Open Access Article
Open Access ArticleThe antioxidant potential of peptides obtained from the spotted babylon snail (Babylonia areolata) in treating human colon adenocarcinoma (Caco-2) cells
Putcha Petsantada,
Papassara Sangtanoob,
Piroonporn Srimongkolb,
Tanatorn Saisavoeyb,
Onrapak Reamtongc,
Ninnaj Chaitanawisutid and
Aphichart Karnchanatat *b
*b
aProgram in Biotechnology, Faculty of Science, Chulalongkorn University, 254 Phayathai Road, Pathumwan, Bangkok 10330, Thailand
bResearch Unit in Bioconversion/Bioseparation for Value-Added Chemical Production, Institute of Biotechnology and Genetic Engineering, Chulalongkorn University, 254 Phayathai Road, Pathumwan, Bangkok 10330, Thailand. E-mail: Aphichart.K@chula.ac.th
cDepartment of Molecular Tropical Medicine and Genetics, Faculty of Tropical Medicine, Mahidol University, 420/6 Ratchawithi Road, Ratchathewi, Bangkok 10400, Thailand
dAquatic Resources Research Institute, Chulalongkorn University, 254 Phayathai Road, Pathumwan, Bangkok 10330, Thailand
First published on 7th July 2020
Abstract
This research study investigated the free radical-scavenging activities of peptides which were obtained from the protein hydrolysates of the spotted babylon snail using a combination of pepsin and pancreatin proteolysis which can replicate the conditions of gastrointestinal digestion. In this study, spotted babylon protein hydrolysate (SPH) derived from a sequential 3 hour digestion, first with pepsin and then with pancreatin, was examined. SPH was fractionated using molecular weight cut-off membranes for 10 kDa, 5 kDa, 3 kDa, and 0.65 kDa. It was found that the MW < 0.65 kDa fraction provided the greatest levels of 2,2′-azino-bis-3-ethylbenzothiazoline-6-sulphonic acid (ABTS), 2,2-diphenyl-1-picrylhydrazl (DPPH), and nitric oxide (NO) radical scavenging activity. Three subfractions of the MW < 0.65 kDa fraction were then generated via RP-HPLC. The subfraction which subsequently demonstrated the greatest free radical scavenging activity was F3, which was accordingly chosen for further investigation commencing with quadrupole-time-of-flight-electron spin induction-mass spectrometry-based de novo peptide sequencing. This resulted in the identification of a pair of novel peptides: His–Thr–Tyr–His–Glu–Val–Thr–Lys–His (HTYHEVTKH), and Trp–Pro–Val–Leu–Ala–Tyr–His–Phe–Thr (WPVLAYHF). The WPVLAYHF peptide exhibited greater antioxidant activity. The study also confirmed that the F3 sub-fraction was able to prevent hydroxyl radicals from causing DNA damage by conducting tests which involved the pKS, pUC19, and pBR322 plasmids using the Fenton reaction. In addition, cellular antioxidant activity was demonstrated by two synthetic peptides toward the human adenocarcinoma colon (Caco-2) cell line, with the potency of the activity dependent upon the peptide concentration.
1. Introduction
Among the numerous challenges faced by people in today's world is that of pollution, which exposes people to toxic substances which can adversely affect the biochemical balance of the body, resulting in internal chemical reactions which can lead to disorders in the metabolism and an increase in the radicals or free radicals within the body. When cells endure oxidative stress, the outcome is the release of free radical, which can then cause significant damage to the body. This kind of damage can often be seen in patients exhibiting signs of cancer or cardiovascular disease, as well as disorders of the immune system, or even simply aging.1,2 Free radicals are characterized by the presence of a single electron in the outer orbital which provides high levels of energy, making the molecule highly reactive. When these molecules react with others, the outcome is a chain reaction which can break apart the components of cells, including proteins, fatty acids, and nucleic acids.3 One means of preventing these reactions from taking place involves removing the free radicals by allowing them to react instead with natural antioxidant compounds. Studies carried out in recent times have shown that bioactive peptides (BPs) appear capable of scavenging free radicals. These BPs can be obtained from different sources of protein, and it has been shown that scavenging capabilities are at their highest for fractions which have a molecular weight not exceeding 3 kDa. Researchers have therefore focused attention of peptides of low molecular weight which also serve to provide the benefits of antioxidant activity.4–6There have also been growing levels of interest in the way the human diet can influence health. Among the general public, there is an increasing awareness not only of the desirability of a healthy lifestyle, but also that when functional foods are used in combination with such a lifestyle, there can be a positive effect on health and well-being. There is strong consumer demand today for foods which promise improved health, longer lives, and the potential to inhibit the possibility of disease.7 It is likely that BPs will be obtained from the proteins found in various marine organisms, which themselves account for half of worldwide biodiversity and are known as a rich source of biofunctional components displaying a wide range of structural types. Those which are proteinaceous in nature can supply the proteins, peptides, and amino acids which serve as excellent starting materials to create BPs derived from protein. Some of the BPs which result are understood to offer nutraceutical benefits which can enhance human health. BPs can vary in size from 2–20 amino acids, and are protein fragments which are activated upon release during the processes of enzymatic hydrolysis, gastrointestinal transit, and fermentation. Studies of the BPs found in proteins derived from marine sources have revealed a number of useful biological qualities, with antioxidative, antidiabetic and hypolipidemic properties reported alongside the inhibition of angiotensin-converting enzymes (ACE), and signs of immunomodulatory activity.8,9
The spotted babylon snail, which is the subject of this study, is identified as Babylonia areolata Link, 1807, and is classified under the phylum Mollusca, class Neogastropoda, order Neogastropoda, and family Buccinidae. There are more than 5000 members of the order Neogastropoda, the vast majority of which comprise marine benthic species and carnivores. The Buccinidae is highly diverse, containing a wide range of predatory gastropods which inhabit the seabed. In the case of B. areolata, it can be recognized by its thin shell, which is shaped as an elongated oval, with a high cone-shaped spire and large, bulbous body, with convex spire whorls, distinctly shouldered underneath the sutures. The operculum which serves to close the shell is both large and thick.10 Snails of the genus Babylonia are found throughout the Indo West Pacific region, from Sri Lanka and the Andaman Islands to the Gulf, and along the Vietnamese and Chinese coast as far as the waters around Taiwan. Along the Thai coast, B. areolata can be found along the sea bed at depths of 5–15 m. Coastal provinces which are home to B. areolata include Trat and Rayong in the East, and Petchaburi, Prachuab Khiri Khan, and Chumphon in the West. Spotted babylon snails flourish when the water salinity is in the range of 28–35 ppt and the temperature is 25–30 °C. They thrive in sandy mud, muddy sand, and both coarse and fine sand, and will typically bury themselves into the sand for most of their lives. Their distribution thus depends upon the availability of the substrate in which they are buried.11 Market prices of these snails lie in the range of 250–500 baht per kilogram, with the lower prices found in markets and higher rates in restaurants. They contain more protein than oysters, mussels or abalone, and are similar to fish or shrimps in having around 20% protein and 2.4% lipids. The content includes around 17 different amino acid types and 20 different types of fatty acids.12–14
We have previously presented a study demonstrating that spotted babylon hydrolysates are able to act as tyrosinase inhibitors through an examination of kinetic inhibition, the inhibition of mushroom tyrosinase, the capacity for the inhibition of cellular tyrosinase obtained from B16F10 murine melanoma cells, as well as the capability to prevent the production of melanin. We also carried out preliminary work to investigate the capacity of spotted babylon snails hydrolysates to induce apoptosis in B16F10 melanoma cells.15 These previous studies did not, however, result in sufficient evidence to suggest that spotted babylon snails might find a productive role in functional foods and similar products. To realize this potential and seek natural antioxidants which are non-toxic, this study has the objective to produce BPs from the protein of spotted babylon snails before analyzing the antioxidant capabilities, which will include ABTS, DPPH, and NO radical scavenging activities. In this study, proteins from the spotted babylon snails underwent hydrolysis using pepsin–pancreatin, whereupon the antioxidant peptides derived from the spotted babylon protein hydrolysate (SPH) entered the process of ultrafiltration and RP-HPLC, with 10, 5, 3, and 0.65 molecular weight cut-off (MWCO) membranes. The fractions were collected and tested to determine the level of antioxidant activity by examining the extent of any radical scavenging. Fractionation and identification of the amino acid sequences of the SPH antioxidant peptides was performed using quadrupole time-of-flight (Q-TOF) liquid chromatography-tandem mass spectrometry (LC-MS/MS), and tests were then carried out to determine the ability of these hydrolysates to reduce oxidative stress in the human colon adenocarcinoma (Caco-2) cells.
2. Materials and methods
2.1 Chemical and biological materials
The spotted babylon snails used in this research were supplied by the Sichang Marine Science Research and Training Station of Chulalongkorn University, which farms the snails of Koh Sichang, which is an offshore island of Chonburi province, Thailand. These snails were carefully washed using running water prior to placement on ice as an anesthetic for 15 minutes. The snail flesh was then removed from the shells and sliced using a knife before a mixer was used to create a homogenized product. From this homogenate, the fat was extracted for removal as proposed by Prakot et al.15 Isopropanol was then added to form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (v/w) mixture with the homogenate, which was then stirred for 60 minutes at room temperature. Vacuum filtration was then carried out to remove the isopropanol, before drying the homogenate in an oven at 60 °C so that it could be ground into a fine powder, henceforth to be known as the spotted babylon powder, and stored in a desiccator until needed.
4 (v/w) mixture with the homogenate, which was then stirred for 60 minutes at room temperature. Vacuum filtration was then carried out to remove the isopropanol, before drying the homogenate in an oven at 60 °C so that it could be ground into a fine powder, henceforth to be known as the spotted babylon powder, and stored in a desiccator until needed.
The principal chemical ingredients were obtained from Sigma-Aldrich, Merck (St. Louis, MO, USA), and included 2,2′-azobis(2-amidinopropane)dihydrochloride (AAPH), L-ascorbic acid, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), curcumin, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), bovine serum albumin (BSA), 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA), 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-1-benzopyran-4-one, 3,3′,4′,5,6-pentahydroxyflavone (quercetin), dithiothreitol (DTT), disodium hydrogen phosphate (Na2HPO4), Dulbecco's Modified Eagle's Medium (DMEM), ferrous sulfate (FeSO4), fetal bovine serum (FBS), hydrogen peroxide (H2O2), N-(1-naphthyl)ethylenediamine dihydrochloride (NED), sodium nitroprusside, sodium tetraborate decahydrate, monosodium dihydrogen orthophosphate (NaH2PO4), and sulphanilamide. All were of analytical grade. In addition, acetic acid, acetonitrile (ACN), formic acid, ethanol, and trifluoroacetic acid (TFA) were supplied by Thermo Fisher Scientific (San Jose, CA, USA) and were all of chromatographic grade.
2.2 The use of pepsin and pancreatin for the hydrolysis of crude protein
In producing the required protein hydrolysate, the crude proteins which were derived from the spotted babylon powder were employed as the substrate as described in the approach put forward by Inthuwanarud et al.16 The technique requires that the spotted babylon powder preparations of 0.1 g mL−1 were first combined with pepsin to create a concentration ratio for the substrate to enzyme (v/w) of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, whereupon the pH value was altered to 2.5 using 1 M HCl. The hydrolysis process was carried out at 37 °C for 3 hours while shaking at 180 rpm. In order to finally terminate the activity, 1 M NaOH was added to adjust the pH value to 7.5. Pancreatin was then introduced to the 20
1, whereupon the pH value was altered to 2.5 using 1 M HCl. The hydrolysis process was carried out at 37 °C for 3 hours while shaking at 180 rpm. In order to finally terminate the activity, 1 M NaOH was added to adjust the pH value to 7.5. Pancreatin was then introduced to the 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/w) substrate to enzyme ratio for 3 hours of shaking at 180 rpm at 37 °C. To stop his enzyme hydrolysis reaction, the temperature was then raised to 80 °C for 20 minutes, and the resulting hydrolysates underwent clarification via centrifugation for 30 minutes at 15
1 (v/w) substrate to enzyme ratio for 3 hours of shaking at 180 rpm at 37 °C. To stop his enzyme hydrolysis reaction, the temperature was then raised to 80 °C for 20 minutes, and the resulting hydrolysates underwent clarification via centrifugation for 30 minutes at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 × g, at a temperature of 4 °C. The ACE inhibitory activity of the supernatant was then evaluated. The two proteases in this study were chosen because of their resemblance to the conditions within the human gastro-intestinal tract.
900 × g, at a temperature of 4 °C. The ACE inhibitory activity of the supernatant was then evaluated. The two proteases in this study were chosen because of their resemblance to the conditions within the human gastro-intestinal tract.
2.3 Determination of the protein content
The protein hydrolysates produced in the study were investigated using the Bradford procedure in order to determine he concentration of the protein using BSA as the standard.17,18 The absorbance at 595 nm (A595) was then measured using a spectrophotometer (Multiskan GO; Thermo Fisher Scientific, Waltham, MA, USA).2.4 Free radical scavenging assay
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) combination of ABTS solution (7 mM) and potassium persulfphate (2.45 mM) was kept in dark conditions at room temperature for 12 hours. Dilution of the resulting ABTS cation radical solution was then performed in order to achieve an absorbance of 0.7 ± 0.02 at 734 nm (A734). The solution was the mixed with the hydrolysate to be tested at the ratio of 1
1 (v/v) combination of ABTS solution (7 mM) and potassium persulfphate (2.45 mM) was kept in dark conditions at room temperature for 12 hours. Dilution of the resulting ABTS cation radical solution was then performed in order to achieve an absorbance of 0.7 ± 0.02 at 734 nm (A734). The solution was the mixed with the hydrolysate to be tested at the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (v/v). This ratio involved 25 μL of the sample being combined with 750 μL of the prepared ABTS cation radical solution. The mixture was then incubated in the dark for 10 minutes before a microplate reader was used to measure A734. Ascorbic acid (100 μg mL−1) was used as the positive control.
30 (v/v). This ratio involved 25 μL of the sample being combined with 750 μL of the prepared ABTS cation radical solution. The mixture was then incubated in the dark for 10 minutes before a microplate reader was used to measure A734. Ascorbic acid (100 μg mL−1) was used as the positive control.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (v/v), such that 80 μL of the sample was combined with 320 μL of DPPH radical solution. Once prepared, this mixture was incubated at room temperature for 15 minutes in darkness, before undergoing centrifugation for 5 minutes at 15
4 (v/v), such that 80 μL of the sample was combined with 320 μL of DPPH radical solution. Once prepared, this mixture was incubated at room temperature for 15 minutes in darkness, before undergoing centrifugation for 5 minutes at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 × g. Absorbance measurements were then taken at 517 nm (A517) using a microplate reader. The experiment used ascorbic acid (100 μg mL−1) to serve as the positive control.
900 × g. Absorbance measurements were then taken at 517 nm (A517) using a microplate reader. The experiment used ascorbic acid (100 μg mL−1) to serve as the positive control.
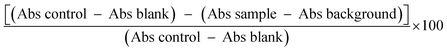 | (1) |
2.5 Isolation and enrichment of BPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0% (v/v) A
0% (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B which was reduced to 90
B which was reduced to 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) A
10 (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B 15 minutes after the process began, and further to 65
B 15 minutes after the process began, and further to 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 (v/v) A
35 (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B at the 30 minute mark, and then to 55
B at the 30 minute mark, and then to 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 (v/v) A
45 (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B ten minutes later, while the flow rate of 0.7 mL min−1 was maintained throughout. Mobile phase A had the composition of 0.1% (v/v) trifluoroacetic acid, whereas B was determined to be 70% (v/v) acetonitrile in 0.05% (v/v) trifluoroacetic acid. Chromatographic analysis was performed using ChromQuest Software (Thermo Fisher Scientific, San Jose, CA, USA) was used to carry out the chromatographic analyses. The eluted peptide peaks were then monitoring at 280 nm (A280) to identify the amino acid sequences of the purified peptides using quadrupole time-of-flight tandem mass spectrometry (Q-TOF-MS/MS).
B ten minutes later, while the flow rate of 0.7 mL min−1 was maintained throughout. Mobile phase A had the composition of 0.1% (v/v) trifluoroacetic acid, whereas B was determined to be 70% (v/v) acetonitrile in 0.05% (v/v) trifluoroacetic acid. Chromatographic analysis was performed using ChromQuest Software (Thermo Fisher Scientific, San Jose, CA, USA) was used to carry out the chromatographic analyses. The eluted peptide peaks were then monitoring at 280 nm (A280) to identify the amino acid sequences of the purified peptides using quadrupole time-of-flight tandem mass spectrometry (Q-TOF-MS/MS).2.6 Protective influence of the enriched F3 sub-fraction on hydroxyl radical-induced DNA damage
2.7 Electrospray Q-TOF-MS/MS and de novo peptide sequencing
The amino acid sequences were identified using Q-TOF-MS/MS, and the molecular mass was also determined in the case of the F3 sub-fraction when the instrument was calibrated appropriately to detect those peptide chains for which the mass lay in the range of 50–25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 m/z (Model Amazon SL, Bruker, Germany). The application of de novo sequencing and mascot allowed the evaluation of the Q-TOF-MS/MS data, and enabled comparisons to be drawn between the sequences obtained and those appearing in the SwissProt database by using BLASTp.
000 m/z (Model Amazon SL, Bruker, Germany). The application of de novo sequencing and mascot allowed the evaluation of the Q-TOF-MS/MS data, and enabled comparisons to be drawn between the sequences obtained and those appearing in the SwissProt database by using BLASTp.
2.8 Synthesis of peptides
The MS/MS method was used to obtain peptide sequences, whereupon those peptides underwent synthesis using a 433A Synergy solid phase peptide synthesizer (Applied Biosystems, CA). This allowed the creation of synthetic peptides including His–Thr–Tyr–His–Glu–Val–Thr–Lys–His (HTYHEVTKH), and Trp–Pro–Val–Leu–Ala–Tyr–His–Phe–Thr (WPVLAYHFT) which could be verified via analytical mass spectrometry (Model Finnigan™ LXQ™, Thermo Fisher Scientific Inc., MA) in combination with a Surveyor HPLC. It was then possible to draw comparisons between the ACE inhibitory activity recorded for the pure synthesized peptides with that of the peptides which were formed through RP-HPLC fractionation.2.9 Cellular antioxidant activity assay (CAA)
| Cell survival (%) = (A540-sample × 100)/(A540-control), | (2) |
| CAA unit = % reduction = (1 − AUCsample/AUCcontrol) × 100 | (3) |
in which AUCsample represents the integrated area under the curve which shows the sample fluorescence versus time, and where AUCcontrol denotes the integrated area under the control curve for quercetin collected from the triplicate outcomes of four different experiments, expressed in terms of mean ± standard error.
2.10 Statistical analysis
The experiments were carried out in triplicate and the results expressed in terms of mean ± standard error for the triplicates. The statistical analysis involved one-way ANOVA with a significance level of P < 0.05, and t-testing which also had a significance level set to P < 0.05. Analyses were performed using the Statistical Package for Social Sciences (SPSS; version 15.0, IBM, NY, USA).3. Results and discussion
3.1 Hydrolysis of spotted babylon protein isolates via pepsin–pancreatin
The properties of numerous food proteins include antioxidant capabilities due to their peptide sequences as well as the structural domains. During the digestive process in the gastrointestinal (GI) tract, the active fragments of these peptides are released, quickly and safely. Antioxidant peptides can thus be produced naturally through the process of digestion by GI enzymes, and the resulting BPs are different from those generated commercially, since they do not degrade further and therefore become inactive after they are ingested orally.23–25 The advantages to human health of dietary proteins might be attributed to the fact that these antioxidant peptide segments can be found in proteins, and thus deliver benefits which exceed those expected in terms of nutrition. When food is digested, pepsin is the principal enzyme which is produced by the body. One of its roles involves endopeptidase activity, and it is capable of cleaving the peptide bonds which exist between hydrophobic and aromatic amino acids, with examples including Phe, Trp, and Tyr. Pancreatin is released by the pancreas, located in the small bowel, and contains proteases which include the endopeptidase, trypsin, which is capable of efficiently cleaving the peptide bonds which are formed between basic amino acids such as Lys and Arg. Along with trypsin is chymotrypsin, another endopeptidase, which is effective in its ability to cleave peptide bonds formed between aromatic amino acids.26The antioxidant abilities of SPH can be assessed by considering the scavenging activity of the ABTS, DPPH, and NO radicals. These findings are presented in Table 1, confirming that those samples which underwent hydrolysis demonstrated greater antioxidant activity than the samples which had not received that treatment. It can be inferred that the enzymatic hydrolysis process allows BPs to be released from the proteins, and the peptides which are released provide hydrogen donors which are able to react with the free radicals present to create stable compounds which serve to end the reaction chain which is fueled by the radicals. This suggests that the antioxidant peptides of SPH which are created through pepsin–pancreatin digestion could possess strong hydrophobic residues within their amino acid sequences, leading to the powerful antioxidant capacity of these peptides. Previous studies have examined the antioxidant capabilities of this type of peptide formed through hydrolysis with pepsin-pancreatin. Rajaram and Nazeer,27 for instance, explain that pepsin hydrolysates obtained from the ribbon and seela fish of the Indian ocean could readily scavenge free radicals. Meanwhile, Nalinanon et al.28 used the pepsin from skipjack tuna to obtain hydrolysates from ornate threadfin bream. Those peptides which had a molecular weight in the range of 1.3 kDa were found to be most effective in ABTS radical scavenging. They were unaffected by in vitro pepsin hydrolysis, but their activity was improved with further digestion with pancreatin.
| MWCO fraction | Free radical scavenging activity (IC50) (μg mL−1) | ||
|---|---|---|---|
| ABTSb | DPPHb | NOc | |
| a Data are expressed in the form of mean ± standard error and the results are generated in triplicate. a–d Those values with different letters appearing in the same row show significant differences (p > 0.05).b The positive control was ascorbic acid, which had an IC50 value of 0.094 ± 0.012, 0.14 ± 0.01 and 98.44 ± 3.79 μg mL−1 respectively for ABTS and DPPH.c The positive control was curcumin, which had an IC50 value of 0.33 ± 0.006 μg mL−1 for NO. | |||
| Crude protein | 21.04 ± 1.47e | 14.71 ± 0.51c | 5.92 ± 0.14b |
| Protein hydrolysate | 5.65 ± 0.71c | 3.79 ± 0.05b | 5.42 ± 1.89b |
| >10 kDa | 14.14 ± 1.58d | 22.98 ± 1.58e | 11.20 ± 0.83c |
| 5–10 kDa | 12.41 ± 2.02d | 20.30 ± 1.26d | 6.39 ± 0.51b |
| 3–5 kDa | 3.83 ± 0.35b | 2.69 ± 0.47b | 1.45 ± 0.21a |
| 0.65–3 kDa | 4.53 ± 0.49b | 2.04 ± 0.26a,b | 1.48 ± 0.43a |
| <0.65 kDa | 0.92 ± 0.10a | 0.51 ± 0.08a | 0.42 ± 0.07a |
3.2 Free radical scavenging activity following SPH size fractionation via ultrafiltration
In order to purify the antioxidant peptides which, come from various sources, including marine invertebrates, the initial process is often membrane ultrafiltration. The approach makes use of membranes made from regenerated cellulose or polyethersulfone, which are designed for specific molecular weight cut-off (MWCO) targets. It is possible, therefore, to separate SPH into different fractions using membranes designed for MWCO 0.65 kDa, 3 kDa, 5 kDa, and 10 kDa, thus covering a range of molecular masses. Having fractionated the SPH via ultrafiltration to generate the five different sub-fractions, these were then examined through the use of ABTS, DPPH, and NO radical scavenging activities assay to determine the extent of the antioxidant activity. Table 1 presents the outcome, confirming that the <0.65 kDa fraction exhibited the highest levels of activity. The calculated IC50 value also indicated that low molecular weight peptides showed superior antioxidant activity compared to heavier varieties. When comparisons were drawn with the scavenging ability of the pepsin–pancreatin hydrolysate fraction, the MW < 0.65 kDa fraction was demonstrated to be 6.1 times greater for ABTS, 7.4 times greater for DPPH, and 26.7 times greater for NO. Based on these results, the MW < 0.65 kDa fraction was accordingly chosen for further separation using a chromatographic approach.The links between antioxidant activity and molecular weight have been of interest to a number of researchers, some of whom have found results similar to those of Wang et al.29 who tested peptides obtained from blue mussel (Mytilus edulis) protein hydrolysates and discovered that those with a molecular weight which did not exceed 3 kDa showed greater antioxidant activity than heavier peptides. It is broadly the case that the antioxidant capacity of a peptide will be determined by the amnio acid composition, peptide sequence, hydrophobicity, and molecular weight, but more detailed knowledge of the SAR (structure–activity relationship) for antioxidant peptides is still required. It is thought that peptides will have greater antioxidant effects if they have a low molecular weight.30,31 When protein hydrolysates undergo fractionation, it has been claimed that the distribution of the molecular is able to influence the hydrolysate antioxidant activity, while peptides of lower molecular weight are more readily able to pass the intestinal barrier when compared to heavier peptides.32,33
3.3 RP-HPLC fractionation of MW < 0.65 kDa and assessment of the free radical scavenging activity
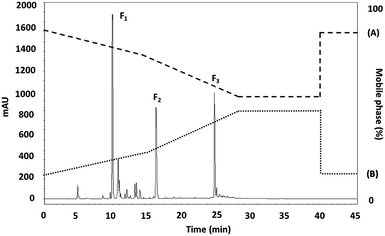
The RP-HPLC technique makes use of a hydrophobic stationary phase while allows interaction with hydrophobic analyses within a polar mobile phase. This approach has been employed widely with protein hydrolysates for fractionation purposes, facilitating both one- and multi-dimensional separation methods. The principal difficulty with RP-HPLC, however, is that it is possible for the peptides to undergo conformational changes during the interactions with the stationary and mobile phases. When fractionation occurs, the way in which peptides can be eluted is dependent upon the extent of their hydrophobicity and also their molecular weight. Those peptides which offer low hydrophobicity as well as a low molecular weight will usually exhibit a low retention time in the column as a result of their amino acid sequence, and will be the first to be eluted.34,35 RP-HPLC was used to separate the MW < 0.65 kDa fraction from the SPH which had been shown to offer the greatest antioxidant activity. A sample solution was prepared which encompassed molecular polarity differences. Fig. 1 then shows the RP-HPLC elution profile for this MW < 0.65 kDa fraction which leads to the production of three further fraction: F1, F2, and F3. The F3 sub-fraction was shown to have an elution time of 25:06 minutes and also demonstrated the greatest level of antioxidant activity. In the ABTS, DPPH, and NO assays, the reported IC50 values were 1.78 ± 0.06, 40.75 ± 0.02, and 20.11 ± 0.03 μg mL−1 (P < 0.05) respectively. The yield of each purification procedure is shown in Table 2.| Fractions | Yield (mg protein) | Yield (%) |
|---|---|---|
| a All the data are given as the mean ± standard deviation of the triplicates. | ||
| Crude extract | 1275.98 ± 9.15 | 100 |
| Crude protein hydrolysate | 1198.16 ± 17.22 | 93.90 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Ultrafiltration | ||
| MW > 10 kDa fraction | 708.65 ± 4.15 | 55.54 |
| MW 5–10 kDa fraction | 534.77 ± 8.21 | 41.91 |
| MW 3–5 kDa fraction | 278.17 ± 1.79 | 21.80 |
| MW 0.65–3 kDa fraction | 119.54 ± 3.71 | 9.37 |
| MW < 0.65 kDa fraction | 85.87 ± 2.87 | 6.74 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| RP-HPLC | ||
| F1 sub-fraction | 5.49 ± 0.02 | 0.43 |
| F2 sub-fraction | 3.51 ± 0.05 | 0.28 |
| F3 sub-fraction | 4.08 ± 0.11 | 0.32 |
It is important to consider the time taken in the hydrophobic chromatography column since this can determine the antioxidative peptides on the basis of their hydrophobic attributes. When the elution time is longer, it can be inferred that the hydrophobic properties are greater, resulting in a corresponding rise in antioxidative activity. The elution time required for the SPH peptides is indicative of the level of antioxidative activity produced by the peptide sequences, as can be confirmed by the antioxidant assay. The concept of using a hydrophobic chromatography column appears in another research study in which loach peptides from Misgurnus anguillicaudatus are shown to have an extended retention time since they exhibit greater quantities of hydrophobic amino acids.36 In one study, Peng et al.37 reported the separation of bombinin-like peptide (BLP) and bombinin hydrophobic-type peptide (BH), necessitating respectively 92 and 147 minutes for elution to be carried out, with BH taking longer as a consequence of its higher degree of hydrophobicity. The two peptides were able to act biologically against both microbes and cancer, as a result of their sequence and α-helical cationic structure, the BH peptide was able to demonstrate superior anticancer capabilities due to its greater hydrophobicity. Due to its high level of radical scavenging performance, the F3 sub-fraction was chosen to be investigated further via Q-TOF-MS/MS in order to assess its potential to prevent DNA damage induced by oxidation.
3.4 Preventing the F3 sub-fraction from causing oxidation-induced DNA damage
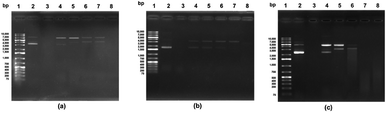
DNA is widely understood to be sensitive to ROS-mediated oxidative damage, since the radicals are readily able to react with any of the DNA molecule components, thus harming the DNA. This type of damage, which increases as the level of attacks on cellular DNA from the free radicals rises, leads to various chronic conditions including cancer, mutagenesis, cancer, or simply aging.38 Hydroxyl radicals are considered to be DNA-damaging agents of physiological significance. For this reason, in vitro tests can readily be conducted using plasmid DNA to provide a simulation of the kind of oxidant damage which DNA can suffer. DNA in its supercoiled form can break down to form open circular DNA in the presence of hydroxyl radicals. When the hydroxyl radical exposure is extended, this circular DNA will undergo further changes to form linear DNA, and in some cases may even be fragmented. Agarose gel electrophoresis can be used to detect each particular form of DNA. The consequences of intracellular oxidation are simulated in vitro, revealing the protective capability of the antioxidant. However, there is a lack of stability in the Fenton reaction system of DNA because of the acidic conditions, so it is important to control the pH levels closely to improve DNA stability while ensuring that hydroxyl radicals are still generated.39The protective effects of the purified antioxidant peptide to counter DNA damage induced by hydroxyl radicals was studied using plasmid DNA samples of pBR322, pKS, and pUC19 for an in vitro evaluation. The F3 sub-fraction was able to provide some defense of the pBR322 plasmid DNA as can be seen in Fig. 2(a). The supercoiled DNA was induced to take the form of linear DNA due to the hydroxyl radical activity (Lanes 4–8). The findings indicate that protection was provided by the F3 sub-fraction against DNA damage triggered by oxidation when concentrations of 15.0, 7.5, 3.75, and 1.88 μg mL−1 were tested (Lanes 4–7). This result matched that of pUC19 plasmid DNA which can be observed in Fig. 2(b), where the quantity of supercoiled DNA declined as linear DNA was formed. The situation for pKS plasmid DNA with the F3 sub-fraction can be seen in Fig. 3(c), where the extent of the protection against oxidation-induced DNA damage can be seen for concentrations of 15.0, 7.5, and 3.75, μg mL−1 (Lanes 4–6). In addition, the findings make apparent the fact that oxidative DNA damage is prevented by the F3 sub-fraction in situations where exposure to OH radicals produced by Fe(II)/H2O2. Fe2+ serves to catalyze the process of converting H2O2 to OH radicals within a physical system. OH radicals are highly reactive and are thus able to severely damage the purine and pyrimidine base as well as the deoxyribose backbone lesion for DNA.40 ROS-mediated oxidative damage also targets DNA and can trigger carcinogenesis or cause pathogenesis in the context of neurodegenerative diseases such as Alzheimer's or Parkinson's disease. For this reason, ROS has been designated a DNA-damaging agent of physiological significance. The biological properties of BPs, including their antioxidative activity, therefore make them useful in the production of industrial pharmaceutical and nutraceutical products.41
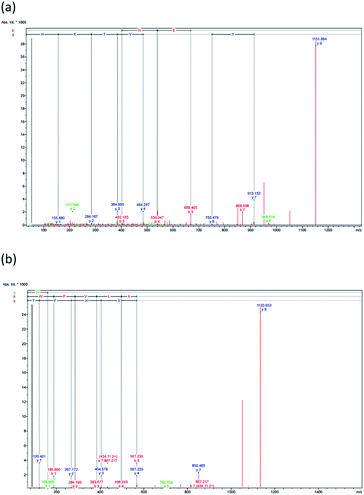 | ||
| Fig. 3 The F3 sub-fraction mass fragmentation spectrum (obtained after RP-HPLC) (see Fig. 1). (a) HTYHEVTKH, and (b) WPVLAYHFT. | ||
It may be the case that differences in capacity can be attributed to the peptides in various different hydrolysates. It was found by Surguladze et al.42 that free radicals could cause the scission of supercoiled DNA strands to a nicked circular form, with nicking proving to be associated with the levels of iron content. The rate of nicking was shown to be reduced by the presence of radical scavengers and chelators. Meanwhile, Je et al.43 revealed that the second step hydrolysates obtained from the livers of tuna could provide better protection from supercoiled DNA conversion than would be the case for the hydrolysates obtained in the first step. It was also pointed out by Sheih et al.21 that purified peptides derived from algae protein waste hydrolysates could potentially protect PET-28a DNA from the threat of damage resulting from oxidation. After damage from hydroxyl radicals through the Fenton reaction (Fe2++ H2O2), it was shown that supercoiled DNA could take the form of open-circular DNA, and this was particularly apparent when the concentrations of the peptides rose from 10.6 to 84.9 μM. It was shown in this study that the F3 sub-fraction was capable both of OH scavenging and Fe2+chelating.
3.5 Peptide identification via mass spectrometry and free radical scavenging comparison of synthetic and enzymatic peptides
The antioxidant properties of peptides are significantly affected by the molecular structure and mass, whereby a peptide of low molecular mass will, in vitro, demonstrate more effective antioxidant capabilities. RP-HPLC was used to derive the F3 sub-fraction from the SPH which and offered the greatest antioxidant activity, and the amino acid sequence was then identified via ESI-Q-TOF-MS/MS analysis. The mass spectra of the antioxidant peptides from this F3 sub-fraction ae presented in Fig. 3(a and b). The analysis revealed the presence of the previously unreported peptides listed as: (1) His–Thr–Tyr–His–Glu–Val–Thr–Lys–His (HTYHEVTKH; m/z = 1151.658), and (2) Trp–Pro–Val–Leu–Ala–Tyr–His–Phe–Thr (WPVLAYHFT; m/z = 1151.659). The fragments were then aligned via the de novo deducing method using the SwissProt database in order to identify the homologous region. In the case of HTYHEVTKH, 100% (9/9) similarity was confirmed for the amino acid sequence to the NADH dehydrogenase subunit 5 (mitochondrion) from Buccinulum vittatum (SwissProt accession number QFG38849.1), and coluporin-14 from Colubraria reticulata (SwissProt accession number AXS67886.1). The WPVLAYHFT peptides were similar to cytochrome oxidase subunit I, partial (mitochondrion), and the amino acid sequence identity was assessed as 100% (9/9) from B. areolata (SwissProt accession number ABS30401.1), so it is potentially the case that this protein may be able to act as a transport protein or signaling protein. The peptide was short, and hence unable to find specific sites of conflict, and therefore this peptide could be replaced in the proteins' polymorphic regions derived from divergent subunit proteins.In order to prepare functional peptides, it is essential first to understand the stabilities and properties of the peptides. The Innovagen server provides much of this information, noting for instance that WPVLAYHFT exhibits poor water solubility, as can be seen in Table 3. In our own research, however, it could be seen that the water solubility of WPVLAYHFT was good on the condition that the concentration did not exceed 5.0 mg mL−1. As the concentration rose above that level, the water solubility began to decline. The two peptides which were identified were then synthesized in order to assess their antioxidant activity, which can be observed in Table 2. While both peptides did indeed exhibit such activity, the results for the ABTS, DPPH and NO assays indicated higher levels of free radical scavenging in the case of WPVLAYHFT, for which the respective IC50 values were 29.94 ± 0.02, 64.21 ± 0.19, and 12.88 ± 0.03 mM. Furthermore, the antioxidative peptide WPVLAYHFT was found to contain high levels of hydrophobic amino acids, such as Trp (W), Pro (P), Val (V), Leu (L), Ala (A), and Phe (F) within WPVLAYHFT. The hydrophobic properties of WPVLAYHFT matched those reported in other studies which had investigated the sardinelle (Sardinella aurita) (Gly–Ala–Trp–Ala, GAWA: hydrophobic: 75%),44 the black pomfret (Parastromateus niger) (Ala–Met–Thr–Gly–Leu–Glu–Ala, AMTGLEA: hydrophobic: 57.14%),45 the Nile tilapia (Oreochromis niloticus) (Asp–Pro–Ala–Leu–Ala–Thr–Glu–Pro–Asp–Pro–Met–Pro–Phe, NPALATEPDPMPF: hydrophobic: 69.23%),46 and also the croaker (Otolithes ruber) (Gly–Asn–Arg–ly–Phe–Ala–Cys–Arg–His–Ala, GNRGFACRHA: hydrophobic: 30%).47
| Synthesized peptides | Free radical scavenging activity (IC50) (mM) | Hydrophobicityb (%) | Water solubilityc | Toxicityd | ||
|---|---|---|---|---|---|---|
| ABTS | DPPH | NO | ||||
| a a,bMean values indicated by the same letter in superscript within the same row or column are not statistically significant (p > 0.05).b Calculations were performed using the peptide property calculator (www.peptide2.com).c The Innovagen server is useful in providing data concerning the solubility of different peptides (www.innovagen.com/proteomics-tools).d Analysis of the toxicity of different peptides is supported by the use of the ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/). | ||||||
| HTYHEVTKH | 58.82 ± 0.30b | 90.48 ± 0.05b | 58.51 ± 0.04b | 11.11 | Good | Non-toxic |
| WPVLAYHFT | 29.94 ± 0.02a | 64.21 ± 0.19a | 12.88 ± 0.03a | 66.67 | Poor | Non-toxic |
Amino acids are understood to have antioxidant properties as a consequence of their composition, structure, and hydrophobic qualities. Research has indicated that there is correlation between the antioxidant activity of certain peptides and the presence of particular amino acid residues. Those amino acids which are hydrophobic amino acids, such as Phe, Ala, and Pro can support higher free radical scavenging activity.48 Similarly, other hydrophobic amino acids such as Val or Leu can be positioned at the N-terminus of the peptide, and will accordingly support the antioxidant activity. It is assumed that this occurs because Leu possesses a long aliphatic side-chain group which readily interacts with the acyl chains which are found in susceptible fatty acids.49 Since amino acids also have aromatic residues such as Trp or Tyr, these are able to induce radicals which are deficient in electrons to donate protons when the interaction involves phenolic and indolic groups.50
Histidine exhibits imidazole qualities which have a positive influence upon the ability to donate protons. A study conducted by Li et al.48 examined 28 synthetic peptides to test their antioxidative properties. The synthetic peptides were designed on the basis of the LLPHH antioxidative peptide which was derived from the proteolytic digestion of soybean protein. The findings suggested that if the C-terminal His residue is removed, the antioxidative activity will be reduced. In contrast, when the N-terminal Leu is removed there is no effect. Meanwhile, Je et al.51 found that His, or peptides containing His offer both chelating ability and lipid radical trapping, because of the presence of the imidazole ring. For instance, the HTYHEVTKH and WPVLAYHFT peptides have residues containing His, and this can explain their antioxidative properties.
One property of hydrophobic amino acids is that they are able to improve the interactions which take place between peptides offering anticancer properties, and the outer leaflets of tumor cell membrane bilayers. This helps them to generate stronger cytotoxic effects, which can be selectively directed against cancer cells. Studies have revealed that cell selectivity, as well as the extent to which cells are susceptible to lysis caused by peptides, depends upon the compositions of the cell membrane bilayers and the phospholipid distribution. Some studies have indicated that if the sequence contains charged (Glu) and heterocyclic amino acid (Pro), the anticancer properties of the peptides may be increased. It was reported by Sae-Leaw et al.52 that when hydrophobic amino acids are present in the gelatin peptides obtained from seabass skins, this can enhance their antioxidant capabilities, because of the improved accessibility of the hydrophobic cellular targets, including polyunsaturated chains of fatty acids, to the hydrophobic antioxidant peptides. This makes interaction with radical species much more likely.
One antiproliferative peptide was identified by Song et al.53 and comprised six amino acid residues, namely Tyr–Ala–Leu–Pro–Ala–His (670.35 Da), and was derived from the peptic hydrolysates of half-fin anchovy. This peptide had a hydrophobic ratio of 50%, which could well be responsible for the antiproliferative attributes. The work of Otani and Suzuki54 involved the isolation of a trio of cytotoxic peptides, Tyr–Lys, Leu–Lys–Lys, and Arg–Pro–Lys from a trypsin digest of bovine αs1-casein. These were able to induce necrosis toward the T and B cells of mouse spleen and also the human leukemic T and B cell lines. It could be inferred that there may be correlation between the positive charge strength in the peptides and the extent of any cytotoxic activity. Meanwhile, greater anticancer activity is reported for peptides of lower molecular weight which boast higher quantities of hydrophobic amino acids. Shorter peptides are more mobile and diffusive at the molecular level, and will therefore offer greater interactivity with the components of cancer cells, thus achieving better anticancer performance. In peptides derived from food products, it may be the case that the molecular weight will account for any differences in cytotoxic activities, and therefore it is important to understand the structural characteristics of peptides when considering anticancer properties.55
3.6 CAA outcomes with Caco-2 cell lines
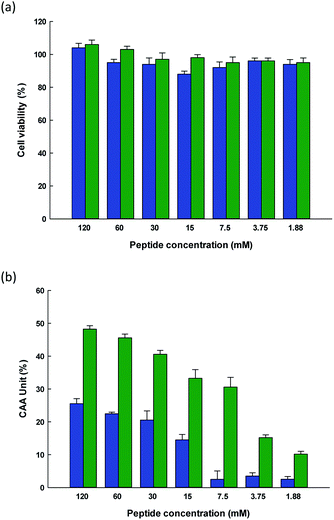
The CAA assay offers one particular advantage, in that while it can measure the antioxidative activity of the substance being tested, it goes further by establishing whether or not it is possible for cells to absorb the substance so that it can act as an antioxidant within the biological system. Many studies have examined human adenocarcinoma colon (Caco-2) cell monolayers using in vitro models to investigate their intestinal permeability in the presence of bioactive components, since they share certain characteristics with intestinal endothelium cells.56 The cells create tight junctions in order to achieve the morphological and functional attributes of enterocytes. Meanwhile, it is possible for smaller peptides to be wholly absorbed across the brush border membrane. Notably, it is said that many of the typical physiological responses to different modulatory agents are produced in a simulation of the damage caused by exposure to both exogenous and endogenous oxidative agents.Tests were conducted with two synthetic peptides to examine their toxicity toward Caco-2 cells. No changes in cell viability for the Caco-2 cells was found with the HTYHEVTKH and WPVLAYHFT peptides, as can be seen in Fig. 4(a), indicating that at the concentrations chosen in the test (1.88 to 120 mM), there was no harm caused to the integrity of the cells during incubation. None of the samples at different concentrations which were tested showed any cytotoxic effects with the Caco-2 cells. Where hydrolysates or fractions were present, cell viability was slightly increased, but this may be due to the presence of nutrients whose balance can aid the survival of cells. It was reported by Kim et al.57 that when soy protein hydrolysate is introduced to the ovary cells of Chinese hamsters, the cell intensity rose, along with the promotion of cell growth. Cell metabolism requires the peptides, short chain proteins, or amino acids which are produced during hydrolysis. Meanwhile, Zhang et al.58 added that whey protein hydrolysates are better able to prevent cell death when the peptide concentrations are higher, in the range of 50–200 μg mL−1. It was also reported by Samaranayaka et al.59 that the protein hydrolysates obtained from Pacific white hake did not exhibit toxicity toward human hepatocellular liver carcinoma cells if the concentrations did not exceed 1 mg mL−1.
This study sought to measure the level of ROS production within cells by employing DCFH to serve as a fluorescent probe. Following the uptake of cells, the DCFH-DA probe undergoes hydrolysis to DCFH via intracellular esterase and is hence oxidized to form the highly fluorescent DCF when ROS are present. The intensity of this resulting fluorescence can therefore be used to quantify the level of oxidation, thus serving as a reasonable indicator of the oxidative stress levels within the cells. In the case of the HTYHEVTKH and WPVLAYHFT peptides, the DCF fluorescent intensity decreased, suggesting that the intensity levels were comparable for these two peptides at any concentration up to 120 mM, as can be observed from Fig. 4(b). This confirms the cellular radical-scavenging effects of the two peptides, and suggests that cells can be protected from ROS-induced oxidative damage by SPH extract, which may become an excellent candidate for further development to act against the formation of cytotoxic ROS which is linked to a number of degenerative conditions. It has been shown that as ROS concentrations increase in the gastrointestinal context, intestinal pathologies a likely to develop, and therefore antioxidants in food are of particular interest as a means to prevent oxidative stress from damaging the intestinal epithelium. Similar protective effects have been linked to the use of casein and whey protein hydrolysates when applied respectively to H2O2-induced hepatic (HepG2) and neuronal (PC-12) cells.60,61 Peptides are multifunctional in their nature, and in vitro studies indicate that they are able to hold down the concentration levels of ROS. Meanwhile, they are considered healthy and safe, offering high levels of activity with a low molecular weight, and can easily be absorbed. Moreover, peptides are able to act in synergy with other forms of antioxidants, which can improve the level of protection they can offer.
4. Conclusions
This research demonstrated that pepsin–pancreatin enzymes hydrolysate which was derived from spotted babylon snail was capable of generating peptides offering the levels of antioxidant activities. The assessment of the activity levels for the various membrane-separated ultrafiltration fractions indicated that low MW peptides (MW < 0.65 kDa) played an essential part in antioxidant activity, with strong performance recorded in ABTS, DPPH and NO assays for free radical scavenging. Additional fractionation via RP-HPLC created the three principal fractions, F1–3, among which the fraction F3 was shown to be the most highly active during the free radical scavenging assay in sufficient quantity for quadrupole-time-of-flight-electron spin induction-mass spectrometry amino acid sequencing, and revealed the sequence of two peptides (HTYHEVTKH, and WPVLAYHFT). Two identified peptides were synthesized, and their antioxidant activity was evaluated. Both synthesized peptides showed antioxidant activity; however, WPVLAYHFT had higher radical scavenging activities. The presence of the hydrophobic amino acids in the peptide sequences are believed to contribute to the high antioxidant activity. In addition, F3 sub-fraction was also shown to inhibit DNA damage caused by hydroxyl radical activity. Furthermore, both synthesized peptides showed cytoprotective effects during the CAA, although this was apparently dependent on the concentration of the peptide. The findings indicate that the protein hydrolysate obtained from the spotted babylon snail might serve as a useful type of bioactive compound with the advantage of a natural source. Moreover, the antioxidant properties have the potential to make the hydrolysate effective against cancer.Funding
Sincere thanks are expressed to the Annual Government Statement of Expenditure (GRB_BSS_99_59_61_06), the Research and Researcher for Industry: MAG (MSD57I0075), and the Ratchadapisek Sompoch Endowment Fund (2019), Chulalongkorn University (762008) in recognition of the financial assistance offered which enabled this research study to be completed.Conflicts of interest
The authors have no conflict of interest. This publication has not been submitted earlier to any journal and is not being considered for publication elsewhere. All of the authors, including the corresponding authors, have read and approved the final submitted manuscript.Acknowledgements
The researchers are grateful to the Institute of Biotechnology and Genetic Engineering at Chulalongkorn University for the facilities they made available during the course of the study. We also thank Dr Robert Butcher (Publication Counseling Unit, Chulalongkorn University) for his constructive comments in preparing this manuscript.References
- M. Valko, D. Leibfritz, J. Moncol, M. T. Cronin, M. Mazur and J. Telser, Int. J. Biochem. Cell Biol., 2007, 39, 44–84 CrossRef CAS PubMed.
- A. Phaniendra, D. B. Jestadi and L. Periyasamy, Indian J. Clin. Biochem., 2015, 30, 11–26 CrossRef CAS PubMed.
- V. Lobo, A. Patil, A. Phatak and N. Chandra, Pharmacogn. Rev., 2010, 4, 118–126 CrossRef CAS PubMed.
- E. B. Daliri, D. H. Oh and B. H. Lee, Foods, 2017, 6(pii), E32 CrossRef PubMed.
- E. B. Daliri, B. H. Lee and D. H. Oh, Crit. Rev. Food Sci. Nutr., 2018, 58, 2273–2284 CrossRef CAS PubMed.
- P. Jeampakdee, S. Puthong, P. Sangtanoo, P. Srimongkol, T. Saisavoey and A. Karnchanatat, Poult. Sci., 2020, 99, 1693–1704 CrossRef PubMed.
- S. Chakrabarti, S. Guha and K. Majumder, Nutrients, 2018, 10, 1738 CrossRef PubMed.
- R. Ghanbari, Int. J. Pept. Res. Ther., 2019, 25, 1187–1199 CrossRef CAS.
- A. Nuchprapha, S. Paisansak, P. Sangtanoo, P. Srimongkol, T. Saisavoey, O. Reamtong, K. Choowongkomon and A. Karnchanatat, RSC Adv., 2020, 10, 12711–12720 RSC.
- N. Chaitanawisuti and A. Kritsanapuntu, Asian Fish. Sci., 1999, 12, 77–82 Search PubMed.
- N. Chaitanawisuti, S. Kritsanapuntu and Y. Natsukari, Aquacult. Res., 2002, 33, 1265–1272 CrossRef.
- S. Kritsanapuntu, N. Chaitanawisuti, W. Santhaweesuk and Y. Natsukari, Aquacult. Int., 2006, 14, 587–594 CrossRef.
- S. Kritsanapuntu, N. Chaitanawisuti, W. Santhaweesuk and Y. Natsukari, Aquacult. Res., 2006, 37, 618–629 CrossRef.
- S. Kritsanapuntu, N. Chaitanawisuti, W. Santhaweesuk and Y. Natsukari, J. Shellfish Res., 2006, 25, 913–918 Search PubMed.
- P. Prakot, N. Chaitanawisuti, P. Sangtanoo, T. Saisavoey and A. Karnchanatat, J. Aquat. Food Prod. Technol., 2018, 27, 811–829 CrossRef CAS.
- K. Inthuwanarud, P. Sangvanich, S. Puthong and A. Karnchanatat, Pak. J. Pharm. Sci., 2016, 6, 1893–1900 Search PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- P. Wongtay, P. Sangtanoo, P. Sangvanich and A. Karnchanatat, Protein J., 2019, 38, 565–575 CrossRef CAS PubMed.
- T. Saisavoey, P. Sangtanoo, C. Chanchao, O. Reamtong and A. Karnchanatat, J. Apic. Res., 2020 DOI:10.1080/00218839.2020.1745434.
- R. Suttisuwan, S. Phunpruch, T. Saisavoey, P. Sangtanoo, N. Thongchul and A. Karnchanatat, Food Biotechnol., 2019, 33, 303–324 CrossRef CAS.
- I. C. Sheih, T. K. Wu and T. J. Fang, Bioresour. Technol., 2009, 100, 3419–3425 CrossRef CAS PubMed.
- K. L. Wolfe and R. H. Liu, J. Agric. Food Chem., 2007, 55, 8896–8907 CrossRef CAS PubMed.
- S. Ketnawa, O. Martínez-Alvarez, S. Benjakul and S. Rawdkuen, Food Chem., 2016, 192, 34–42 CrossRef CAS PubMed.
- A. Hamzeh, W. Wongngam, R. Kiatsongchai and J. Yongsawatdigul, Poult. Sci., 2019, 98, 6138–6148 CrossRef CAS PubMed.
- P. R. Roberts, J. D. Burney, K. W. Black and G. P. Zaloga, Digestion, 1999, 60, 332–337 CAS.
- C. Megías, M. del Mar Yust, J. Pedroche, H. Lquari, J. Girón-Calle, M. Alaiz, F. Millán and J. Vioque, J. Agric. Food Chem., 2004, 52, 1928–1932 CrossRef PubMed.
- D. Rajaram and R. A. Nazeer, Int. J. Biotech. Biochem., 2010, 6, 435–444 Search PubMed.
- S. Nalinanon, S. Benjakul, H. Kishimura and F. Shahidi, Food Chem., 2011, 124, 1354–1362 CrossRef CAS.
- B. Wang, L. Li, C. F. Chi, J. H. Ma, H. Y. Luo and Y. F. Xu, Food Chem., 2013, 138, 1713–1719 CrossRef CAS PubMed.
- T. T. Chai, Y. C. Law, F. C. Wong and S. K. Kim, Mar. Drugs, 2017, 15(pii), E42 CrossRef PubMed.
- T. Sangtitanu, P. Sangtanoo, P. Srimongkol, T. Saisavoey, O. Reamtong and A. Karnchanatat, Food Funct., 2020, 11, 4927–4939 RSC.
- J. Chen, Y. Wang and Q. Zhong, Peptides, 2012, 33, 52–58 CrossRef CAS PubMed.
- Y. Li and J. Yu, J. Med. Food, 2015, 18, 147–156 CrossRef CAS PubMed.
- C. Acquah, Y. W. Chan, S. Pan, D. Agyei and C. C. Udenigwe, J. Food Biochem., 2019, 43, e12765 CrossRef PubMed.
- I. D. Nwachukwu and R. E. Aluko, J. Food Biochem., 2019, 43, e12761 CrossRef PubMed.
- L. You, M. Zhao, R. H. Liu and J. M. Regenstein, J. Agric. Food Chem., 2011, 59, 7948–7953 CrossRef CAS PubMed.
- X. Peng, C. Zhou, X. Hou, Y. Liu, Z. Wang, X. Peng, Z. Zhang, R. Wang and D. Kong, Amino Acids, 2018, 50, 241–253 CrossRef CAS PubMed.
- J. Cadet, T. Delatour, T. Douki, D. Gasparutto, J. P. Pouget, J. L. Ravanat and S. Sauvaigo, Mutat. Res., 1999, 424, 9–21 CrossRef CAS PubMed.
- H. J. You, D. H. Oh, C. Y. Choi, D. G. Lee, K. S. Hahm, A. R. Moon and H. G. Jeong, Biochim. Biophys. Acta, 2002, 1573, 33–38 CrossRef CAS.
- G. R. Martinez, A. P. Loureiro, S. A. Marques, S. Miyamoto, L. F. Yamaguchi and J. Onuki, Mutat. Res., 2003, 554, 115–127 Search PubMed.
- R. Abuine, A. U. Rathnayake and H. G. Byun, Fish. Aquat. Sci., 2019, 22, 1–14 CrossRef CAS.
- N. Surguladze, K. M. Thompson, J. L. Beard, J. R. Connor and M. G. Fried, J. Biol. Chem., 2004, 279, 14694–14702 CrossRef CAS PubMed.
- J. Y. Je, K. H. Lee, M. H. Lee and C. B. Ahn, Food Res. Int., 2009, 42, 1266–1272 CrossRef CAS.
- B. Ali, N. A. Naima, L. Manni, R. Ravallec, B. Ahmed and G. Didier, Food Chem., 2010, 118, 559–565 CrossRef.
- R. Jaiganesh, R. A. Nazeer and N. S. Sampath Kumar, Food Sci. Biotechnol., 2011, 20, 1087–1094 CrossRef CAS.
- D. H. Ngo, Z. J. Qian, B. M. Ryu, J. W. Park and S. K. Kim, J. Funct. Foods, 2010, 2, 107–117 CrossRef CAS.
- N. S. Sampath Kumar, R. A. Nazeer and R. Jaiganesh, Amino Acids, 2012, 42, 1641–1649 CrossRef CAS PubMed.
- B. Li, F. Chen, X. Wang, B. Ji and Y. Wu, Food Chem., 2007, 102, 1135–1143 CrossRef CAS.
- R. J. Elias, S. S. Kellerby and E. A. Decker, Crit. Rev. Food Sci. Nutr., 2008, 48, 430–441 CrossRef CAS PubMed.
- N. Rajapakse, E. Mendis, H. G. Byun and S. K. Kim, J. Nutr. Biochem., 2005, 16, 562–569 CrossRef CAS PubMed.
- J. Y. Je, P. J. Park and S. K. Kim, Food Res. Int., 2005, 38, 45–50 CrossRef CAS.
- T. Sae-Leaw, S. Karnjanapratum, Y. C. O'Callaghan, M. B. O'Keeffe, R. J. FitzGerald, N. M. O'Brien and S. Benjakul, J. Food Biochem., 2017, 41, 1–11 CrossRef CAS.
- R. Song, R. B. Wei, B. Zhang, Z. S. Yang and D. F. Wang, Mar. Drugs, 2011, 9, 1142–1156 CrossRef CAS PubMed.
- H. Otani and H. Suzuki, Anim. Sci. J., 2003, 74, 427–435 CrossRef CAS.
- M. Chalamaiah, W. Yu and J. Wu, Food Chem., 2018, 245, 205–222 CrossRef CAS PubMed.
- M. E. Kellett, P. Greenspan and R. B. Pegg, Food Chem., 2018, 244, 359–363 CrossRef CAS PubMed.
- S. Y. Kim, J. Y. Je and S. K. Kim, J. Nutr. Biochem., 2007, 18, 31–38 CrossRef CAS PubMed.
- Q. X. Zhang, Y. F. Ling, Z. Sun, L. Zhang, H. X. Yu, S. M. Kamau and R. R. Lu, Biotechnol. Lett., 2012, 34, 2001–2006 CrossRef CAS PubMed.
- A. G. Samaranayaka, D. D. Kitts and E. C. Y. Li-Chan, J. Agric. Food Chem., 2010, 58, 1535–1542 CrossRef CAS PubMed.
- T. Li, B. Chen, M. Du, J. Song, X. Cheng, X. Wang and X. Mao, Nutrients, 2017, 9(pii), E31 CrossRef PubMed.
- Q. X. Zhang, Y. F. Ling, Z. Sun, L. Zhang, H. X. Yu, S. M. Kamau and R. R. Lu, Biotechnol. Lett., 2012, 34, 2001–2006 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |