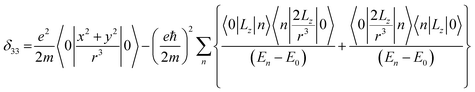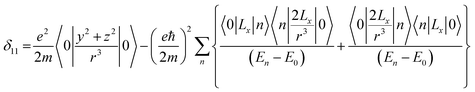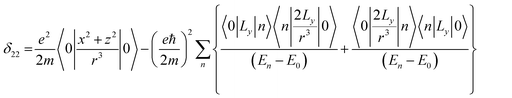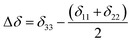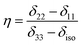Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
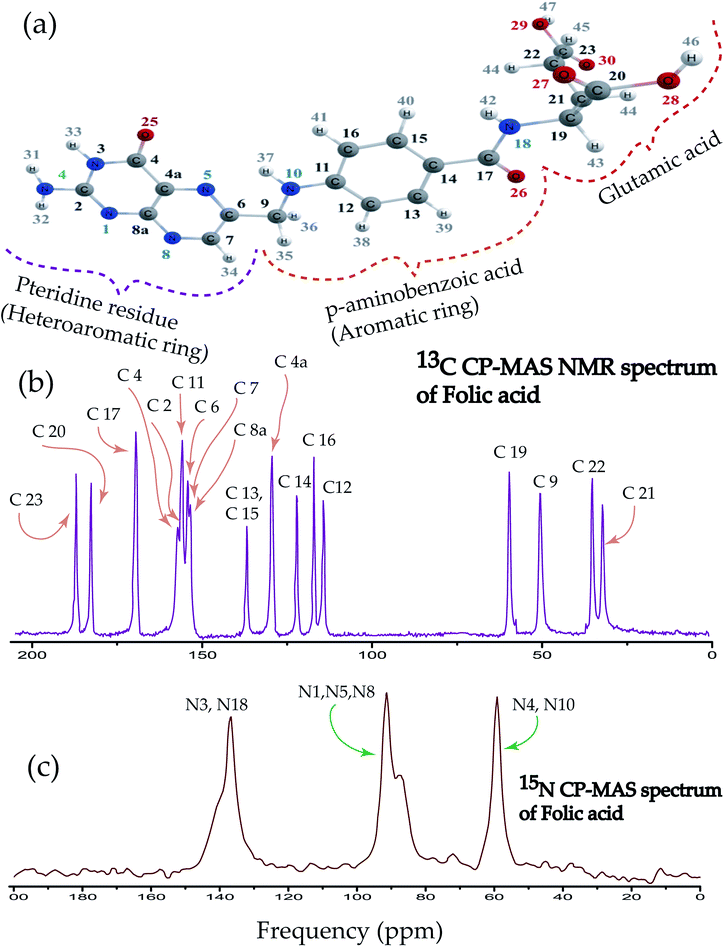
An atomic resolution description of folic acid using solid state NMR measurements
Manasi Ghosh *a,
Shovanlal Gayen
*a,
Shovanlal Gayen b and
Krishna Kishor Dey
b and
Krishna Kishor Dey *a
*a
aDepartment of Physics, Dr Harisingh Gour Central University, Sagar-470003, MP, India. E-mail: manasi.ghosh@gmail.com; dey.12@buckeyemail.osu.edu
bDepartment of Pharmaceutical Sciences, Dr Harisingh Gour Central University, Sagar-470003, MP, India
First published on 1st July 2020
Abstract
The chemical shift anisotropy tensor and site-specific spin-lattice relaxation time of folic acid were determined by a 13C 2DPASS CP-MAS NMR experiment and Torchia CP experiment respectively. The molecular correlation time at various carbon nuclei sites of folic acid was evaluated by assuming that the 13C spin-lattice relaxation mechanism is mainly governed by chemical shift anisotropy interaction and hetero-nuclear dipole–dipole coupling. CSA parameters are larger for the carbon nuclei residing at the heteroaromatic ring and aromatic ring, and those attached to double-bonded electronegative oxygen atoms. It is comparatively low for C9, C19, C21, and C22. The molecular correlation time is of the order of 10−4/10−5 s for C9, C19, C21 and C22 carbon nuclei, whereas it is of the order of 10−3 s for the rest of the carbon nuclei sites. Spin lattice relaxation time varies from 416 s to 816 s. For C23 and C14, the value is 816 s, and it is 416 s for C7 nuclei. The correlation between structure and dynamics on an atomic scale of such an important drug as folic acid can be visualized by these types of extensive spectroscopic measurements, which will help to develop an advanced drug for DNA replication.
1. Introduction
Folic acid (vitamin B9) is indispensable for DNA replication, growth, and development of the fetus. This synthetic form of folate is used as a substrate for enzymatic reactions associated with amino acid synthesis and vitamin metabolism. Deficiency of folic acid can cause congenital abnormalities, anemia, and peripheral neuropathy. The body cannot synthesize folic acid; it can be obtained from diet or supplementation. Dietary supplement of folic acid during pregnancy decreases the hazard of neural tube defects (NTD) in the offspring, reduces the risk of preterm birth (PTB) and protects against inborn heart defects.1–5 Folic acid and its derivatives play a vital role in the process of formation of purine and thymidine, which are considered as essential ingredients for the formation of nucleic acids. Folic acid also plays a pivotal role in the synthesis of the methylation agent S-adenosyl-methionine, which is essential for protein and lipid production.4Nuclear magnetic resonance (NMR) spectroscopic technique plays a central role to elucidate the three-dimensional structure and dynamics of a molecule with atomic resolution at sufficient sensitivity. NMR interactions can be classified into two categories-spin-field interactions and spin–spin interaction. Zeeman, chemical shift, magnetic susceptibility, and radio-frequency field interactions are labeled as spin-field interactions. Whereas homo-nuclear dipole–dipole coupling, hetero-nuclear dipole–dipole coupling, J-coupling, and quadrupole coupling are designated as spin–spin interactions. Although at high value of the magnetic field, the strongest interaction is the Zeeman interaction, it failed to provide any direct information about the three-dimensional structure and spin dynamics of a molecule, because its value is the same for all the nuclei of the same kind. The effective magnetic field experienced by the nucleus or the Larmor precession frequency of the nucleus is modified due to the presence of the electron cloud surrounding the nucleus – this is referred as nuclear shielding or de-shielding effect and the change in the nuclear resonance frequency due to this effect is known as chemical shift. The electronic distribution around a nucleus has low-symmetry. Hence, the chemical shift has an anisotropic part as well as an isotropic part. The anisotropic part depends on the molecular conformation and orientation of the molecule with respect to the external magnetic field. Chemical shift anisotropy can be represented mathematically by a second-rank tensor associated with nine components. In principal axis system (PAS) only the diagonal components restored. The expressions of those diagonal components of the chemical shift anisotropy tensor (δ11, δ22 and δ33) are given by6,7
where Lx, Ly and Lz are the components of angular momentum along the x, y and z directions respectively. ‘r’ is the position of the electron with respect to the nucleus. The first part of these three equations arises from spherically symmetric electronic charge distribution i.e. when the electrons reside in the ground state (the ‘s’ orbital state). The second term arises due to the distortions in the symmetry of the charge distribution when electrons are in excited states. δ11 corresponds to minimum nuclear shielding with maximum nuclear resonance frequency. Conversely, δ33 corresponds to maximum nuclear shielding with minimum nuclear resonance frequency molecule has no preferred direction of orientation for liquid sample, it can orient in any direction. Hence, the averaging of CSA tensor over all possible directions of orientation generates the isotropic chemical shift δiso = (δ11 + δ22 + δ33)/3. Hence, for solid sample it is possible to extract information about the electronic distribution surrounding the nucleus by determining the values of δ11, δ22, and δ33.
There are several techniques to determine CSA parameters – two dimensional MAS/CSA NMR experiment;8 SUPER (separation of undistorted powder patterns by effortless recoupling);9 ROCSA (recoupling of chemical shift anisotropy);10 RNCSA(γ-encoded RNnν-symmetry based chemical shift anisotropy);11 2DMAF (two-dimensional magic angle flipping) experiment;12–15 2DMAT (two-dimensional magic angle turning) experiment;16 and two-dimensional phase adjusted spinning side-band (2DPASS) cross-polarization(CP) magic angle spinning (MAS) SSNMR experiment.17,18 2DPASS CP-MAS SSNMR technique was applied to elucidate the structure and dynamics of glass, biopolymers, biomedicine.19–25
5,6,7,8-Tetrahydrofolate is an important cofactor for the synthesis of thymidylate from deoxyuridylate. If the body fails to synthesize thymidylate, DNA synthesis and cell division would not be possible. Hence, blockade of tetrahydrofolate formation eventually leads to cell death. Folic acid reduces to 7,8-dihydrofolate and tetrahydrofolate step by step within the human body by the action of the enzyme dihydrofolate reductase (DHFR).26 This is a fascinating system for developing anticancer agents. It is necessary to have in-depth information about the structure and dynamic of folic acid. The aim of this work is to study the structure and dynamics of this essential dietary supplement by extracting chemical shift anisotropy (CSA) parameters by 2DPASS CP-MAS NMR experiment; spin-lattice relaxation time at chemically different carbon nuclei positions by Torchia CP method; and determination of molecular correlation time at numerous carbon nuclei position. This type of research will enlighten the path of inventing dietary supplements, which can reduce the risk of birth defects.
2. Experimental
2.1 NMR measurements
Folic acid, purchased from Sigma Aldrich, was used for the NMR experiment. The purity of the sample was 99.98%. NMR experiments were performed on a JEOL ECX 500 NMR spectrometer. Nearly 50 mg folic acid was tightly packed in 3.2 mm JEOL double resonance MAS probe for performing solid state NMR experiments. 13C CP-MAS and 15N CP-MAS experiments were done at magic angle spinning speed 10 kHz. The CP contact time was 2 ms to satisfy the state of cross-polarization (CP). SPINAL-64 1H decoupling was used during acquisition. External referencing for 15N was done by NH4Cl. 13C spin-lattice relaxation time was measured by the Torchia CP method.35 The number of scans for 13C CP MAS experiment scan was 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 768 and for 13C 2DPASS CP MAS experiment number of scans was 4030. For measuring 13C relaxation by the Torchia CP method, the number of scans was 2048.
768 and for 13C 2DPASS CP MAS experiment number of scans was 4030. For measuring 13C relaxation by the Torchia CP method, the number of scans was 2048.
2.2 CSA measurements
Chemical shift anisotropy parameters of folic acid were evaluated by 2DPASS CP MAS SSNMR experiment.17,18 Thirteen steps cogwheel phase cycling was used. Sixteen data points were acquired in the indirect dimension as the numbers of sidebands were less than sixteen. π pulse spacing was given in Tabular form by Antzutkin et al.18 90° pulse length for 13C was 3.3 us. The Relaxation delay was 10 s. The number of scans for the experiment were 4030 (integral multiple of 13). The coherence transfer pathway diagram for 2D PASS NMR experiment was given in Ghosh et al.20 With the help of this experiment, the isotropic dimension can be correlated with anisotropic dimension by shearing transformation and two dimensional Fourier Transformation. The overlapping signals can be identified by the 2DPASS CP MAS SSNMR experiment, as their CSA parameters are different. CSA eigenvalues can be determined by using the sideband intensities of spinning CSA sideband pattern of chemically different carbon nuclei.27 2DPASS CP MAS SSNMR experiment is associated with five π pulses.18 The time evolution of theses five π pulses is varied according to the PASS equation, but the total time duration of all the PASS sequences are same. 2DPASS experiments were performed at MAS speed 600 Hz and 2 kHz.3. Results and discussion
3.1 NMR spectral analysis
Folic acid is 2S-2-{[4-(2-amino-4hydroxypteridin-6-yl)-methyl]amino}phenyl formamido pentanedioic acid. Fig. 1(a) shows that the folic acid molecule is composed of pteridine, p-aminobenzolate, and glutamate moieties. Aminobenzoyl moiety acts as a connector between pteridine and glutamate moieties and it is strongly bound to the receptor. Hence, it acts like ‘anchors’ between the ligand and the receptor. Glutamate moiety is formed by two isonizable carboxylic acid functional groups, which can be used to append with other molecules or to constitute salt-bridges with receptor residues.4 The biologically inactive folic acid becomes biologically active within the liver by forming dihydrofolic and terahydrofolic acids. Fig. 1(b) shows 13C CP-MAS NMR spectrum of folic acid. The isotropic chemical shift is very high for C23, C20, C17, C4 carbon atom associated with double bonded oxygen, and it is lowest for C21, C22, C9, and C19. Chemical shift is highly correlated with the presence of electronegative atoms in the neighbourhood. Electronegative atom attracts the electron cloud around the carbon nucleus. As a consequence, the electron cloud density is reduced and the effective field experienced by the carbon nuclei is increased, leads to the higher values of chemical shift. The nucleus C9 resides on the symmetry axis of a rigid molecule with a strong magnetic susceptibility experienced the magnetic field, which is lower than the applied magnetic field – shielding effect. On the contrary, the local magnetic field experienced by the nuclei C4a, C8a, C6, C7 reside in the plane of the rigid molecule is higher than the applied magnetic field – known as deshielding effect. This phenomenon is also known as the ring-current shift. In the next section, it will be discussed that the chemical shift anisotropy (CSA) parameters are also very high for C4a, C8a, C6, C7 compared to C9 nucleus. Three well resolved resonance peaks are observed in 15N CP-MAS NMR spectrum of folic acid (Fig. 1(c)). The overlapping resonance line near 140 ppm originates from N3 and N18 nuclei. The resonance line near 90 ppm comes from N1, N5, N8 nuclei resides on pteridine ring. Resonance line near 60 ppm comes from N4 and N10 nuclei.3.2 Chemical shift anisotropy parameters
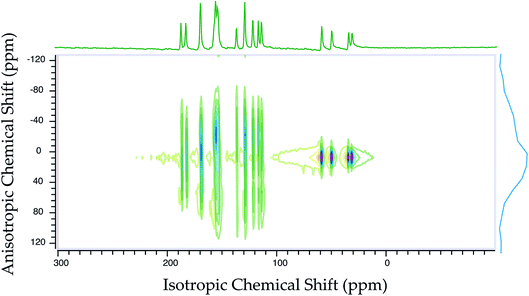
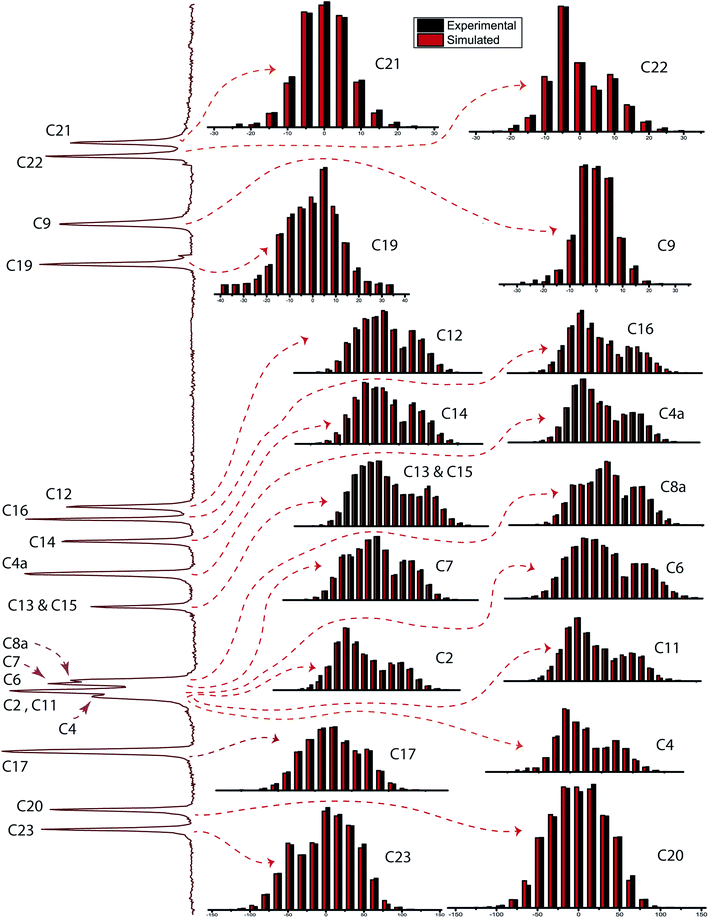
Fig. 2 shows the 13C 2DPASS CP-MAS spectrum of folic acid. The direct dimension of the two-dimensional spectrum represents an infinite spinning speed spectrum and the indirect dimension represents the anisotropic spectrum. Table 1 represents the CSA parameters at crystallographically and chemically different carbon nuclei sites of folic acid. Different values of CSA parameters suggest that the molecular orientation, conformation, and electron cloud density varies significantly at different location of the molecule. For example, C19, C20, C21, C22, C23 carbon nuclei reside in glutamate moieties – but the CSA parameters are remarkably high for C20 and C23carbon nuclei compared to C19, C21, and C22. The center of gravity of the spinning CSA sideband pattern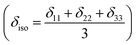 is the isotropic chemical shift.27–32 Small changes in the position of the center of gravity of spinning CSA sideband pattern furnish larger changes in chemical shift anisotropy parameters.31 The width of the spinning CSA sideband pattern is defined as span (Ω = δ11 − δ33). The anisotropy (Δδ) and asymmetry (η) parameters are defined as
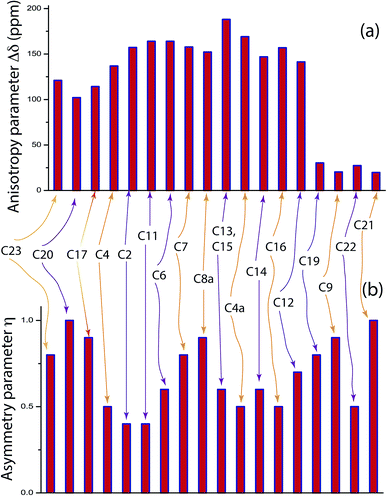
is the isotropic chemical shift.27–32 Small changes in the position of the center of gravity of spinning CSA sideband pattern furnish larger changes in chemical shift anisotropy parameters.31 The width of the spinning CSA sideband pattern is defined as span (Ω = δ11 − δ33). The anisotropy (Δδ) and asymmetry (η) parameters are defined as 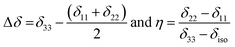 followed by Haeberlen convention.37 The magnitude of anisotropy parameter says the distance of largest separation of the spinning CSA sideband pattern from the position of the center of gravity, and its sign represents on which side of the center of gravity the distance of separation is maximum. Spinning CSA sideband patterns of numerous carbon nuclei of folic acid are shown in Fig. 3. Sideband pattern is axially symmetric (η = 0) when δ22 is equal to δ33 or δ11. Hence, asymmetry parameter monitors the digression of the spinning CSA side band pattern from its axially symmetric shape. Table 1 says that the asymmetry parameter is greater than and equal to 0.4 for all nuclei of folic acid, and for C7, C8a, C9, C17, C19, C20, C21 and C23 carbon nuclei the value is equal or greater than 0.8, indicating that the side-band pattern is highly asymmetric for those carbon nuclei.
followed by Haeberlen convention.37 The magnitude of anisotropy parameter says the distance of largest separation of the spinning CSA sideband pattern from the position of the center of gravity, and its sign represents on which side of the center of gravity the distance of separation is maximum. Spinning CSA sideband patterns of numerous carbon nuclei of folic acid are shown in Fig. 3. Sideband pattern is axially symmetric (η = 0) when δ22 is equal to δ33 or δ11. Hence, asymmetry parameter monitors the digression of the spinning CSA side band pattern from its axially symmetric shape. Table 1 says that the asymmetry parameter is greater than and equal to 0.4 for all nuclei of folic acid, and for C7, C8a, C9, C17, C19, C20, C21 and C23 carbon nuclei the value is equal or greater than 0.8, indicating that the side-band pattern is highly asymmetric for those carbon nuclei. 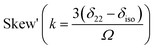 defines the amount of orientation of the asymmetric pattern. From Fig. 3, it is clear that there is no carbon nucleus in a folic acid molecule whose spinning CSA sideband pattern is highly symmetric (η = 0). Fig. 4(a) and (b) portrayed vividly how anisotropy and asymmetry parameters vary at different sites of carbon nuclei. Chemical shift anisotropy parameters are remarkably high for carbon nuclei resides in heteroaromatic (C2, C4, C4a, C6, C7, C8a) and aromatic rings(C11, C12, C13, C14, C15, C16) and carbonyl group(C23, C20, C17, C4). The values of CSA parameters are comparatively lower for C9, C19, C21, and C22 carbon nuclei. For heteroaromatic and aromatic rings, the existence of π-electrons gives rise to magnetic shielding and deshielding effect. When π-electron rotates in a clockwise direction-the induced magnetic field is along the direction of the external magnetic field. As a consequence, the effective magnetic field experienced by the nucleus is increased – this is called the deshielding effect. On the other hand, the magnetic field generates by the π-electron revolves in the anticlockwise direction, is along the opposite direction of the applied magnetic field. Hence, the effective magnetic field experienced by the nucleus is decreased, this phenomenon is known as the shielding effect. As a consequence, the chemical shift anisotropy (CSA) parameters for those carbon nuclei surrounded by nonbonded electrons are substantially large.
defines the amount of orientation of the asymmetric pattern. From Fig. 3, it is clear that there is no carbon nucleus in a folic acid molecule whose spinning CSA sideband pattern is highly symmetric (η = 0). Fig. 4(a) and (b) portrayed vividly how anisotropy and asymmetry parameters vary at different sites of carbon nuclei. Chemical shift anisotropy parameters are remarkably high for carbon nuclei resides in heteroaromatic (C2, C4, C4a, C6, C7, C8a) and aromatic rings(C11, C12, C13, C14, C15, C16) and carbonyl group(C23, C20, C17, C4). The values of CSA parameters are comparatively lower for C9, C19, C21, and C22 carbon nuclei. For heteroaromatic and aromatic rings, the existence of π-electrons gives rise to magnetic shielding and deshielding effect. When π-electron rotates in a clockwise direction-the induced magnetic field is along the direction of the external magnetic field. As a consequence, the effective magnetic field experienced by the nucleus is increased – this is called the deshielding effect. On the other hand, the magnetic field generates by the π-electron revolves in the anticlockwise direction, is along the opposite direction of the applied magnetic field. Hence, the effective magnetic field experienced by the nucleus is decreased, this phenomenon is known as the shielding effect. As a consequence, the chemical shift anisotropy (CSA) parameters for those carbon nuclei surrounded by nonbonded electrons are substantially large.
| CSA parameters of folic acid | ||||||||
|---|---|---|---|---|---|---|---|---|
| Carbon from different chemical environment | δ11(ppm) | δ22(ppm) | δ33(ppm) | Span (ppm) | Skew | δiso(ppm) | Anisotropy (ppm) | Asymmetry |
| C23 | 258.7 | 194.8 | 105.8 | 152.9 | 0.2 | 186.4 | −121 | 0.8 |
| C20 | 250.2 | 181.8 | 114.4 | 135.8 | 0 | 182.1 | 102.1 | 1 |
| C17 | 245 | 163.5 | 98 | 147 | −0.1 | 168.9 | 114.2 | 0.9 |
| C4 | 248.4 | 133.5 | 89.5 | 158.9 | −0.4 | 157.1 | 136.9 | 0.5 |
| C2 | 260.7 | 127.4 | 79.7 | 181 | −0.5 | 155.9 | 157.2 | 0.4 |
| C11 | 264.5 | 124.9 | 76.1 | 188.3 | −0.5 | 155.2 | 164 | 0.4 |
| C6 | 263.2 | 133.5 | 65.2 | 198 | −0.3 | 154 | 163.8 | 0.6 |
| C7 | 258.4 | 141.3 | 60 | 198 | −0.2 | 153.2 | 157.7 | 0.8 |
| C8a | 254.3 | 146 | 58.1 | 196.2 | −0.1 | 152.8 | 152.2 | 0.9 |
| C13, C15 | 261.8 | 109.2 | 38.2 | 223.6 | −0.4 | 136.4 | 188.1 | 0.6 |
| C4a | 241.7 | 100.3 | 44.9 | 196.8 | −0.4 | 128.9 | 169.1 | 0.5 |
| C14 | 219.1 | 103.3 | 41.1 | 178 | −0.3 | 121.1 | 146.9 | 0.6 |
| C16 | 221.1 | 88.5 | 39.9 | 181.2 | −0.5 | 116.5 | 156.9 | 0.5 |
| C12 | 207.5 | 101.1 | 31.3 | 176.2 | −0.2 | 113.3 | 141.3 | 0.7 |
| C19 | 76.3 | 60.2 | 68 | 38.3 | 0.2 | 58.1 | −30.3 | 0.8 |
| C9 | 62.7 | 48.6 | 36.1 | 26.6 | 0 | 49.2 | 20.3 | 0.9 |
| C22 | 52.1 | 29 | 20.6 | 31.5 | −0.5 | 33.9 | 27.3 | 0.5 |
| C21 | 43.9 | 30.6 | 17.8 | 26 | 0 | 30.8 | 19.7 | 1 |
The carbonyl group does not have any axis of symmetry. Naturally, the electronic charge distribution is not spherically symmetric. By the action of electrostatic interaction, there induced polarization on the electron cloud surrounding the nucleus. This polarization affects the strength of the induced magnetic field. As a result, the strength of the magnetic field is varied in different directions.33 In the principal axes system (PAS), the magnetic susceptibilities (χx, χy, χz) have different values along three mutually perpendicular directions. The McConnell equation of magnetic anisotropy for carbonyl group is34
δanis = {Δχ‖(3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos2 cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ1 − 1) + Δχ⊥(3 θ1 − 1) + Δχ⊥(3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos2 cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ2 − 1)}/3R3 θ2 − 1)}/3R3
| (1) |
The existence of hydrogen bonds between the water molecule and the oxygen atom connected with C4 carbon and N10 atoms of the pteridine group brings stability to the molecular structure.26 The existence of intermolecular and intramolecular hydrogen bonding in the pteridine group plays the foremost role to change molecular orientation and electron density. This is also another reason for the high values of CSA parameters of the carbon nuclei reside at the pteridine ring.
3.3 Spin-lattice relaxation time and molecular correlation time
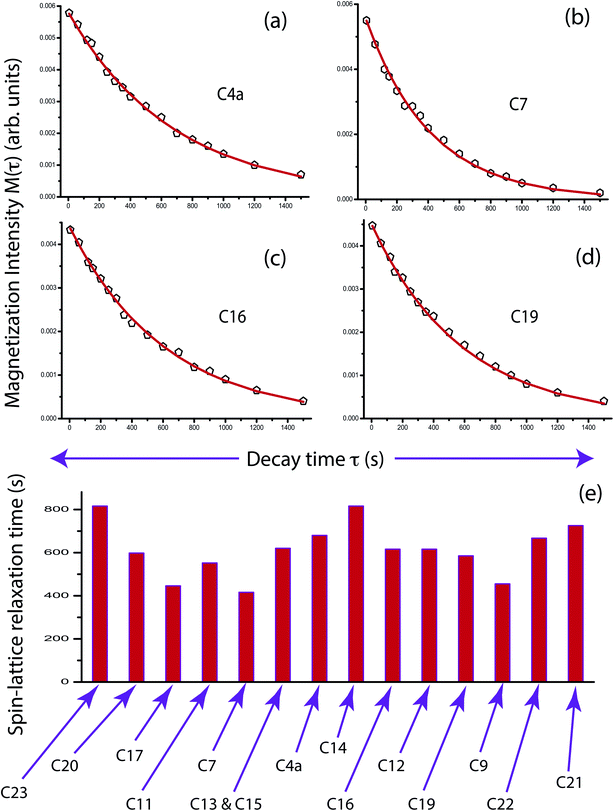
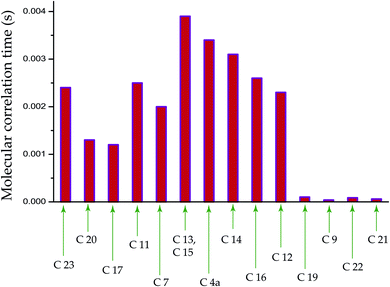
The population of stationary states follows the Boltzmann distributions when the equilibrium is re-established among spin and lattice systems with the withdrawal of perturbation. As a consequence, the off-diagonal elements of the density operator are averaged out, and the diagonal elements are restored. That means all coherence transfer pathways have vanished. This is referred as spin-lattice relaxation. Spin-lattice relaxation mechanism is governed by homo-nuclear dipole–dipole coupling, hetero-nuclear dipole–dipole coupling, electric quadrupolar coupling, anisotropic chemical shift (CSA), and scalar coupling. Although for 13C nuclei, the heteronuclear dipole–dipole coupling and chemical shift anisotropy interactions play a fundamental role in the spin-lattice relaxation mechanism. The relaxation mechanism of nonprotonated 13C nuclei is predominated by CSA interaction. For proton relaxation, the contribution of CSA is negligible.Fig. 5 shows 13C spin-lattice relaxation decay curves of folic acid at (a) C4a, (b) C7, (c) C16, and (d) C19 carbon nuclei sites. Fig. 5(e) bar-diagram and Table 2 say that spin-lattice relaxation time at numerous carbon nuclei sites of folic acid is very high and it varies from 816 s to 416 s. This significant variation of the spin-lattice relaxation time indicates that the motional degrees of freedom are different at various portions of the molecule and the motion of different molecular moieties is independent of each other. For C23 and C14 the value of spin-lattice relaxation time exceeds 800 s. Each molecule of folic acid is bonded with two other symmetry-related folic-acid molecules and water molecules by strong hydrogen bonding. Two pairs of hydrogen bonding are observed in folic acid. One pair is formed by hydrogen atom connected with N4 with N1 atom and with the alpha carboxyl group of the glutamic acid end of another symmetry-related folic acid molecule.26 Another pair of hydrogen bonds is fabricated between the hydrogen atom connected with N3 and the oxygen atom connected to C4 of the pteridine ring and the gamma carboxyl group of the glutamic acid end of a second symmetry-related folic acid. The second symmetry-related folic acid molecule resides in such a way that the position of the p-aminobenzoyl group is just below the pteridine ring of the first one. The separation between these two parallel rings of two folic acid molecule is 3.28 Å.26 These types of stacking arrangement of pteridine and phenyl rings and the presence of two pairs of hydrogen bonding make folic acid a robust molecule. These may be the reason for the high value of the spin-lattice relaxation time of the carbon nuclei of folic acid. The molecular correlation time (Table 4 and Fig. 6) is of the order of 10−3 s for most of the carbon nuclei, except C9, C19, C21, C22.
| Carbon from different chemical environment | Spin-lattice relaxation time (s) | Carbon from different chemical environment | Spin-lattice relaxation time (s) |
|---|---|---|---|
| C23 | 816 ± 20 | C20 | 598 ± 20 |
| C17 | 446 ± 10 | C11 | 552 ± 20 |
| C7 | 416 ± 10 | C13 and C15 | 620 ± 20 |
| C4a | 680 ± 20 | C14 | 816 ± 20 |
| C16 | 616 ± 20 | C12 | 616 ± 20 |
| C19 | 585 ± 20 | C9 | 455 ± 10 |
| C22 | 667 ± 20 | C21 | 725 ± 20 |
The delocalized π-electrons of the pteridine heteroaromatic ring of one molecule interact significantly with the p-aminobenzoyl rings of the other molecule. The stacking interaction between two molecules plays an important role in the binding of folates at the enzyme dihydrofolate reductase (DHFR) active site. If it is possible to develop folic acid antagonists, which can exploit the difference between the DHFR enzyme of normal and tumour cells, then it would be used as a valuable drug in cancer treatment.26 For developing such antagonists, it is necessary to have information about the structure and dynamics of folic acid in atomic scale resolution.
The local magnetic field experienced by the same nuclei situated in different chemical environments is different. The local magnetic field, experienced 13C nuclei, is anisotropic in nature. It fluctuates with the reorientation of the molecule with respect to the applied magnetic field, and the fluctuating local magnetic field is the source of spin-lattice relaxation. Naturally, the modification of the electronic environments with the fluctuation of the local magnetic fields can be monitored by measuring CSA parameters. CSA interaction plays a significant role in the relaxation mechanism for nuclei associated with a large value of chemical shift like carbon, nitrogen, and phosphorous for the biological molecule. The function of chemical shift anisotropy interaction in spin-lattice relaxation mechanism is expressed as28–31
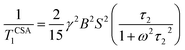 | (2) |
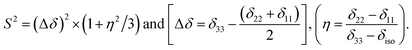
The function of heteronuclear dipole–dipole coupling on spin-lattice relaxation mechanism is expressed as31
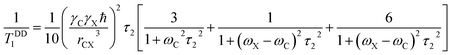 | (3) |
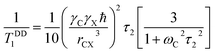 | (4) |
| Bond | Bond distance | Bond | Bond distance |
|---|---|---|---|
| N1–C2 | 1.304 | C22–C23 | 1.511 |
| N1–C8a | 1.371 | C2–N24 | 1.368 |
| C2–N3 | 1.378 | N24–H31 | 1.009 |
| N3–C4 | 1.408 | N24–H32 | 1.011 |
| C4–C4a | 1.475 | N3–H33 | 1.014 |
| C4a-C8a | 1.417 | C4–O25 | 1.219 |
| C4a-N5 | 1.343 | C7–H34 | 1.090 |
| N5–C6 | 1.323 | C9–H35 | 1.102 |
| C6–C7 | 1.417 | C9–H36 | 1.104 |
| C7–C8 | 1.321 | N10–H37 | 1.013 |
| N8–C8a | 1.356 | C12–H38 | 1.085 |
| C6–C9 | 1.514 | C13–H39 | 1.085 |
| C9–N10 | 1.438 | C15–H40 | 1.086 |
| N10–C11 | 1.373 | C16–H41 | 1.087 |
| C11–C12 | 1.413 | C17–O26 | 1.234 |
| C12–C13 | 1.390 | N18–H42 | 1.012 |
| C13–C14 | 1.402 | C19–H43 | 1.098 |
| C14–C15 | 1.409 | C20–O27 | 1.215 |
| C15–C16 | 1.386 | C20–O28 | 1.350 |
| C16–C11 | 1.415 | O28–H46 | 0.973 |
| C14–C17 | 1.491 | C21–H44 | 1.093 |
| C17–N18 | 1.377 | C22–H44 | 1.096 |
| N18–C19 | 1.453 | C22–H45 | 1.098 |
| C19–C20 | 1.520 | C23–O30 | 1.213 |
| C19–C21 | 1.549 | C23–O29 | 1.360 |
| C21–C22 | 1.529 | O29–H47 | 0.972 |
The spin-lattice relaxation rate for 13C can be written as
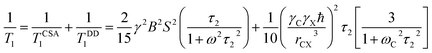 | (5) |
Molecular correlation time at numerous carbon nuclei sites of folic acid can be calculated by using this expression.
The bar-diagram of molecular correlation time at numerous carbon sites of folic acid is shown in Fig. 6 and Table 4. For C9, C21, and C22, molecular correlation time is of the order of 10−5 s. For C19 it is 10−4 s, but for the rest of the nuclei sites of folic acid, the molecular correlation time is of the order of 10−3 s. This indicates that the molecular motion is associated with different degrees of freedom.
| Carbon nuclei | Molecular correlation time(s) | Carbon nuclei | Molecular correlation time(s) |
|---|---|---|---|
| C23 | 2.4 × 10−3 | C20 | 1.3 × 10−3 |
| C17 | 1.2 × 10−3 | C11 | 2.5 × 10−3 |
| C7 | 2 × 10−3 | C13, C15 | 3.9 × 10−3 |
| C4a | 3.4 × 10−3 | C14 | 3.1 × 10−3 |
| C16 | 2.6 × 10−3 | C12 | 2.3 × 10−3 |
| C19 | 1 × 10−4 | C9 | 3.8 × 10−5 |
| C22 | 8.6 × 10−5 | C21 | 6 × 10−5 |
3.4 Structural dynamics of folic acid and its interaction with the receptor
The present study by using 13C 2DPASS CP-MAS NMR experiment and Torchia CP experiment the chemical shift anisotropy parameters, spin-lattice relaxation time as well as molecular correlation time at different nuclei were calculated for folic acid, whose deficiency is linked to diseases like fetal neural tube defects, cardiovascular disease, and cancers etc. These parameters help us to get a picture of the structure and dynamics of the molecule. It will be very interesting if similar dynamics are also present in folic acid in its interaction with the receptor in the binding pocket (Fig. 7(a)). The heteroaromatic (C2, C4, C4a, C6, C7, C8a) carbon nuclei show high CSA parameters whereas C9, C19, C21, and C22 carbon nuclei show comparatively lower values of CSA parameters as mentioned previously. The reason for the large values of the CSA parameter of these carbon nuclei of the heteroaromatic pteridine ring is due to the presence of π electrons as well as the existence of hydrogen bond interactions between C4 carbon and N10 atom. Interestingly, in the receptor bound conformation, the pteridine ring of folic acid forms hydrogen bond interactions with amino acids D81, R103, R106, H135, S174 as shown in Fig. 7(b). The spin-lattice relaxation time at numerous carbon nuclei sites varies from 816 s to 416 s and showed different motional degrees of freedom at different parts of the molecule. The crystal structure of folic acid points out the involvement of the stacking arrangement of the pteridine and phenyl rings, and the presence of two pairs of hydrogen bonding at both ends of the molecule. Similar dynamics are also observed in the case of receptor bound conformation of folic acid. The one end involving the pteridine ring forms a hydrogen bond with five amino acids as mentioned before and the other end i.e. the glutamic acid moiety forms another hydrogen bond with W102, K136, W138 as well as W140.36,38 The similar kind stacking interactions involving the pteridine ring is also found in receptor bound conformations of folic acid where amino acids Y85, W171 as well as W102 and W140 are playing an important role in π–π stacking interactions with the pteridine ring and the phenyl ring of folic acid respectively (Fig. 7(c)).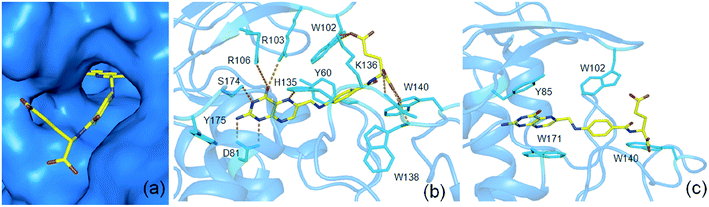 | ||
| Fig. 7 Surface representation (a) and important amino acids of folate receptor for hydrogen bond (b) and π–π stacking interactions (c) for interactions with folic acid (pdb id 4LRH) | ||
4. Conclusion
Folic acid is a crucial nutrient required for DNA replication, growth, and development of the fetus. It is also used as a substrate for enzymatic reactions associated with amino acid synthesis and vitamin metabolism. The structure and dynamics of this dietary supplement are exploited by extracting CSA parameters, site-specific relaxation time, and molecular correlation time at various carbon nuclei sites. At crystallographically different sites of folic acid, the spin-lattice relaxation time varies from 416 to 816 s and the molecular correlation time varies from 10−3 to 10−5 s, which indicates that the motional degrees of freedom are different at the various portion of the molecule. CSA parameters of carbon nuclei reside in the pteridine ring, p-aminobenzoic ring are substantially high due to the magnetic shielding/deshielding effect. Besides, the presence of intermolecular and intramolecular hydrogen bonding in the pteridine group plays a vital role to change molecular orientation and electron density. This is also another reason for the high values of chemical shift anisotropy. In stacking arrangement two consecutive folic acid molecules are oriented with each other in such a way that the position of the p-aminobenzoyl group of the second molecule is just below the pteridine ring of the first one. Hence, the delocalized π-electrons of the pteridine hetero-aromatic ring of the first molecule interact significantly with the p-aminobenzoyl rings of the second molecule. This is also another reason for high values of CSA parameters of the carbon nuclei reside at pteridine ring, p-aminobenzoic ring. The hydrophobic interaction between two molecules plays a vital role in the binding of folic acid at the DHFR active site. CSA parameters of carbonyl group carbon are also high due to the existence of magnetic anisotropy. Elucidation of the structure and dynamics of folic acid like this way will illuminate the path of inventing advanced drugs in Pharmaceutical Industries.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The author Manasi Ghosh is indebted to Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India (file no. EMR/2016/000249) and DST PURSE II Dr Harisingh Gour Central University for financial support.References
- J. A. Greenberg, S. J. Bell, Y. Guan and Y.-h. Yu, Folic acid supplementation and pregnancy: more than just neural tube defect prevention, Reviews in Obsterics and Gynecoligy, 2011, 4, 52–59 Search PubMed.
- D. Braga, L. Chelazzi, F. Grepioni, L. Maschio, S. Nanna and P. Taddei, Folic acid in the solid state: a synergistic computational, spectroscopic, and structural approach, Cryst. Growth Des., 2016, 16, 2218–2224 CrossRef CAS.
- M. A. Dessie, E. G. Zeleke, S. B. Workie and A. W. Berihum, Folic acid usage and associated factors in the prevention of neural tube defects among pregnant women in Ethiopia: cross-sectional study, BMC Pregnancy Childbirth, 2017, 17, 313 CrossRef PubMed.
- F. Abyar and I. Novak, Conformational analysis and electronic structure of folic acid: a theoretical study, J. Mol. Liq., 2019, 276, 819–825 CrossRef CAS.
- A. S. Wibowo, M. Singh, K. M. Reeder, J. J. Carter, A. R. Kovach, W. Meng, M. Ratnam, F. Zhang and C. E. Dann III, Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition, Proc. Natl. Acad. Sci. U. S. A., 2013, 17, 15180–15188 CrossRef PubMed.
- N. F. Ramsey, Magnetic Shielding of Nuclei in Molecules, Phys. Rev., 1950, 78, 699–703 CrossRef CAS.
- N. F. Ramsey, Chemical effects in nuclear magnetic resonance and in diamagnetic susceptibility, Phys. Rev., 1952, 86, 243–246 CrossRef CAS.
- R. Tycko, G. Dabbagh and P. A. Mirau, A. Determination of chemical shift anisotropy lineshapes in a two-dimensional magic angle spinning NMR experiment, J. Magn. Reson., 1989, 85, 265–274 CAS.
- S. F. Liu, J. D. Mao and K. Schmidt-Rohr, A robust technique for two-dimensional separation of undistorted chemical shift anisotropy powder patterns in magic angle spinning NMR, J. Magn. Reson., 2002, 155, 15–28 CrossRef CAS PubMed.
- J. C. C. Chan and R. Tycko, Recoupling of chemical shift anisotropies in solid state NMR under high speed magic angle spinning and in uniformly 13C labelled systems, J. Chem. Phys., 2003, 118, 8378–8389 CrossRef CAS.
- G. Hou, In-Ja L. Byeon, J. Ahn, A. M. Gronenborn and T. Polenova, Recoupling of chemical shift anisotropy by R-symmetry sequences in magic angle spinning NMR spectroscopy, J. Chem. Phys., 2012, 137, 134201–134210 CrossRef PubMed.
- Ad Bax, N. M. Szeverenyi and G. E. Maciel, Correlation of isotropic shifts and chemical shift anisotropies by two-dimensional Fourier-transform magic angle hopping NMR spectroscopy, J. Magn. Reson., 1983, 52, 147–152 CAS.
- Ad Bax, N. M. Szeverenyi and G. E. Maciel, Chemical shift anisotropy in powdered solids studied by 2D FT CP/MAS NMR, J. Magn. Reson., 1983, 51, 400–408 CAS.
- Ad Bax, N. M. Szeverenyi and G. E. Maciel, Chemical shift anisotropy in powdered solids studied by 2D FT NMR with flipping of the spinning axis, J. Magn. Reson., 1983, 55, 494–497 CAS.
- Ad Bax, N. M. Szeverenyi and G. E. Maciel, Correlation of isotropic shifts and chemical shift anisotropies by two-dimensional Fourier-transform magic angle hopping NMR spectroscopy, J. Magn. Reson., 1983, 52, 147–152 CAS.
- Z. Gan, High-resolution chemical shift and chemical shift anisotropy correlation in solids using slow magic angle spinning, J. Am. Chem. Soc., 1992, 114, 8307–8309 CrossRef CAS.
- W. T. Dixon, Spinning-sideband-free and spinning-sideband-only NMR spectra in spinning samples, J. Chem. Phys., 1982, 77, 1800–1809 CrossRef CAS.
- O. N. Antzutkin, S. C. Shekar and M. H. Levitt, Two-dimensional sideband separation in magic angle spinning NMR, J. Magn. Reson., Ser. A, 1995, 115, 7–19 CrossRef CAS.
- B. J. Walder, K. K. Dey, D. C. Kaseman, J. H. Baltisberger and P. J. Grandinetti, Sideband separation experiments in NMR with phase incremented echo train acquisition, J. Chem. Phys., 2013, 138, 174203-1–174203-12 CrossRef PubMed.
- M. Ghosh, S. Sadhukhan and K. K. Dey, Elucidating the internal structure and dynamics of α-chitin by 2DPASS-MAS-NMR and spin-lattice relaxation measurements, Solid State Nucl. Magn. Reson., 2019, 97, 7–16 CrossRef CAS PubMed.
- M. Ghosh, B. P. Prajapati, N. Kango and K. K. Dey, A comprehensive and comparative study of the internal structure and dynamics of natural β-keratin and regenerated β-keratin by solid state NMR spectroscopy, Solid State Nucl. Magn. Reson., 2019, 101, 1–11 CrossRef CAS PubMed.
- M. Ghosh, N. Kango and K. K. Dey, Investigation of the internal structure and dynamics of cellulose by 13C-NMR relaxometry and 2DPASS-MAS-NMR measurements, J. Biomol. NMR, 2019, 73, 601–616 CrossRef CAS PubMed.
- K. K. Dey and M. Ghosh, Understanding the effect of deacetylation on chitin by measuring chemical shift anisotropy tensor and spin lattice relaxation time, Chem. Phys. Lett., 2020, 738, 136782 CrossRef.
- K. K. Dey, S. Gayen and M. Ghosh, Investigation of the detailed internal structure and dynamics of itraconazole by solid-state NMR measurements, ACS Omega, 2019, 4, 21627–21635 CrossRef CAS PubMed.
- K. K. Dey, S. Gayen and M. Ghosh, Understanding the correlation between structure and dynamics of clocortolone pivalate by solid state NMR measurement, RSC Adv., 2020, 10, 4310–4321 RSC.
- D. Mastropaolo, A. Camerman and N. Camerman, Folic acid: crystal structure and implications for enzyme binding, Science, 1980, 210, 334–336 CrossRef CAS PubMed.
- J. Herzfeld and A. E. Berger, Sideband intensities in NMR spectra of samples spinning at the magic angle, J. Chem. Phys., 1980, 73, 6021–6030 CrossRef CAS.
- M. P. Nicholas, E. Eryilmaz, F. Ferrage, D. Cowburn and R. Ghose, Nuclear spin relaxation in isotropic and anisotropic media, Prog. Nucl. Magn. Reson. Spectrosc., 2010, 57, 111–158 CrossRef CAS PubMed.
- A. M. Orendt and J. C. Facelli, Solid state effects on NMR chemical shifts, Annu. Rep. NMR Spectrosc., 2007, 62, 115–178 CrossRef CAS.
- N. Tjandra, A. Szabo and Ad. Bax, Protein backbone dynamics and 15B chemical shift anisotropy from quantitative measurement of relaxation interference effects, J. Am. Chem. Soc., 1996, 118(29), 6986–6991 CrossRef CAS.
- P. Dais and A. Spyros, 13C nuclear magnetic relaxation and local dynamics of synthetic polymers in dilute solution and in the bulk state, Prog. Nucl. Magn. Reson. Spectrosc., 1995, 27, 555–633 CrossRef CAS.
- F. A. L. Anet and D. J. O'Leary, The shielding tensor Part II: Understanding its strange effect on relaxation, Concepts Magn. Reson., 1992, 4, 35–52 CrossRef.
- R. J. Abraham, M. Mobli and R. J. Smith, 1H chemical shifts in NMR: Part 19† Carbonyl anisotropies and steric effects in aromaticaldehydes and ketones, Magn. Reson. Chem., 2003, 41, 26–36 CrossRef CAS.
- H. M. McConnell, Theory of Nuclear Magnetic Shielding in Molecules: Long-Range Dipolar Shielding of protons, J. Chem. Phys., 1957, 27, 226 CrossRef CAS.
- D. A. Torchia, The measurement of proton-enhanced carbon-13 T1 values by method which suppresses artifacts, J. Magn. Reson., 1978, 30, 613 CAS.
- C. Chen, J. Ke, X. E. Zhou, W. Yi, J. S. Brunzelle, J. Li, Eu-L. Yong, H. Eric Xu and K. Melcher, Structural basis for molecular recognition of folic acid by folate receptors, Nature, 2013, 500(7463), 486–489 CrossRef CAS PubMed.
- U. Haeberlen, High Resolution NMR in Solids: Selective Averaging, Academic Press, New York, 1976 Search PubMed.
- B. R. Sahoo, J. Maharana, M. C. Patra, G. K. Bhoi, S. K. Lenka, P. K. Dubey, S. Goyel, B. Dehury and S. K. Pradhan, Structural and dynamic investigation of bovine folate receptor alpha(FOLR1), and role of ultra-high temperature processing on conformational and thermodynamic characteristics of FOLR1-folate complex, Colloids Surf., B, 2014, 121, 307–318 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |