, m), 3.76 (1H, s), 3.30 (2H, m), 2.77 (1H, m), 2.13 (1H, m). 13C NMR (CDCl3): δ 158.7, 149.2 (J = 21 Hz, dd), 145.3, 136.3 (J = 3.9 Hz, d), 130.1, 130.0, 128.1, 127.2, 126.3, 112.6, 86.1 (J = 7.6 Hz, d), 82.5, 82.4, 71.8, 64.7, 54.4, 37.0. 15N NMR (CDCl3): δ 146.7, 128.5. ESIMS m/z 573 [M + Na]+. ESI-HRMS calcd for C29H2915N3NaO8 [M + Na]+: 573.1763; found 573.1777.Compound 7a (α-form)
1H NMR (CDCl3): δ 7.43 (2H, m), 7.30 (6H, m), 7.12 (1H, J = 7.0 Hz, t), 6.81 (4H, m), 6.71 (1H, J = 2.0, 3.3, 10.8 Hz, ddd), 4.36 (1H, J = 3.6 Hz, t), 4.27 (1H, J = 8.0 Hz, d), 3.77 (6H, s), 3.12 (2H, J = 4.6, 9.6, 30 Hz, ddd), 2.81 (1H, m), 2.21 (1H, br). 13C NMR (CDCl3): δ 158.6, 154.8 (J = 12.0 Hz, t), 153.7 (J = 14.8 Hz, t), 136.2 (J = 8.6 Hz, d), 130.1, 128.3, 127.9, 126.7, 113.3, 88.5, 86.4, 74.2, 64.7, 55.2, 45.1, 39.6. 15N NMR (CDCl3): δ 150.1, 145.9.Compound 8
1H NMR (CDCl3): δ 7.42 (2H, J = 6.8 Hz, d), 7.31 (4H, J = 2.8, 8.8 Hz, dd), 7.20 (2H, J = 7.6 Hz, t), 7.14 (1H, J = 7.6 Hz, d), 6.76 (4H, m), 6.58 (1H, m), 4.56 (1H, m), 4.09 (1H, m), 3.73 (8H, m), 3.49 (2H, m), 3.30 (2H, m), 3.05 (2H, J = 7.4, 14.4 Hz, dd), 2.80 (1H, m), 2.55 (2H, J = 6.2 Hz, t), 2.32 (1H, m), 1.20 (3H, J = 7.4 Hz, t), 1.11 (6H, J = 6.4 Hz, d), 0.99 (6H, J = 7.2 Hz, d). 13C NMR (CDCl3): δ 158.2 (J = 3.8 Hz, d), 150.7 (J = 12.0 Hz, t), 150.7 (J = 16.2 Hz, t), 150.0 (J = 16.7 Hz, t), 145.0, 136.3 (J = 7.6 Hz, d), 130.2, 130.2, 128.3, 127.6, 126.5, 117.6, 112.9, 86.0, 85.1, 85.0, 82.5, 82.4, 77.2, 73.9, 73.7, 64.3, 58.3, 58.1, 55.2, 44.6, 43.2, 43.0, 37.1, 24.6, 24.6, 24.4, 24.4, 20.3, 20.3. 15N NMR (CDCl3): δ 147.8, 134.2. 31P NMR (CDCl3): δ 149.2. ESIMS m/z 749 [M − H]−. ESI-HRMS calcd for C38H4514N215N3O9P [M − H]−: 749.2866; found 749.2837.Oligodeoxynucleosides
Prior to introduction to a standard automatic DNA synthesizer, 5′-O-dimethoxytritylation and 3′-O-phosphoramidation of the cyanuryl nucleoside were performed. DNA synthesis incorporating the cyanuryl nucleoside into the oligonucleotide sequences was performed by the BEX corporation (Tokyo, Japan). The synthetic oligonucleotide was purified on an OPC column, reversed phase HPLC, and then freeze-dried immediately (BEX Co., Ltd).Melting temperature analysis
Thermal studies were performed in Teflon-stoppered 1 cm path length quartz cells under an atmosphere of nitrogen using a JASCO V-630 Bio UV-Vis recording spectrophotometer (JASCO, Tokyo, Japan) equipped with a thermoprogrammer. Absorbance was monitored at 260 nm. The temperature was raised from 0 to 80 °C at a rate of 1.0 °C min−1. The solutions for the thermal denaturation studies of duplexes and triplexes were prepared at an oligomer concentration of 15 μM for the base-pairing studies in a pH 6.8 buffer containing 0.20 M NaCl and 10 mM sodium phosphate. Experiments performed under triplex formation conditions were carried out with 0.8 μM per chain of triplex and 1.0 M of NaCl in 10 mM Mcllvaine buffer (phosphate–citric acid, pH 5.8). All Tm values were determined from the highest point of the transition in plots of dA/dT versus temperature curves.NMR sample preparation of the triple helix
A mixture of 90% H2O and 10% D2O was used for experiments examining exchangeable protons. The final concentration was 2.0 mM per base for the oligonucleotide in 400 μL of solution. For experiments carried out in D2O, the DNA sample was lyophilized three times from D2O and then re-dissolved in D2O. The signal of H2O was used as the chemical shift reference, employing temperature correction (4.7 ppm at 7 °C).NMR experiments of the triple helix
NMR spectra of the triple helix were measured with an AVANCE 600 spectrometer (Bruker) at a probe temperature of 283 K. The 15N-labeled cyanuryl nucleoside DNA sample was dissolved in 300 μL of 20 mM potassium phosphate buffer (pH 5.9) with 5% D2O. The sample concentration was 0.2 mM for each strand. NMR data were processed with the TopSpin program (Bruker) and analyzed with the Sparky program.36 A NOESY spectrum measured with a mixing time of 150 ms was used to assign signals and measure NOE volumes.Structure calculation and molecular dynamics simulation of the triple helix
The structure of the triple helix was calculated based on NMR information obtained primarily from the imino proton signals. Fifty-five distance constraints were derived from the NOE volumes. Seventy distances for hydrogen bonding, 19 plane restraints for anti-parallel, and 14 plane restraints for parallel base pairs were used to form the 14 base-triples and 5 base-pairs. Because no strong NOE was observed between the base proton and H1′ in the NOESY spectra, all residues were assumed to adopt an anti-conformation, and 52 torsion restrains were used. Ninety-six backbone and ribose torsion restraints were used for the region of Watson–Crick base pairs to form the B-DNA conformation. Structures were calculated via the conventional method37 with the CNS_SOLVE program.38 Sixty-five structures were accepted and 10 structures with lowest energy were used to calculate the minimized averaged structure. NMR constraints and structural statistics are summarized in Table 3.A molecular dynamics (MD) simulation was performed with AMBER12.39 The ff12SB force field was used. The coordinates of the cyanuryl nucleoside were prepared using the UCSF Chimera40 program, and atomic charges were calculated with the ANTECHAMBER program in the AMBER12 suite. A 10 ns constant volume simulation was performed in the presence of 46 sodium ions and 7760 water molecules using the minimized averaged structure as the starting model. The MD trajectory was processed with the ptraj program in the AMBER12 suite. Molecular images were generated with the UCSF Chimera program.40
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. Masatoshi Kidowaki at the Shibaura Institute of Technology for the ESI mass spectroscopy analysis. The authors would like to thank NAI (https://www.nai.co.jp/) for the English language review. This work was supported in part by the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan. The urea compound incorporating two stable 15N isotopes was supplied by the Taiyo Nippon Sanso Corp.Notes and references
- G. Felsenfeld, D. R. Davis and A. Rich, J. Am. Chem. Soc., 1957, 79, 2023–2024 CrossRef CAS.
- T. Le Doan, L. Perrouault, D. Praseuth, N. Habhoub, J. L. Decout, N. T. Thuong, J. Lhomme and C. Hélène, Nucleic Acids Res., 1987, 15, 7749–7761 CrossRef CAS PubMed.
- H. E. Moser and P. B. Dervan, Science, 1987, 238, 645–650 CrossRef CAS PubMed.
- K. M. Vasquez and J. H. Wilson, Trends Biochem. Sci., 1998, 23, 4–9 CrossRef CAS PubMed.
- P. P. Chan and P. M. Glazer, J. Mol. Med., 1997, 75, 267–282 CrossRef CAS PubMed.
- N. T. Thuong and C. Hélène, Angew. Chem., Int. Ed., 1993, 32, 666–690 CrossRef.
- S. M. Mirkin, V. I. Lyamichev, K. N. Drushlyak, V. N. Dobrynin, S. A. Filippov and M. D. F. Kamenetskii, Nature, 1987, 330, 495–497 CrossRef CAS PubMed.
- M. Darren and F. R. Keith, Nucleic Acids Res., 1999, 27, 1569–1577 CrossRef PubMed.
- M. Riley, B. Maling and M. J. Chamberlin, J. Mol. Biol., 1966, 20, 359–389 CrossRef CAS PubMed.
- S. Arnott and E. Selsing, J. Mol. Biol., 1974, 88, 509–512 CrossRef CAS PubMed.
- L. Lavelle and J. R. Fresco, Nucleic Acids Res., 1995, 23, 2692–2705 CrossRef CAS PubMed.
- C. Y. Hung, G. Bi and P. S. Miller, Nucleic Acids Res., 1996, 24, 2606–2613 CrossRef PubMed.
- K. Hoogsteen, Acta Crystallogr., 1959, 12, 822 CrossRef CAS.
- D. W. Ussery and R. R. Sinden, Biochemistry, 1993, 32, 6206–6213 CrossRef CAS PubMed.
- R. Macaya, E. Wang, P. Schultze, V. Sklenář and J. Feigon, J. Mol. Biol., 1974, 225, 755–773 CrossRef.
- H. U. Stilz and P. B. Dervan, Biochemistry, 1993, 32, 2177 CrossRef CAS PubMed.
- D. M. Gowers and K. R. Fox, Nucleic Acids Res., 1999, 27, 1569–1571 CrossRef CAS PubMed.
- S. P. Parel and C. J. Leumann, Nucleic Acids Res., 2001, 29, 2260 CrossRef CAS PubMed.
- D. Rusling, V. E. C. Powers, R. T. Ranasinghe, Y. Wang, S. D. Osborne, T. Brown and K. R. Fox, Nucleic Acids Res., 2005, 33, 3025–3032 CrossRef CAS PubMed.
- Y. Hari, M. Akabane and S. Obika, Chem. Commun., 2013, 49, 7421–7423 RSC.
- A. Ohkubo, K. Yamada, Y. Ito, K. Yoshimura, K. Miyauchi, T. Kanamori, Y. Masaki, K. Seio, H. Yuasa and M. Sekine, Nucleic Acids Res., 2015, 43, 5675–5686 CrossRef CAS PubMed.
- S. Sasaki, Y. Taniguchi, R. Takahashi, Y. Senko, K. Kodama, F. Nagatsugi and M. Maeda, J. Am. Chem. Soc., 2004, 126, 516–528 CrossRef CAS PubMed.
- H. Okamura, Y. Taniguchi and S. Sasaki, Angew. Chem., Int. Ed., 2016, 55, 12445–12449 CrossRef CAS PubMed.
- S. Obika, Y. Hari, M. Sekiguchi and T. Imanishi, Chem.–Eur. J., 2002, 20, 4796–4802 CrossRef.
- Y. Hari, S. Obika and T. Imanishi, Eur. J. Org. Chem., 2012, 6381–6391 Search PubMed.
- T. L. Trapane, M. S. Christopherson, C. D. Roby, P. O. P. Ts'o and D. Wang, J. Am. Chem. Soc., 1994, 116, 8412–8413 CrossRef CAS.
- T. L. Trapane and P. O. P. Ts'o, J. Am. Chem. Soc., 1994, 116, 10437–10449 CrossRef CAS.
- V. S. Rana, D. A. Barawkar and K. N. Ganesh, J. Org. Chem., 1996, 61, 3578–3579 CrossRef CAS PubMed.
- V. S. Rana and K. N. Ganesh, Org. Lett., 1999, 1, 631–634 CrossRef CAS PubMed.
- V. S. Rana and K. N. Ganesh, Nucleic Acids Res., 2000, 28, 1162–1169 CrossRef CAS PubMed.
- D. Gasparutto, D. C. Cruz, A.-G. Bourdat, M. Jaquinod and J. Cadet, Chem. Res. Toxicol., 1999, 12, 630–638 Search PubMed.
- S. Burney, J. C. Niles, P. C. Dedon and S. R. Tannenbaum, Chem. Res. Toxicol., 1999, 12, 513–520 Search PubMed.
- R. Vysabhatta and K. N. Ganesh, Tetrahedron Lett., 2008, 49, 1314–1318 CrossRef.
- D.-M. She, H.-L. Yu, Q.-L. Hung, F.-M. Li and C.-J. Li, Molecules, 2010, 15, 1898–1902 CrossRef CAS PubMed.
- S. Chen, D. Lin, Z. Jiang, J. Zhao, B. Gao, X. Mei, J. Ning and D. She, J. Labelled Compd. Radiopharm., 2013, 56, 305–306 CrossRef CAS PubMed.
- T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco Search PubMed.
- T. Sakamoto, M. Otsu and G. Kawai, NMR Studies on RNA, in Experimental Approaches of NMR Spectroscopy, ed. The Nuclear Magnetic Resonance Society of Japan, 2018 Search PubMed.
- A. T. Brünger, P. D. Adams, G. M. Clore, W. L. Delano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, N. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson and G. L. Warren, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1998, 54, 905–921 CrossRef PubMed.
- D. A. Case, T. A. Darden, T. E. Cheatham III, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, R. C. Walker, W. Zhang, K. M. Merz, B. Roberts, S. Hayik, A. Roitberg, G. Seabra, J. Swails, A. W. Götz, I. Kolossváry, K. F. Wong, F. Paesani, J. Vanicek, R. M. Wolf, J. Liu, X. Wu, S. R. Brozell, T. Steinbrecher, H. Gohlke, Q. Cai, X. Ye, J. Wang, M. J. Hsieh, G. Cui, D. R. Roe, D. H. Mathews, M. G. Seetin, R. S. Ferrer, C. Sagui, V. Babi, T. Luchko, S. Gusaro, A. Kovalenko and P. A. Kollman, AMBER 12, University of California, San Francisco, 2012 Search PubMed.
- Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311), E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03889j |
| This journal is © The Royal Society of Chemistry 2020 |