Open Access Article
Open Access ArticleA sensitive and specific method for microRNA detection and in situ imaging based on a CRISPR–Cas9 modified catalytic hairpin assembly†
Yang Liu *,
Shihong Li,
Likun Zhang,
Qian Zhao,
Nuo Li and
Yuxin Wu
*,
Shihong Li,
Likun Zhang,
Qian Zhao,
Nuo Li and
Yuxin Wu
Department of Clinical Laboratory, The Third Affiliated Hospital of Jinzhou Medical University, Jinzhou, Liaoning Province 121000, China. E-mail: jykly43@163.com
First published on 28th July 2020
Abstract
We report here a method for the molecular detection of miRNAs in exosomes and imaging in living cells based on CRISPR–Cas9 and catalytic hairpin assembly. With the cascade signal amplification of catalytic hairpin assembly and high specific recognition of CRISPR–Cas9, the detection method showed a sensitivity of 23.5 fM and was applied for accurate exosomal miR-21 detection and in situ miR-21 imaging.
MicroRNAs (miRNAs) are small, non-coding RNAs that play vital roles in regulating gene expression and related pathological process.1,2 As an important biomarker, the distribution and expression of miRNAs in cells have an intimate relationship with many diseases, especially cancers. Therefore, not only the in vitro detection, but also the in situ imaging of miRNAs will benefit disease diagnosis.3 Recently, exosomes, which are small vehicles with a diameter of about 30–150 nm contained several different biomolecules including proteins, lipids, and both mRNA and noncoding RNAs. Exosomes were also considered as important in cell–cell communication medium since they can release their contents, particularly miRNAs, to both neighbouring and distal cells.4–6 Therefore, exosomal miRNAs are regarded as a promising biomarker for disease diagnosis and pathologic studies.
Many miRNA detection methods, such as real-time quantitative polymerase chain reaction (qRT-PCR), northern blotting, microarray, were reported to achieve sensitive miRNAs detection in solutions or in cell lysates.7,8 Even though, they are also criticized for time-consuming steps, complicated procedures and expensive costs, impeding their wide applications.7,9,10 Many attempts have been made to deal with these issues and thus perfect the existing miRNA detection methods, such as electrochemical methods, surface-enhanced Raman spectroscopy, colorimetric assays.9,11,12 Even though, few of them could be applied into in situ miRNA detection. CHA (catalytic hairpin assembly), as a non-enzymatic autonomous assemble based signal amplification strategy has received extensive attention due to its potential for in vitro and in vivo identification of DNA, RNA, small molecules and nucleic acid assembly compounds.13–15 Besides, signal output modes of CHA are also multiple like effective electrochemical, fluorescence and colorimetric commonly with high signal-to-noise ratio and strong universality. Even though, the specificity of CHA method still needs further improvement.
CRISPR/Cas (clustered regularly interspaced short palindromic repeats), as an RNA-based acquired immune system of most bacteria and archaea, has become a powerful gene editing tool and is widely used in gene function research, gene modification and treatment.16,17 Among all the Cas enzymes discovered, CRISPR–Cas9, which is capable of binding and cutting dsDNA (double chain DNA) target under the guidance of crRNA, attracted abundant attention as a new tool for nucleic acid detection.18 Many CRISPR–Cas9 based methods that integrate specific cleavage activity of the CRISPR–Cas9 system and the in vitro amplification strategy for dsDNA and ssDNA detection have developed.19–22 Even though, it remained to be improved when applied in in situ miRNA imaging.
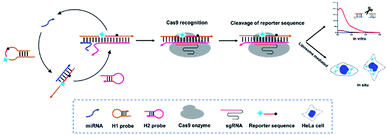
Integration of CHA and CRISPR–Cas9 provides new ideas for solving the above problems. Herein, a CRISPR–Cas9 modified fluorescence biosensor was constructed for both exosomal miRNA detection and imaging in living cells. In this method, a specially designed H1 probe including CRISPR–Cas9 recognition region (PAM) in the middle and two complementary regions at both ends is introduced. Besides, the florescence Cy3 and corresponding quenching moiety BHQ-1 is labelled at the two terminals, whose initial florescence is quenched due to the fluorescence resonance energy transfer (FRET). This H1 probe with a hairpin structure formed by the partially complementary sequence at both ends can be recognized by the miRNA target, basically with miRNA-21 as the proof-of-concept demonstration target and opened through strand replacement from one end. Therefore, a dsDNA of unequal length is formed, consisting of open H1 and miRNA-21. Subsequently, the H2 probe gradually hybridizes with the H1–miRNA21 complex from the exposed end of H1 probe and forms a double-stranded DNA complex, thus displaces miRNA-21, which can enter the next cycle to open H1 probes. As a result, there will be a large amount of H1–H2 complex in the system. Meanwhile, the CRISPR–Cas9 system recognizes the H1–H2 complexes through the PAM site on the H1 probe under the guidance of sgRNA, and thus triggers the Cas9 enzyme to arbitrarily cut amounts of Reporters existed in the surrounding environment and generate fluorescent signal (Scheme 1).
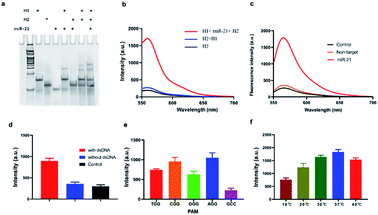
In order to verify the efficiency of the constructed CHA signal amplification system, H1 probes and H2 probes annealed were mixed with the targets (the final concentration was 1 μM), incubating for 30 min. Then 12% PAGE electrophoresis was carried with H1 probe, H2 probe, miR-21, H1 and target sequence, H2 and target sequence. The results showed that an extra band came out, probably at a position of 50 bp, when H1 and miRNA-21 were mixed, suggesting that miRNA-21 turned on the H1 probe and hybridized, while no obvious strand appeared in the mixture of H2 and miRNA. More importantly, when H1, H2 and miRNA-21 were mixed, another band appeared, which was located at about 65 bp, and is considered to be H1–H2 complex (Fig. 1a). In addition, we further investigated the signal amplification of CHA by labelling the fluorophore (Cy3) and the corresponding quenching group (BHQ-1) on both ends of the H2 probe. The results showed that when H1 and H2 were present, the fluorescence signal did not increase significantly compared with the control group, while a considerable high fluorescence signal, which was almost 7.2 time higher than that in control group, was obtained when H1, H2 and miRNA-21 were present (Fig. 1b). We then investigated the specificity of CHA by mixing miRNA-21 and let-7a, respectively. The results showed a significantly higher fluorescence signal when miRNA-21 present than that of let-7a, suggesting a higher CHA specificity for detection of different miRNAs (Fig. 1c).
To investigate the accurate cleavage activity of Cas9 enzyme, target dsDNA and non-specific dsDNA were carefully designed with Cy3 and BHQ-1 labelled at both terminals. The result showed that fluorescence signal increased significantly when dsDNA target existed, while remained the same as that in control when absence of it, suggesting that dsDNA target can be recognized by CRISPR/Cas9 thus trigger trans-cleavage of Reporter (Fig. 1d). According to previous reports, Cas9 requires a 5′-NGG-3′ PAM site to cleave target dsDNA.23 To further optimize the PAM sequence, we designed different sequences (5′-AGG-3′, 5′-CGG-3′, 5′-TGG-3′, 5′-GGG-3′ and 5′-GCC-3′). As shown in Fig. 1e, the specifically recognition of the PAM sequence could thereby initiate the following Cas9-mediated accurate-cleavage. Furthermore, the PAM sequences of 5′-AGG-3′ and 5′-CGG-3′ induced a higher specificity which could be reflected by higher fluorescence signals. Considering that fluorescence intensity of 5′-AGG-3′ group was more stable than that in 5′-CGG-3′ group, 5′-AGG-3′ was added in H1. We further investigated the effect of temperature on the trans-cleavage properties of CRISPR–Cas9. The results show that the trans-cleavage activity is lower when in 10 °C. As the temperature increased, the activity of the Cas9 enzyme gradually increased and reached its peak at 37 °C (Fig. 1f). This indicates that the method is temperature sensitive and can be applied to in situ imaging in cells.
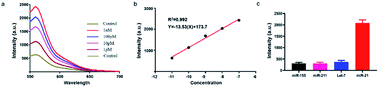
Subsequently, we investigated the sensitivity of the established method by analysing target miRNA-21 with a series of concentrations. The fluorescence curves linearly proportional to the logarithmic value of the concentration of amplified target DNA ranging from 1 pM to 10 nM (Fig. 2a). The correlation equation was obtained as Y = −13.53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 173.7, with R2 = 0.9875 (Fig. 2b). The detection limit was calculated as low as 23.5 fM, which is enough for not only the in vitro detection but also the in vivo imaging.
C + 173.7, with R2 = 0.9875 (Fig. 2b). The detection limit was calculated as low as 23.5 fM, which is enough for not only the in vitro detection but also the in vivo imaging.
We then investigated the specificity of the proposed method by distinguishing completely matched target (miRNA-21) from three different miRNAs (miR-211, miR-155, let-7a). Results showed that miRNA-21 induced much stronger fluorescence signal (>4 times) than the other three types of miRNAs (Fig. 2c), indicating that this method had an outstanding specificity in the discrimination of complementary target owing to the accurate identification capability of CRISPR–Cas9.
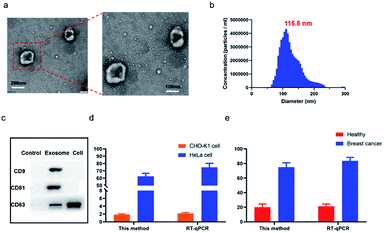
We then applied the proposed method for exosomal miRNAs detection. In order to quantify the exosomal miRNA-21, we firstly extracted the exosomes from the HeLa cell lines culture medium through sequential centrifugation and characterized the obtained exosomes through TEM, nanoparticle tracking analysis (NTA) and western blotting. As shown in Fig. 3a, the obtained purified exosomes exhibited round structures and the sizes distribution varied from 30 to150 nm, which agrees with the former reports. The NTA results demonstrated the distribution of the obtained exosomes ranging between 5 nm and 278 nm in diameter with a peak of 110.5 nm (Fig. 3b). Western blot was used to verify the extracted exosomes through surface protein in exosomes and cells (Fig. 3c).
We then utilized the proposed method and rt-qPCR to quantify exosomal miRNA-21 extracted from HeLa cell and CHO-K1 cell. The result of the two methods showed that a relative high miRNA-21 expression in the HeLa cell (Fig. 3d). Furthermore, we utilized the two methods to quantify exosomal miRNA-21 extracted from the serum of breast cancer patients and that of healthy volunteer, respectively. As shown in Fig. 3e, exosomal miR-21 extracted from serum samples of breast cancer patients was 3 times higher than that of normal controls, indicating overexpression of exosome miR-21 in breast cancer patients. These results are consistent with cellular analysis, revealing the potential of this method to diagnose breast cancer by exosome miR-21. The obtained miRNA-21 quantification results from cell culture supernatant and clinical sample were consistent with former reports, suggesting that this method can provide a good diagnostic reference for miRNA-21 detection of breast cancer in vitro.
We then applied the proposed method for the dynamic in vivo imaging of miRNA-21 in Hela cells. We utilized miR-21 inhibitor, a modified single-stranded RNA molecule, to selectively decrease intracellular miR-21 concentration and miR-21 mimic, a double-stranded RNA mimicking miR-21, to increase miR-21 concentration in Hela cells. From the result in Fig. 4, we can clearly see that a higher fluorescence signals were observed in the Hela cells treated with miR-21 mimic, while a distinguishable decreased fluorescence signals wad observed in the miR-21 inhibitor treated Hela cells, indicating the potential application of the proposed method for dynamic monitoring the intracellular miRNAs.
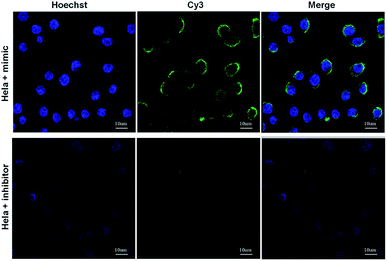 | ||
| Fig. 4 Florescence imaging results of the intracellular miR-21 fluctuations. HeLa cells were pre-treated with miR-21 mimic and miR-21 inhibitor. Scale bar: 10 μm. | ||
The proposed approach has provided a unique avenue for the in vitro and in situ detection of miRNA with the following outstanding features: (1) a high specificity ensured by the CRISPR–Cas9 system and (2) direct detection of miRNAs without additional reverse transcription procedure inevitable in conventional CRISPR-based nucleic acid detection methods. We have then compared the features of the proposed strategy with the former reporter conventional ones (Table 1). Despite that the proposed strategy showed a satisfying result in the sensitivity and specificity, we will conduct more experiments to verify the clinical application potential of the proposed strategy.
We reported here a novel miRNA detection method by integrating the advantages of powerful CHA amplification and CRISPR–Cas9-mediated specific cleavage. Based on this, the sensitivity of this method was determined to be 23 fM. More importantly, this method can be applied in situ imaging of intracellular miRNAs, that opening a new avenue for applications in bioanalysis and disease diagnostics.
Ethical approval statement
Informed consent was obtained from all participants in the clinical trial and protocols were approved by The Third Affiliated Hospital of Jinzhou Medical University Ethics Committee. Approval is given on understanding that unnecessary risks are avoided, patients' safety and privacy are protected and the research plan of this article is in accordance with the principles of medical ethics and the requirements of Declaration of Helsinki.Conflicts of interest
All authors declare that they have no competing interests.Acknowledgements
This work was financially supported by the Science and Technology Project of Liaoning Province (Grant No. 2019-ZD-0815).Notes and references
- M. I. Qadir and A. Faheem, Crit. Rev. Eukaryotic Gene Expression, 2017, 27, 197–204 CrossRef PubMed.
- A. Vishnoi and S. Rani, Methods Mol. Biol., 2017, 1509, 1–10 CrossRef CAS PubMed.
- B. Martinez and P. V. Peplow, Neural Regener. Res., 2020, 15, 606–619 CrossRef PubMed.
- K. Ren, Odontology, 2019, 107, 271–284 CrossRef CAS PubMed.
- X. Zhao, C. Luo, Q. Mei, H. Zhang, W. Zhang, D. Su, W. Fu and Y. Luo, Anal. Chem., 2020 DOI:10.1021/acs.analchem.0c00141.
- X. Zhao, L. Zeng, Q. Mei and Y. Luo, ACS Sens., 2020 DOI:10.1021/acssensors.0c00944.
- R. Deng, K. Zhang and J. Li, Acc. Chem. Res., 2017, 50, 1059–1068 CrossRef CAS.
- T. Tian, J. Wang and X. Zhou, Org. Biomol. Chem., 2015, 13, 2226–2238 RSC.
- K. Guk, S. G. Hwang, J. Lim, H. Y. Son, Y. Choi, Y. M. Huh, T. Kang, J. Jung and E. K. Lim, Chem. Commun., 2019, 55, 3457–3460 RSC.
- H. Kutluk, R. Bruch, G. A. Urban and C. Dincer, Biosens. Bioelectron., 2019, 148, 111824 CrossRef PubMed.
- S. Wang, L. Wang, X. Xu, X. Li and W. Jiang, Anal. Chim. Acta, 2019, 1063, 152–158 CrossRef CAS PubMed.
- N. Zhang, S. Ye, Z. Wang, R. Li and M. Wang, ACS Sens., 2019, 4, 924–930 CrossRef CAS PubMed.
- J. Liu, Y. Zhang, H. Xie, L. Zhao, L. Zheng and H. Ye, Small, 2019, e1902989, DOI:10.1002/smll.201902989.
- T. Yao and X. Hun, Chem. Commun., 2019, 55, 10380–10383 RSC.
- Y. Zang, J. Lei, P. Ling and H. Ju, Anal. Chem., 2015, 87, 5430–5436 CrossRef CAS PubMed.
- B. Koo, D.-e. Kim, J. Kweon, C. E. Jin, S.-H. Kim, Y. Kim and Y. Shin, Sens. Actuators, B, 2018, 273, 316–321 CrossRef CAS PubMed.
- X. Zhao, W. Zhang, X. Qiu, Q. Mei, Y. Luo and W. Fu, Anal. Bioanal. Chem., 2020, 412, 601–609 CrossRef CAS PubMed.
- K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan, N. M. Daringer, I. Bosch, D. M. Dudley, D. H. O'Connor, L. Gehrke and J. J. Collins, Cell, 2016, 165, 1255–1266 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- W. Deng, X. Shi, R. Tjian, T. Lionnet and R. H. Singer, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11870–11875 CrossRef CAS PubMed.
- Y. Fu, P. P. Rocha, V. M. Luo, R. Raviram, Y. Deng, E. O. Mazzoni and J. A. Skok, Nat. Commun., 2016, 7, 11707 CrossRef CAS PubMed.
- B. Zhang, Q. Wang, X. Xu, Q. Xia, F. Long, W. Li, Y. Shui, X. Xia and J. Wang, Anal. Bioanal. Chem., 2018, 410, 2889–2900 CrossRef CAS PubMed.
- R. Wang, X. Zhao, X. Chen, X. Qiu, G. Qing, H. Zhang, L. Zhang, X. Hu, Z. He, D. Zhong, Y. Wang and Y. Luo, Anal. Chem., 2020, 92, 2176–2185 CrossRef CAS PubMed.
- A. Avino, C. S. Huertas, L. M. Lechuga and R. Eritja, Anal. Bioanal. Chem., 2016, 408, 885–893 CrossRef CAS PubMed.
- A. M. James, M. B. Baker, G. Bao and C. D. Searles, Theranostics, 2017, 7, 634–646 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03603j |
| This journal is © The Royal Society of Chemistry 2020 |