Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rubia tinctorum root extracts: chemical profile and management of type II diabetes mellitus
Enas E. Eltamany a,
Mohamed S. Nafie
a,
Mohamed S. Nafie b,
Dina M. Khodeerc,
Aya H. H. El-Tanahyd,
Maged S. Abdel-Kader*ef,
Jihan M. Badr
b,
Dina M. Khodeerc,
Aya H. H. El-Tanahyd,
Maged S. Abdel-Kader*ef,
Jihan M. Badr a and
Reda F. A. Abdelhameed
a and
Reda F. A. Abdelhameed a
a
aDepartment of Pharmacognosy, Faculty of Pharmacy, Suez Canal University, Ismailia 41522, Egypt
bDepartment of Chemistry, Faculty of Science, Suez Canal University, Ismailia 41522, Egypt
cDepartment of Pharmacology & Toxicology, Faculty of Pharmacy, Suez Canal University, Ismailia 41522, Egypt
dDamietta Health Affairs Directorate, Damietta, Egypt
eDepartment of Pharmacognosy, College of Pharmacy, Prince Sattam Bin Abdulaziz University, 173, AlKharj 11942, Saudi Arabia. E-mail: mpharm101@hotmail.com; Tel: +966545539145
fDepartment of Pharmacognosy, Faculty of Pharmacy, Alexandria University, Alexandria 21215, Egypt
First published on 24th June 2020
Abstract
The chemical and biological profiling of the root extracts of Rubia tinctorum was performed. The activities of different extracts were determined considering the antidiabetic effect against type II diabetes mellitus together with anti-obesity and hepatoprotective effects and lipid profile. The methanolic extract of Rubia tinctorum exhibited significant results in decreasing body weight, improving lipid profile, normalizing hyperglycaemia, insulin resistance, hyperinsulinemia. Additionally, it showed enhancement of liver tissue structure and function. The methanolic extract, being the most significant one, was subjected to LC-HRMS analysis to determine its chemical constituents. Finally, the chemical constituents were evaluated by molecular docking study that was carried out to identify the interaction of a panel of 45 compounds in silico and to correlate the structures to their anti-diabetic activity. Among the tested compounds, 1-hydroxy-2-hydroxymethyl anthra-quinone and naringenin-7-O-glucoside showed the most potent activity as α-amylase inhibitors.
1. Introduction
Diabetes mellitus and obesity are important health problems worldwide. They contribute to the development of various pathological processes such as hypertension, cardiovascular diseases, hyperlipidaemia, certain types of cancer and even Alzheimer's.1,2 There is a strong correlation between diabetes and obesity, where adipose tissue has an important role in diabetes, a disease characterized by hyperglycemia, insulin hyposecretion, and insulin resistance.3,4 The exploitation of phyto-medicine as a therapy for diabetes as well as obesity is a crucial issue for the development of safer alternatives to pharmaceuticals which not only decrease blood glucose but also enhance the antioxidant system.5 The genus Rubia belongs to the family Rubiaceae and comprises about 70 species.6 Previous research on Rubia species yielded the isolation and chemical identification of about 250 compounds with different chemical classes which possess diverse pharmacological activities including anti-bacteria, antithrombic, anticancer, anti-inflammatory and anti-oxidant.7–10Rubia tinctorum L. (Madder root) was used as a source of red dyes in ancient Egypt.11 In addition, several ethnobotanical surveys have reported its use for treatment of various ailments, such as cardiovascular disease,12,13 liver pain, diarrhea,14,15 rheumatism16 and kidney stones.17,18 Moreover numerous biological studies have been extensively conducted on R. tinctorum and proved its therapeutic potential as antiplatelet aggregation,19 antitumor,20 hepatoprotective,21 vasoconstriction and protective effect on aorta.22 The current work was designed to evaluate the antiobesity, antidiabetic and hepatoprotective efficacy of different extracts of R. tinctorum along with profiling of potent bioactive components responsible for the antidiabetic activity. In our strategy, we used LC-HRMS technique to identify the phytochemicals in the root extract of R. tinctorum then to determine the binding mode of the tested compounds with 1HX0 as α-amylase inhibitor by using molecular docking tool.
2. Materials and methods
2.1. In vitro determination of hypoglycemic activity, liver enzymes and lipid profile
Portion of liver tissue was removed from the biggest hepatic lobe, fixed in formaldehyde and finally stained with hematoxylin and eosin (H&E).
2.1.6.1. Liver enzymes. Spectrophotometrically method was done with marketable kits (Biocon Diagnostic, Germany) to evaluate serum activity of liver enzymes; alanine transaminase enzyme (ALT) (EC 2.6.1.2), aspartate transaminase enzyme (AST) (EC 2.6.1) in accordance with the protocol reported by the manufacturer.26
2.1.6.2. Lipid profile. A spectrophotometric assay kits (Spinreact, Spain) were used to measure serum total cholesterol (TC)(CHOD-POD), triglycerides (TGs)(GPO-POD, Líquido), high-density lipoprotein (HDL) (HDLc-P) and low-density lipoprotein (LDL)(LDLc-D) according to the manufacturer's protocol.27,28
2.1.6.3. Insulin & leptin ELISA kits. The level of serum insulin and leptin were determined by a rat insulin and leptin ELISA kits (PELOBIOTECH GmbH-Am Klopferspitz 19-82152 planning-Germany) following the manufacturer's protocol. Insulin resistance was determined using the homeostasis model assessment index for insulin resistance (HOMA-IR) index.29
2.2. Preparation of the sample and LC-HRMS analysis
The mobile phase working solution (MP-WS) was prepared from DI-water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile (50
acetonitrile (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25). One mL of MP-WS was added to 50 mg weighted dry methanolic extract, vortex for 2 min. This step was followed by ultra-sonication for 10 min then centrifugation for 10 min at 10
25). One mL of MP-WS was added to 50 mg weighted dry methanolic extract, vortex for 2 min. This step was followed by ultra-sonication for 10 min then centrifugation for 10 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. 20 μL stock (50/1000 μL) was diluted with 1000 μL reconstitution solvent. Finally, the injected concentration was 1 μg μL−1 where 10 μLs were injected on positive mode. Also, 10 μL MP-WS were injected as a blank sample. The used mobile phase consisted of (A): 5 mM ammonium formate buffer pH 3 containing 1% methanol and (B): 100% acetonitrile. The flow rate was 0.3 mL min−1. The used pre-column was in-line filter disks (Phenomenex, 0.5 μm × 3.0 mm) and the column was X select HSS T3 (Waters, 2.5 μm, 2.1 × 150 mm). Data processing was via MS-DIAL3.52. Master view was used for feature (peaks) extraction from total ion chromatogram based on the following criteria: features should have signal-to-noise greater than 5 (non-targeted analysis) and features intensities of the sample-to-blank should be greater than 5.
000 rpm. 20 μL stock (50/1000 μL) was diluted with 1000 μL reconstitution solvent. Finally, the injected concentration was 1 μg μL−1 where 10 μLs were injected on positive mode. Also, 10 μL MP-WS were injected as a blank sample. The used mobile phase consisted of (A): 5 mM ammonium formate buffer pH 3 containing 1% methanol and (B): 100% acetonitrile. The flow rate was 0.3 mL min−1. The used pre-column was in-line filter disks (Phenomenex, 0.5 μm × 3.0 mm) and the column was X select HSS T3 (Waters, 2.5 μm, 2.1 × 150 mm). Data processing was via MS-DIAL3.52. Master view was used for feature (peaks) extraction from total ion chromatogram based on the following criteria: features should have signal-to-noise greater than 5 (non-targeted analysis) and features intensities of the sample-to-blank should be greater than 5.
2.3. Molecular docking
Molecular modelling study was carried out to study the interaction of a panel of 28 anthraquinone and 17 flavonoids in silico and to correlate the structures to their anti-diabetic activity. Molecular docking study was conducted on a computational software basis using the Molecular Operating Environment (MOE 2014.09 Chemical Computing Group, Canada). The three-dimensional structures of 1HX0 completed with AC1 as alpha-amylase inhibitor was freely accessible from the protein data bank (https://www.rcsb.org/structure/1HX0).30 Compounds were chemically optimized and energetically minimized, while the receptor was prepared and manipulated using routine protocol according to Nafie et al.31 The active sites were defined using grid boxes of appropriate sizes around the co-crystallized ligands. These compounds were docked into the receptor active site, each ligand-receptor complex was tested for binding energy using MOE and interaction analysis using Chimera as a visualizing software.3. Results and discussion
3.1. Effect of different R. tinctorum extracts and pioglitazone (10 mg kg−1) on percent change in body weight and obesity index on type II diabetic rats
Treatment with high fat diet followed with STZ (30 mg kg−1) in diabetic group resulted in a significant increase in final body weight (352 ± 7.5), % change in body weight (152 ± 14.1) and obesity index (5.9 ± 0.5) versus normal group (210 ± 10) (42.5 ± 9) and (0.83 ± 0.07) respectively at p ≤ 0.05 (Table 1). Treatment with pioglitazone (10 mg kg−1) for four weeks after induction of diabetes induced significantly decrease in final body weight, % change in body weight and obesity index when compared with diabetic group at p ≤ 0.05. Pioglitazone is one member from thiazolidines, as it is PAPR-gamma agonist in particular is known to favorably influence the majority of the components of insulin resistance characteristic of type 2 diabetes mellitus including adiposity, dyslipidaemia, hyperglycaemia and changes in liver and ovaries.32 However, its effect in weight gain was previously discussed.33,34 On the other hand, its role in decreasing weight gain, enhancing lipid profile and stimulation of lipid mobilization from visceral part to the lower part of body was also reported.35,36| Group | Base line body weight (g) | Final body weight (g) | % change in body weight | Obesity index |
|---|---|---|---|---|
| a Results are expressed as mean ± S. E. M. and analysed using one-way ANOVA followed by Bonferroni's test for multiple comparisons. aP ≤ 0.05 versus normal group. bP ≤ 0.05 versus diabetic group. cP ≤ 0.05 versus diabetic + pioglitazone (10 mg kg−1) group. dP ≤ 0.05 diabetic + RM (200 mg kg−1) group, eP ≤ 0.05 diabetic + RH (200 mg kg−1) group. n = 5. | ||||
| Normal | 147.5 ± 2.5 | 210 ± 10 | 42.5 ± 9 | 0.83 ± 0.07 |
| Diabetic | 140 ± 5 | 352 ± 7.5a | 152 ± 14.1a | 5.9 ± 0.5a |
| Diabetic + pioglitazone (10 mg kg−1) | 143.5 ± 1.5 | 227.5 ± 17.6b | 58 ± 13.6b | 2 ± 0.36b |
| Diabetic + RM (200 mg kg−1) | 167.5 ± 7.5 | 308 ± 8a,b,c | 84 ± 3.4a,b | 2.5 ± 0.89a,b |
| Diabetic + RH (200 mg kg−1) | 140 ± 2 | 333.5 ± 16.6a,c,e | 138 ± 8.2a,c,d | 5.1 ± 0.17a,c,d |
| Diabetic + RC (200 mg kg−1) | 141 ± 4 | 291.5 ± 18.6a–c | 106 ± 7.1a–c | 3.7 ± 0.1a–c |
| Diabetic + RE (200 mg kg−1) | 145 ± 10 | 297.3 ± 3a–c | 106 ± 15.9a–c | 5.3 ± 0.03a,c–e |
The current results are in agreement with these articles, as pioglitazone treatment reduced total body weight and decreased liver-fat resulting in elevation of insulin sensitivity in these tissues. Additionally, the effect of pioglitazone is related to the correct choice of its dose as lower and higher doses of pioglitazone may exert no or adverse action like sodium water retention and weight gain.37 So, the dose of pioglitazone should be monitored and well selected.23 In the current study the selected dose was (10 mg kg−1) which is considered to be medium dose and has significant effect in all measured parameters.
Similarly, diabetic rats treated with RM, RC or RE (each 200 mg kg−1) extracts showed a significant decrease in final body weight and % change in body weight, however, only diabetic rats treated with RM (200 mg kg−1) and RC (200 mg kg−1) showed a significant improvement in obesity index compared with diabetic group. On the other hand, the group treated with the extract RH (200 mg kg−1) couldn't show any significant enhancement in final body weight, % change in body weight or obesity index in comparison with diabetic group at p ≤ 0.05. Moreover, the results achieved by the treatment with the extract RM (200 mg kg−1) were the best in improving the decrease % change in body weight and obesity index (Table 1).
3.2. Effect of different R. tinctorum extracts and pioglitazone (10 mg kg−1) on blood glucose level, serum insulin, insulin resistance and serum leptin level on type II diabetic rats
The current results showed significant increases in blood glucose level (mM L−1), serum insulin level (ng L−1), HOMA-IR and serum leptin level (ng L−1) in diabetic group in comparison with normal group at p ≤ 0.05 (Table 2). However, treatment with pioglitazone (10 mg kg−1) significantly decreased blood glucose level (mM L−1), serum insulin level (ng L−1), HOMA-IR and serum leptin level (ng L−1) when compared to diabetic group at p ≤ 0.05. Additionally, treatment with RM, RH, RC and RE (each of 200 mg kg−1) significantly induced a decrease in blood glucose level (mM L−1), serum insulin level (ng L−1), HOMA-IR and serum leptin level (ng L−1) when compared to diabetic group at p ≤ 0.05. However, the most significant results were obtained from the group treated with RM (200 mg kg−1) concerning normalization of the level of serum leptin (ng L−1) at p ≤ 0.05 (Table 2).| Group | Blood glucose level (mM L−1) | Serum insulin (ng L−1) | HOMA-IR | Serum leptin (ng L−1) |
|---|---|---|---|---|
| a Results are expressed as mean ± S. E. M. and analysed using one-way ANOVA followed by Bonferroni's test for multiple comparisons. aP ≤ 0.05 versus normal group. bP ≤ 0.05 versus diabetic group. cP ≤ 0.05 versus diabetic + pioglitazone (10 mg kg−1) group. dP ≤ 0.05 diabetic + RM (200 mg kg−1) group. n = 5. | ||||
| Normal | 97 ± 3.9 | 2.4 ± 0.05 | 14.3 ± 0.87 | 3.8 ± 0.3 |
| Diabetic | 172 ± 24a | 7.6 ± 0.29a | 74.79 ± 16.7a | 14 ± 1a |
| Diabetic + pioglitazone (10 mg kg−1) | 110 ± 5.6b | 3.9 ± 0.02a,b | 26.5 ± 1.5b | 4.9 ± 0.3b |
| Diabetic + RM (200 mg kg−1) | 99 ± 2b | 4 ± 0.06a,b | 29.8 ± 0.99b | 5.6 ± 0.1b |
| Diabetic + RH (200 mg kg−1) | 125 ± 7.6b | 4.1 ± 0.1a,b | 34.6 ± 2.8b | 10.5 ± 0.5a–d |
| Diabetic + RC (200 mg kg−1) | 111 ± 2.0b | 4.2 ± 0.16a,b | 30.14 ± 1.6b | 8.8 ± 0.7a–d |
| Diabetic + RE (200 mg kg−1) | 102 ± 0.66b | 4 ± 0.6a,b | 25.5 ± 0.58b | 8.8 ± 0.15a–d |
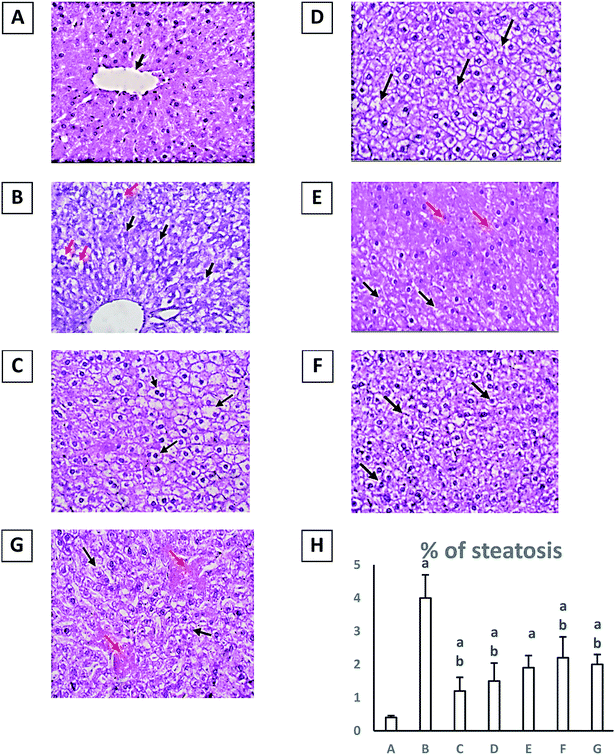
3.3. Effect of different R. tinctorum extracts and pioglitazone (10 mg kg−1) on liver index, serum liver enzymes level, hepatic tissue histopathological changes and percent of steatosis on type II diabetic rats
In the current study, diabetic group showed a significant increase in liver index with a value (3.5 ± 0.2) versus (2.3 ± 0.1) in normal group at p ≤ 0.05 (Table 3). Diabetic group showed elevation in serum liver enzymes AST and ALT in comparison with normal group at p ≤ 0.05. Furthermore, treatment of diabetic rats with only pioglitazone (10 mg kg−1) or the extract RM (200 mg kg−1) for four weeks could significantly normalize the liver index in comparison with diabetic group at p ≤ 0.05. Additionally, treatment with pioglitazone (10 mg kg−1), RM, RH, RC and RE (each 200 mg kg−1) significantly induced a decrease in both two serum liver enzymes ALT and AST in comparison with diabetic group at p ≤ 0.05.| Group | Liver index | AST (U L-1) | ALT (U L-1) |
|---|---|---|---|
| a Results are expressed as mean ± S. E. M. and analyzed using one-way ANOVA followed by Bonferroni's test for multiple comparisons. aP ≤ 0.05 versus normal group. bP ≤ 0.05 versus diabetic group. cP ≤ 0.05 versus diabetic + pioglitazone (10 mg kg−1) group. dP ≤ 0.05 diabetic + RM (200 mg kg−1) group, eP ≤ 0.05 diabetic + RH (200 mg kg−1) group. n = 5. | |||
| Normal | 2.3 ± 0.1 | 39 ± 1 | 34.5 ± 1.5 |
| Diabetic | 3.5 ± 0.2a | 92.5 ± 0.5a | 78 ± 2.02a |
| Diabetic + pioglitazone (10 mg kg−1) | 2.5 ± 0.3b | 50.5 ± 1.5a,b | 26.5 ± 2.5b |
| Diabetic + RM (200 mg kg−1) | 2.5 ± 0.02b | 35 ± 1.01b,c | 30 ± 1.01b |
| Diabetic + RH (200 mg kg−1) | 3.9 ± 0.07a,c,d | 72 ± 1.01a–d | 55 ± 1.01a–d |
| Diabetic + RC (200 mg kg−1) | 3.07 ± 0 0.04a,c–e | 65.5 ± 2.5a–e | 46.5 ± 1.5a–e |
| Diabetic + RE (200 mg kg−1) | 3.35 ± 0.02a,c–e | 66 ± 1.01a–e | 46 ± 1.01a–e |
Finally, diabetic group showed evidence of injury; hydropic degeneration (black arrows) and a significant increase in percent of steatosis (red arrows) (H&E, 40×) when compared to normal group at p ≤ 0.05 (Fig. 1). Treatment with either pioglitazone (10 mg kg−1) or any of the Rubia extracts showed enhancement in liver architecture and a significant decrease in percent of steatosis with a significant reduction in hydropic degeneration of hepatocytes (black arrows) and many hepatocytes show uniform morphology (red arrows) (H&E, 40×) in comparison with diabetic group at p ≤ 0.05 (Fig. 1). However, the results obtained by treatment with the extract RM were the best and closer to the normal group (Table 3).
3.4. Effect of different R. tinctorum extracts and pioglitazone (10 mg kg−1) on lipid profile, serum triglycerides (TG), total cholesterol (TC), high-density lipoprotein (HDL) and low-density lipoprotein (LDL) on type II diabetic rats
Treatment with high fat diet followed with STZ (30 mg kg−1) in diabetic group resulted in a significant increase in serum triglycerides (TG) (mg dL−1), total cholesterol (TC) (mg dL−1) and low-density lipoprotein (LDL) (mg dL−1) and a significant decrease in high-density lipoprotein (HDL) (mg dL−1) in comparison with normal group at p ≤ 0.05 (Table 4). Treatment with pioglitazone (10 mg kg−1) resulted in a significant decrease in serum triglycerides (TG) (mg dL−1), total cholesterol (TC) (mg dL−1) and low-density lipoprotein (LDL) (mg dL−1) and a significant increase in high-density lipoprotein (HDL) (mg dL−1) in comparison with diabetic group at p ≤ 0.05.| Group | Serum TG (mg dL−1) | Serum TC (mg dL−1) | HDL (mg dL−1) | LDL (mg dL−1) |
|---|---|---|---|---|
| a Results are expressed as mean ± S. E. M. and analyzed using one-way ANOVA followed by Bonferroni's test for multiple comparisons. aP ≤ 0.05 versus normal group. bP ≤ 0.05 versus diabetic group. cP ≤ 0.05 versus diabetic + pioglitazone (10 mg kg−1) group. dP ≤ 0.05 diabetic + RM (200 mg kg−1) group, eP ≤ 0.05 diabetic + RH (200 mg kg−1) group. n = 5. | ||||
| Normal | 54.5 ± 3.5 | 63 ± 3.03 | 45 ± 11 | 24.5 ± 0.5 |
| Diabetic | 161 ± 16.1a | 116.5 ± 3.5a | 20 ± 2.02a | 55.5 ± 4.5a |
| Diabetic + pioglitazone (10 mg kg−1) | 42.5 ± 2.5b | 88.5 ± 1.6a,b | 40.5 ± 3.5b | 30 ± 2.02b |
| Diabetic + RM (200 mg kg−1) | 68 ± 1.01b,c | 72 ± 1.01a–c | 36 ± 0.5b | 23 ± 1.01b |
| Diabetic + RH (200 mg kg−1) | 98.5 ± 0.5a–d | 89.5 ± 0.5a,b,d | 30.5 ± 0.5a | 46.5 ± 0.5a–d |
| Diabetic + RC (200 mg kg−1) | 85 ± 2.02a–c | 82.5 ± 0.5a–e | 26 ± 1.01a,c | 45.5 ± 3.5a–d |
| Diabetic + RE (200 mg kg−1) | 80.5 ± 0.5a–c | 76.5 ± 0.5a–c,e | 26.5 ± 0.5a | 39.5 ± 0.5a–d |
Furthermore, all extracts induced a significant decrease in serum triglycerides (TG) (mg dL−1), total cholesterol (TC) (mg dL−1) and low-density lipoprotein (LDL) (mg dL−1) at p ≤ 0.05 (Table 4).
Moreover, treatment with RM (200 mg kg−1) resulted in a significant improvement and normalization in high-density lipoprotein (HDL) serum level (mg dL−1) in comparison with diabetic group at p ≤ 0.05 (Table 4).
Accordingly, the results obtained from treatment of the diabetic rats with the methanolic extract of Rubia tinctorum were the best and more close to normal group and pioglitazone treated group either in decreasing body weight, obesity, improving lipid profile, normalization hyperglycaemia, insulin resistance, hyperinsulinemia or in enhancing liver tissue structure and function.
3.5. LC-HRMS analysis
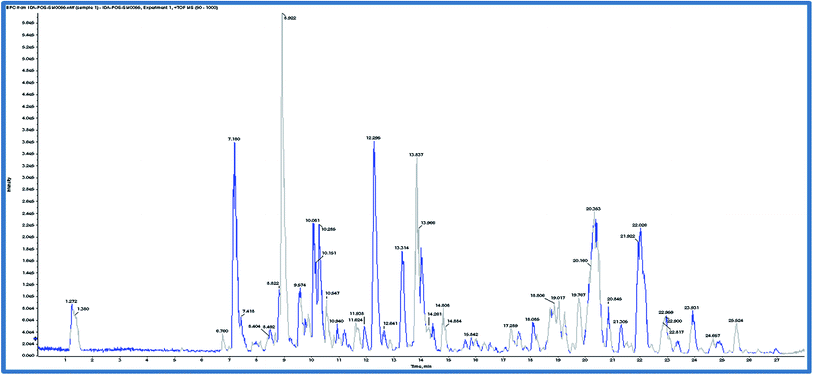
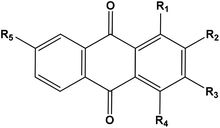
Based on the results of the biological activities of different extracts of R. tinctorum which revealed that the best extract was the methanolic one, accordingly, the methanolic extract was subjected to LC-HRMS analysis to detect its chemical constituents (Fig. 2). The results showed 45 hits as indicated in Tables 5 and 6.| Rt (min) | Molecular formula | m/z [M + H]+ | Name of the compound | Substituents | ||||
|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | R5 | ||||
| a *, **, ***, ****, #, ##, ###: interchangeable values. | ||||||||
| 7.18 | C26H28O14 | 565.1557 | Lucidin-3-O-primveroside | OH | CH2OH | O-primverose | H | |
| 8.82 | C25H26O13 | 535.1451 | Ruberythric acid | OH | O-Glucose (6-1)xylose | H | H | |
| 8.92 | C15H8O7 | 301.0348 | Pseudopurpurin | OH | COOH | OH | OH | H |
| 9.57 | C15H8O6 | 285.0399 | Munjistin | OH | COOH | OH | H | |
| 10.08* | C14H8O5 | 257.045 | Purpurin | OH | OH | H | OH | H |
| 10.28* | C14H8O5 | 257.045 | Anthragallol | OH | OH | OH | H | H |
| 12.29** | C14H8O4 | 241.0501 | Alizarin | OH | OH | H | H | H |
| 12.31** | C14H8O4 | 241.0501 | Xanthopurpurin | OH | H | OH | H | H |
| 12.64 | C15H10O5 | 271.0606 | 1,4-Dihydroxy-2-hydroxymethyl anthraquinone | OH | CH2OH | H | OH | H |
| 12.64 | C14H8O3 | 225.0551 | 2-Hydroxy anthraquinone | H | OH | H | H | H |
| 13.31 | C15H10O4 | 255.0657 | Rubiadin | OH | CH3 | OH | H | H |
| 13.96 | C17H14O5 | 299.0919 | 1,4-Dihydroxy-2-ethoxymethyl anthraquinone | OH | CH2OC2H5 | H | OH | H |
| 13.77*** | C15H10O5 | 271.0606 | Anthragallol-3-methylether | OH | OH | OCH3 | H | H |
| 13.83*** | C15H10O5 | 271.0606 | Lucidin | OH | CH2OH | OH | H | H |
| 18.08**** | C15H10O3 | 239.0708 | 1-Hydroxy-2-methyl AQ | OH | CH3 | H | H | H |
| 18.75**** | C15H10O3 | 239.0708 | 7-Hydroxy-2-methyl AQ | H | CH3 | H | H | OH |
| 18.80# | C15H10O4 | 255.0657 | Xanthopurpurin-3-methylether | OH | H | OCH3 | H | H |
| 19.02# | C15H10O4 | 255.0657 | Alizarin-1-methyl ether | OCH3 | OH | H | H | H |
| 19.53# | C15H10O4 | 255.0657 | Alizarin -2-methyl ether | OH | OCH3 | H | H | H |
| 19.77# | C15H10O4 | 255.0657 | Xanthopurpurin-1-methylether | OCH3 | H | OH | H | H |
| 20.16 | C15H10O4 | 255.0657 | 1-Hydroxy-2-hydroxymethyl anthraquinone | OH | CH2OH | H | H | H |
| 20.5 | C16H12O5 | 285.0763 | Anthragallol-2,3-dimethylether | OH | OCH3 | OCH3 | H | H |
| 20.84 | C15H10O3 | 239.0708 | 2-Methoxy- anthraquinone | H | OCH3 | H | H | H |
| 21.30## | C16H12O4 | 269.0814 | Alizarin-dimethyl ether | OCH3 | OCH3 | H | H | H |
| 21.40## | C16H12O4 | 269.0814 | Xanthopurpurin dimethylether | OCH3 | H | OCH3 | H | H |
| 23.9### | C16H12O3 | 253.0864 | 1-Methoxy methyl anthraquinone | CH2OCH3 | H | H | H | H |
| 24.12### | C16H12O3 | 253.0864 | 1-Methoxy-2-methyl anthraquinone | OCH3 | CH3 | H | H | H |
| 25.52 | C17H14O3 | 266.0943 | 2-Ethoxymethyl anthraquinone | H | CH2OC2H5 | H | H | H |
The anthraquinones were previously reported to be major components of Rubia species specially and family Rubiaceae generally.38 Table 5 revealed the presence of 28 anthraquinones. All of the detected compounds were previously reported to be isolated from the Rubia tinctorum.38–41
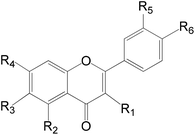
On the other hand, a number of anthraquinones were previously reported to be isolated from the same plant as for example nor damnacanthal and lucidin methyl ether42 were not recorded her in the present study. Additionally, family Rubiaceae revealed the presence of a number of flavonoids of which derivatives of quercetin, rhamnetin, isorhamnetin, apigenin and kaempferol are the most common.42 In this work a number of 17 flavonoids were recorded as minor constituents of Rubia tinctorum (Table 6). On the other hand, rutin which was previously reported to be isolated from R. tinctorum38 was not detected here, and instead, quercetrin (which could be considered as a secondary glycoside of rutin) was recorded.
3.6. Docking study
α-Amylase has been considered as an important therapeutic target for the management of type 2 diabetes mellitus, hence, we aimed to elucidate the binding mode of the tested compounds with 1HX0 as α-amylase inhibitor.43 We performed induced fit molecular docking studies with the compounds under investigation. The docking results with docking scores, and the hydrogen bonded residues are given in Table 7. Additionally, 3D representative images of one of the high binding affinities of both anthraquinone and flavonoids compared to AC1 as the co-crystallized ligand are shown in Fig. 3.| Group | Binding affinity | Identified compound | Binding energy (kcal mol−1) | Ligand–receptor interactions with |
|---|---|---|---|---|
| a #Highly-bonded interactive docked compounds in the same way like AC1. *The rest of compounds of both groups weren't able to bind with the receptor pocket. | ||||
| Anthraquinone* | High | 1-Hydroxy-2-hydroxymethyl AQ# | −13.92 | 2 HB with Gly 106 and Val 163 |
| Ruberythric acid | −21.03 | 2 HB with Val 163 | ||
| Mild | 1,4-Dihydroxy-2-ethoxymethyl anthraquinone | −9.05 | 1 HB with either Val 163 or Gly 106 | |
| 2-Hydroxy anthraquinone | −9.36 | |||
| 2-Methoxy-anthraquinone | −10.43 | |||
| Lucidin-3-O-primveroside | −24.9 | |||
| Lucidin | −11.9 | |||
| Purpurin | −14.78 | |||
| Xanthopurpurin dimethylether | −11.34 | |||
| Xanthopurpurin | −11.68 | |||
| Rubiadin | −15.45 | |||
| Flavonoids* | High | Baicalein-7-O-glucuronide | −19.66 | 2 HB with Val 163 |
| Kaempferol-3-O-L-rhamnoside | −20.95 | 2 HB with Val 163 | ||
| Naringenin-7-O-glucoside# | −16.16 | 2 HB with Gly 106 and Val 163 | ||
| Neohesperidin dihydrochalcon | −23.56 | 2 HB with Gly 106 | ||
| Mild | Acacetin 7-rutinoside | −21.21 | 1 HB with either Val 163 or Gly 106 | |
| Apigenin | −16.59 | |||
| Hesperetin | −20.98 | |||
| Hesperetin-7-O-neohesperidoside | −25.20 | |||
| Isorhamnetin-3-O-glucoside | −22.21 | |||
| Kaempferol-7-neohesperidoside | −17.04 | |||
| Luteolin | −18.08 | |||
| Luteolin-3-7-diglucoside | −18.38 | |||
| Luteolin-7-O-glucoside | −27.45 | |||
| Naringenin | −17.67 | |||
| Quercetin-3-D-xyloside | −19.95 | |||
| Quercetin | −20.89 | |||
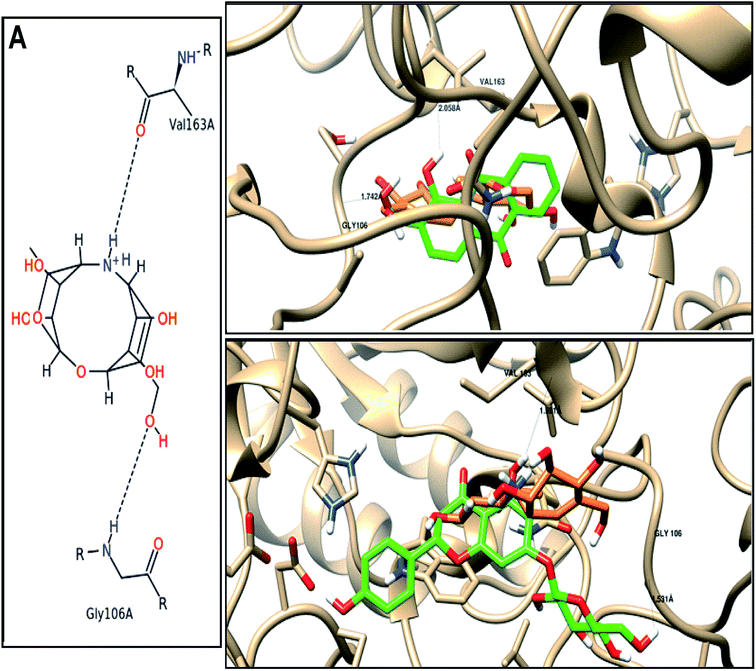 | ||
| Fig. 3 Binding disposition and ligand–receptor interactions of (A) co-crystallized ligand (AC1), and the two high affinity. Docked compounds; (B) 1-hydroxy-2-hydroxymethyl AQ, and (C) naringenin-7-O-glucoside inside the 1HX0 binding site as alpha-amylase inhibitor. Co-crystallized ligand (orange), and docked compounds (green). | ||
As shown in Table 7, most of the tested derivative were docked and bound to amino acids of the receptor binding site with high and mild binding affinities (energies), while other derivatives couldn't be docked. In reference to the co-crystallized ligand (AC1) forms two major interactions with the Val 163 and Gly 106 as the key amino acids residues, we found two of the anthraquinones with high binding affinity (−13.92–21.03 kcal mol−1), and four flavonoids (−16.16–23.56 kcal mol−1) towards α-amylase inhibition by forming the same key interactions. Nine of the anthraquinones, and twelve of flavonoids showed mild binding affinities (−9.05–15.45 kcal mol−1) and (−16.59–27.45 kcal mol−1), respectively, as they form only one hydrogen bond with either Val 163 or Gly 106.
As shown in Fig. 3, three-dimensional representation of two highly docked compounds as two active leads relative to the AC1 with moieties of ligand and receptor involved in the interaction, interaction-type, bond-length for each docking procedure. Among anthraquinone, 1-hydroxy-2-hydroxy-methyl anthraquinone forms two hydrogen bonds through the hydroxyl groups as H-donor with Val 163 with bonds length 2.05 A, and H-acceptor with Gly 106 with bonds length 1.74 A. Among flavonoids, naringenin-7-O-glucoside forms two hydrogen bonds through the hydroxyl groups as H-donors with Val 163 and Gly 106 with bonds length 1.59 and 1.53 Å.
From docking study, we conclude the good affinity of compounds under investigation through their hydroxyl and carbonyl active groups that bind inside the tested target (α-amylase inhibition compared to AC1), which is correlated to anti-diabetic activity.
In addition to α-amylase inhibition activity, most of the compounds detected in the extract proved to possess antioxidant and anti-inflammatory activities.44–46 It is well known that both activities play an important role in treatment of diabetes mellitus.47,48 This also can justify the activity of the methanolic extract of Rubia tinctorum which accumulate a great number of compounds showing antioxidant, anti-inflammatory and α-amylase inhibition activities.
4. Conclusions
The current study proved that the methanolic extract of Rubia tinctorum showed significant results in decreasing body weight, improving lipid profile, normalizing hyperglycaemia, insulin resistance, hyperinsulinemia in addition to enhancing liver tissue structure and function. There activities could be attributed to its chemical constituents that exert antioxidant, anti-inflammatory and α-amylase inhibition activities. As indicated by the docking study, 1-hydroxy-2-hydroxymethyl anthraquinone and naringenin-7-O-glucoside were the most potent as α-amylase inhibitors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This publication was supported by the Deanship of Scientific Research at Prince Sattam Bin Abdulaziz University, Alkharj, Saudi Arabia as well as Suez Canal University, Ismailia, Egypt.References
- J. R. Petrie, T. J. Guzik and R. M. Touyz, Can. J. Cardiol., 2018, 34(5), 575–584 CrossRef PubMed.
- S. S. Khalaf, M. M. Hafez, E. T. Mehanna, N. M. Mesbah and D. M. Abo-Elmatty, Int. J. Res. Pharm. Biomed. Sci., 2019, 3, 1–14 Search PubMed.
- B. Saad, H. Zaid, S. Shanak and S. Kadan, in Anti-diabetes and Anti- obesity Medicinal Plants and Phytochemicals, Springer Nature, Switzerland, 2017, vol. 1, pp. 3–19 Search PubMed.
- Y. Shi and F. B. Hu, Lancet, 2014, 383, 1947–1948 CrossRef.
- W. J. Sandborn and W. A. Faubion, Curr. Gastroenterol. Rep., 2000, 2, 440–445 CrossRef CAS PubMed.
- K. N. Abdel-Khalik, M. M. El-Ghani and A. R. Elkordy, Turk. J. Bot., 2008, 32, 353–359 Search PubMed.
- N. H. Tan and J. Zhou, Chem. Rev., 2006, 106, 840–895 CrossRef CAS PubMed.
- S. M. Zhao, B. Kuang, J. T. Fan, H. Yan, W. Y. Xu and N. H. Tan, Chimia, 2011, 65, 952–956 CrossRef CAS PubMed.
- K. Xu, P. L. Wang, B. Yuan, Y. T. Cheng, Q. Li and H. M. Lei, Chem. Cent. J., 2013, 7, 81–110 CrossRef PubMed.
- K. Xu, P. L. Wang, L. Wang, C. M. Liu, S. X. Xu, Y. T. Cheng, Y. H. Wang, Q. Li and H. M. Lei, Chem. Biodiversity, 2014, 11, 341–363 CrossRef CAS PubMed.
- H. E. Ahmed, I. F. Tahoun, I. Elkholy, A. B. Shehata and Y. Ziddan, Dyes Pigm., 2017, 145, 486–492 CrossRef CAS.
- J. Bellakhdar, R. Claisse, J. Fleurentin and C. Younos, J. Ethnopharmacol., 1991, 35, 123–143 CrossRef CAS PubMed.
- H. Jouad, M. Haloui, H. Rhiouani, J. El Hilaly and M. Eddouks, J. Ethnopharmacol., 2001, 77, 175–182 CrossRef CAS PubMed.
- M. Eddouks, M. Maghrani, A. Lemhadri, L. M. Ouahidi and H. Jouad, J. Ethnopharmacol., 2002, 82, 97–103 CrossRef CAS PubMed.
- M. El Haouari and J. A. Rosado, Phytother. Res., 2016, 30, 1059–1071 CrossRef CAS PubMed.
- M. Adams, C. Berset, M. Kessler and M. Hamburger, J. Ethnopharmacol., 2009, 121, 343–359 CrossRef PubMed.
- R. T. Gunther, J. Goodyer, The Greek herbal of Dioscorides. New York: Hafner Publishing, 1959 Search PubMed.
- K. Agarwal and R. Varma, J. Ethnopharmacol., 2015, 174, 17–24 CrossRef PubMed.
- F. Z. Marhoume, M. A. Laaradia, Z. Younes, J. Laadraoui, S. Oufkir, R. Aboufatima, A. Chait and A. Bagri, J. Ethnopharmacol., 2019, 241, 111971 CrossRef PubMed.
- L. Rashan, H. Lukmanul, F. Heinz, K. Gerhardt, M. Irmgard, A. Mohammed and H. Sidgi, Jordan J. Biol. Sci., 2018, 11, 489–494 CAS.
- F. Z. Marhoume, Z. Younes, H. Boufous, N. Errafiy, M. Ait Laaradia, J. Laadraoui, A. Hakmaoui, B. Abdellah and C. Abderrahman, Eur. J. Med. Plants, 2017, 20, 1–10 CrossRef.
- H. Nejat, K. Sedaghat, A. Vakili, M. Jarrahi and M. Z. Khorasani, Jundishapur J. Nat. Pharm. Prod., 2017, 12(3), e64319, DOI:10.5812/jjnpp.64319.
- D. M. Khodeer, S. A. Zaitone, N. E. Farag and Y. M. Moustafa, Can. J. Physiol. Pharmacol., 2016, 94, 463–476 CrossRef CAS PubMed.
- Z. A. Saad, D. M. Khodeer, S. A. Zaitone, A. A. M. Ahmed and Y. M. Moustafa, Life Sci., 2020, 253, 117725, DOI:10.1016/j.lfs.2020.117725.
- S. Zaitone, N. Hassan, N. El-Orabi and E.-S. El-Awady, Eur. J. Pharmacol., 2011, 662, 70–77 CrossRef CAS PubMed.
- M. Pan, Y. L. Song, J. M. Xu and H. Z. Gan, J. Pineal Res., 2006, 41, 79–84 CrossRef CAS PubMed.
- C. C. Allain, L. S. Poon, C. S. Chan, W. Richmond and P. C. Fu, Clin. Chem., 1974, 20, 470–475 CrossRef CAS.
- P. Fossati and L. Prencipe, Clin. Chem., 1982, 28, 2077–2080 CrossRef CAS.
- D. R. Matthews, J. P. Hosker, A. S. Rudenski, B. A. Naylor, D. F. Treacher and R. C. Turner, Diabetologia, 1985, 28, 412–419 CrossRef CAS PubMed.
- M. Qian, V. Nahoum, J. Bonicel, H. Bischoff, B. Henrissat and F. Payan, Biochemistry, 2001, 40, 7700–7709 CrossRef CAS PubMed.
- M. S. Nafie, M. A. Tantawy and G. A. Elmgeed, Steroids, 2019, 152, 108485 CrossRef CAS PubMed.
- S. Banerjee, J. Assoc. Physicians India, 2012, 60, 62–66 Search PubMed.
- J. Rosenstock, S. W. Kim, M. A. Baron, R.-P. Camisasca, F. Cressier, A. Couturier and S. Dejager, Diabetes, Obes. Metab., 2007, 9, 175–185 CrossRef CAS PubMed.
- D. Russell-Jones, R. M. Cuddihy, M. Hanefeld, A. Kumar, J. G. González, M. Chan, A. M. Wolka and M. K. Boardman, Diabetes Care, 2012, 35, 252–258 CrossRef CAS PubMed.
- N. Kodama, N. Tahara, A. Tahara, A. Honda, Y. Nitta, M. Mizoguchi, H. Kaida, M. Ishibashi, T. Abe, H. Ikeda, J. Narula, Y. Fukumoto, S. Yamagishi and T. Imaizumi, J. Clin. Endocrinol. Metab., 2013, 98, 4438–4445 CrossRef CAS PubMed.
- P. K. Shah, S. Mudaliar, A. R. Chang, V. Aroda, M. Andre, P. Burke and R. R. Henry, Diabetes, Obes. Metab., 2011, 13, 505–510 CrossRef CAS PubMed.
- D. K. McGuire and S. E. Inzucchi, Circulation, 2008, 117, 440–449 CrossRef PubMed.
- G. C. H. Derksen and T. A. van.Beek, Stud. Nat. Prod. Chem., 2002, 26, 629–684 CAS.
- G. Cuoco, C. Mathe, P. Archier, F. Chemat and C. Vieillescazes, Ultrason. Sonochem., 2009, 16, 75–82 CrossRef CAS PubMed.
- L. Ford, C. M. Rayners and R. S. Blackburn, Phytochemistry, 2015, 117, 168–173 CrossRef CAS PubMed.
- I. Essaidi, A. Snoussi, H. Ben Haj Koubaier, H. Casabianca and N. Bouzouita, Pigm. Resin Technol., 2017, 46, 379–384 CrossRef CAS.
- D. Martins and C. V. Nunez, Molecules, 2015, 20, 13422–13495 CrossRef CAS PubMed.
- C. Proença, M. Freitas, D. Ribeiro, S. M. Tomé, E. F. T. Oliveira, M. F. Viegas, A. N. Araújo, M. J. Ramos, A. M. S. Silva, P. A. Fernandes and E. Fernandes, J. Enzyme Inhib. Med. Chem., 2019, 34, 577–588 CrossRef PubMed.
- P. Rathee, H. Chaudhary, S. Rathee, D. Rathee, V. Kumar and K. Kohli, Inflammation Allergy: Drug Targets, 2009, 8, 229–235 CrossRef CAS PubMed.
- P. G. Pietta, J. Nat. Prod., 2000, 63, 1035–1042 CrossRef CAS PubMed.
- Y. Gow-Chin, D. Pin -Der and C. Da-Yon, Food Chem., 2000, 70, 437–441 CrossRef.
- J. L. Evans, I. D. Goldfine, B. A. Maddux and G. M. Grodsky, Diabetes, 2003, 52, 1–8 CrossRef CAS PubMed.
- S. Gothai, P. Ganesan, S.-Y. Park, S. Fakurazi, D.-K. Choi and P. Arulselvan, Nutrients, 2016, 8, 461 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |