Open Access Article
Open Access ArticleEvaluation of the antimicrobial activity of silver nanoparticles obtained by microwave-assisted green synthesis using Handroanthus impetiginosus (Mart. ex DC.) Mattos underbark extract†
Renata Pascoal Illanes Tormena a,
Eliane Vieira Rosa
a,
Eliane Vieira Rosa ab,
Bruna de Fátima Oliveira Motac,
Juliano Alexandre Chakera,
Christopher William Fagg
ab,
Bruna de Fátima Oliveira Motac,
Juliano Alexandre Chakera,
Christopher William Fagg d,
Daniel Oliveira Freiree,
Paula Melo Martinsc,
Izabel Cristina Rodrigues da Silvad and
Marcelo Henrique Sousa
d,
Daniel Oliveira Freiree,
Paula Melo Martinsc,
Izabel Cristina Rodrigues da Silvad and
Marcelo Henrique Sousa *a
*a
aGreen Nanotechnology Group, Faculty of Ceilandia, University of Brasilia, Centro Metropolitano, Ceilandia, Brasilia, DF 72220-900, Brazil. E-mail: mhsousa@unb.br; mhsqui@gmail.com; Tel: +55 61 3107 8933
bFederal Institute of Education, Science and Technology Goiano, Ceres, GO 76300-000, Brazil
cDepartment of Pharmacy, Faculty of Ceilandia, University of Brasilia, Brasilia, DF 72220-900, Brazil
dGraduate Program in Health Sciences and Technologies, Faculty of Ceilandia, University of Brasilia, Brasilia, DF 72220-900, Brazil
eMicrobiology Laboratory, LS Educational, Brasilia, DF 72020-111, Brazil
First published on 1st June 2020
Abstract
We describe here a green method for the preparation of silver nanoparticles (AgNPs), by a microwave-assisted synthesis route using Handroanthus impetiginosus underbark extract, with antibacterial activity. After optimizing the synthesis parameters with a Box–Benhken designed experiment, samples were characterized by powder XRD, TEM, UV-Vis spectroscopy, FTIR and zetametry. Using the overall optimized conditions of synthesis – time of reaction 15 min at 200 °C and plant extract/AgNO3 volume ratio equal to 10% – highly crystalline ∼13.4 nm-sized spherical AgNPs in a well-dispersed colloidal state were obtained. It was also proved that the plant extract compounds act as reductant and capping agents during synthesis to functionalize AgNPs, resulting in a negatively charged surface with high values of zeta potential in a wide range of pH, from acidic to alkaline media. Biological activity tests against Staphylococcus aureus and Escherichia coli and cell viability experiments showed that synthesized AgNPs were not toxic to HaCaT mammalian cells and presented a high efficiency against Gram-positive bacteria (S. aureus). This was associated with the synergistic combination of AgNP silver cores with the capping layer containing natural compounds with antimicrobial properties and considered an alternative to the AgNPs commonly obtained from conventional routes that present antibacterial effectiveness preferentially against Gram-negative strains.
Introduction
In recent years, the number of microorganisms acquiring resistance to the conventionally used antibiotics has reached critical levels, accelerated by the indiscriminate use of these drugs. However, with the increasing advances of nanotechnology in the medical field, some new nanodrugs – such as silver nanoparticles (AgNPs) – have proven to show a broad spectrum of antimicrobial activities, at low concentrations and negligible toxicity, against multidrug-resistant strains.1Traditional synthesis protocols for the obtention of nanometals (including AgNPs) normally utilize harsh and/or toxic raw materials, are costly, harmful, and not environment-friendly. Therefore, greener synthetic strategies to prepare these nanomaterials using benign reagents have been proposed, toward eliminating or at least minimizing waste generation.2,3 In this regard, the use of benign, renewable materials (as plant extracts) has been studied extensively in the literature and authors show that almost all phenolics (flavonoids, catechins, tannins), sugars, and vitamins present in these extracts can reduce Ag+ in solution to form AgNPs, in view of the low Ag+/Ag0 reduction potential.2–10 Moreover, these natural molecules can act as capping agents during synthesis to functionalize the surface of AgNPs, enhancing their colloidal stability, decreasing their toxicity and improving their antimicrobial activity.4,8,9,11 Besides, simple sugars or hydroxylated glycosides and glycerol (non-toxic waste from biodiesel industry) can also work as reducing and capping agents in AgNPs preparation.12,13
The heating of reactions with microwaves has been successfully reported as a practical method for the greener synthesis of nanomaterials – the main advantages are shorter reaction times, reduced energy consumption, and better product yields which prevents the agglomeration of the particles formed. Particularly, microwave heating is better than a conventional procedure when it comes to consistently yielding nanostructures with smaller sizes, narrower size distributions, and a higher degree of crystallization.2,3,5,7,14,15
Handroanthus impetiginosus, in the Bignoniaceae family, is a native tree to tropical rain and gallery forests throughout Central and South America. Popularly called ‘ipe-roxo’ in Brazil, this family of trees is known for its beauty and use in folk medicine, often associated to their antioxidant, anti-inflammatory, antibacterial and antifungal properties.16 Therefore, the change of the reducing (and capping) agents from plethora of these extractives could increase our knowledge about materials that can be used to produce nanosilver.
Herein, a green route by microwave-assisted synthesis, using the underbark extract of Handroanthus impetiginosus as a natural reductant and capping agent, was exploited to obtain AgNPs in minimal time. The prepared AgNPs show excellent colloidal stability, low toxicity (with HaCaT mammalian cells) and antibacterial effectiveness against Gram-positive bacteria (S. aureus).
Experimental
Plant extract
The underbarks of Handroanthus impetiginosus (Mart. ex. DC.) Mattos (purple ipe) were collected in the morning in Brasília, Brazil (latitude: −15.937266 and longitude: −47.959666) and voucher (Fagg CW 2528) deposited at University of Brasilia (UnB) Herbarium and according to the license number A7B10E5-SISGEN. Botanical material was dried at 50 °C, in drying oven for three days and then ground in a knife mill. Thereafter, 4.0 g of the ground powder was added to 100 mL of ultrapure boiling water and extracted for 15 min. Finally, the suspension was filtered using filter paper to obtain the aqueous plant extract that was stored at −4 °C for further use.Total solid content was 1.21 ± 0.13 wt% and the total polyphenol content, determined through the Folin–Ciocalteu method, was 0.93 ± 0.005 mg mL−1. Moreover, using standard methods of prospection, a preliminary phytochemical analysis of this extract identified the presence of the following principal secondary metabolite groups: phenols, tannins, anthocyanins, flavonoids, triterpenoids, saponins, alkaloids and quinones.17
AgNPs synthesis
In a typical synthesis of AgNPs, the aqueous plant extract was mixed with a 1.0 × 10−3 mol L−1 AgNO3 solution using a specific proportion to give a final volume of 20.0 mL in a 30 mL borosilicate glass vial (G30, Anton Paar). The mixture was then heated in microwave-assisted solvothermal conditions at different temperature and times, using an Anton-Paar Monowave 300 reactor, under magnetic agitation (heat to temperature in time mode, 800 W). After synthesis, we used acetone to induce precipitation of AgNPs and centrifuged the slurries for 10 min at 8000 rpm. The supernatant was discarded, the residual acetone was evaporated and the AgNPs were redispersed in water. To obtain sample powders, AgNPs sols were lyophilized.Characterization
X-ray diffractometry (XRD): lyophilized samples were deposited on the surface of a zero-background sample holder and analyzed in an X-ray Miniflex 600 diffractometer (Rigaku), using Cu-Kα radiation (λ = 1.541 Å) and operating at 40 kV and 30 mA. Transmission electron microscopy (TEM): samples were analyzed in a JEM-2100 (JEOL) microscope (operating at 200 kV). TEM specimens were prepared by dispersing AgNPs in ultrapure water under ultrasonication and further by dropping this suspension on a carbon-lacey copper grid. UV-Vis spectrophotometric analysis: UV-Vis spectra were obtained using a Hitachi 3900H UV-Vis spectrometer in a quartz cell with a 1.0 cm optical path. Suspensions were diluted 10 times in ultrapure water prior to analysis. Fourier transform infrared spectroscopy (FTIR): spectra were recorded with KBr pellets in the region of 4000 to 400 cm−1 on a Varian FTIR spectrophotometer with a resolution of 2 cm−1. Zetametry: the zeta potential (ζ) of AgNPs was estimated from electrophoretic experiments as a function of pH, using a Nano ZS ZetaSizer coupled to a MPT-2 Titrator with a DTS 1070 disposable cuvette (Malvern). The results of electrophoretic mobilities were converted in zeta potential values using the Henry equation.18Minimum inhibitory concentration of AgNPs to control the growth of bacteria (MIC)
The bacteria used were Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922. Both strains were grown in Müller–Hinton Broth (MHB) at 37 °C overnight, resulting in approximately 5 × 108 CFU mL−1. The MICs of S. aureus and E. coli were determined by the broth microdilution method, according to Clinical and Laboratory Standards Institute (CLSI).19Cell viability assay
To study the cytotoxicity of AgNPs, HaCat keratinocyte lineage of human origin was used. Cells were thawed from their storage in liquid nitrogen one week before the assay for acclimatization in a culture flask containing DMEM (Dulbecco modified Eagle's Medium) culture medium, supplemented with 1% antibiotic solution (penicillin and streptomycin) and 10% fetal bovine serum (FBS), and maintained in a humidified incubator at 37 °C and 5% CO2. When the cells reached an ideal confluence, they were removed from the flasks (trypsin EDTA solution – Gibco, USA) and seeded in culture plates per the assay's need.After treating the cells with AgNPs and plant extracts at different concentrations, cell viability was evaluated according to the MTT assay, as described before.20 Briefly, the HaCat cells were transferred to a 96-well culture plate (3000 cells per well) and treated with varying product concentrations, starting at 1 μg mL−1 concentration, and incubated for 24 hours. After the incubation period, the medium was removed, and 150 μL of 0.5 mg mL−1 MTT solution (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) was added. After a 3 hours incubation in a humidified incubator (37 °C and 5% CO2), the MTT was removed, and 200 μL of DMSO was added to each well to dissolve the formed product. The cell viability quantification was determined after reading the absorbance at a 570 nm wavelength.
Repeatability analyzes were performed by calculating the 95% intraclass correlation interval and IC50 calculations using the sigmoidal dose–response curve adjustment method. The statistical programs used were SPSS version 22.0 and GraphPad prism version 6.0. The significance level adopted was 5%.
Results and discussion
Synthesis and characterization of AgNPs
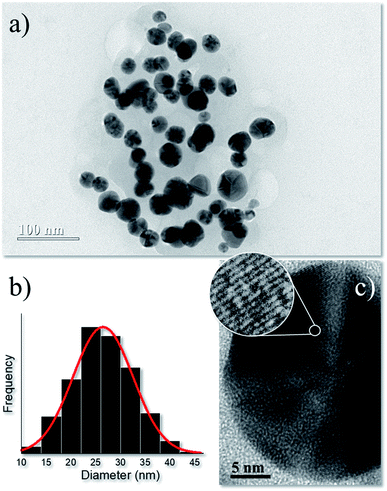
Synthesis parameters were firstly optimized using a Box–Benhken design for three factors (time, temperature, and pressure). XRD data (Fig. S1†) was utilized as the dependent variable for the statistical analysis (Fig. S2†), which indicated an overall optimized synthesis condition (15 min reaction time at 200 °C using a plant extract/AgNO3 volume ratio, CPE = 10%) to obtain ∼13.4 nm-sized metallic nanosilver of face-centered cubic (fcc) structure. Therefore, this sample was utilized in in vitro nanotoxicological tests (cell viability) and in antibacterial activity experiments. Details about XRD characterization and synthesis optimization are shown in ESI.†The TEM micrograph of optimized synthesis in Fig. 1a displays roughly-spherical AgNPs with typical multi-faceted aspect.21 The size distribution of Fig. 1b associated with the AgNPs counted in the TEM images was adjusted by a normal function with a narrow size distribution of 23.5 ± 6 nm. Fig. 1c shows the high-resolution TEM image of a single AgNP and the selected circle displays the lattice fringes with interplanar distance ∼0.24 nm which agrees with the first (111) lattice spacing of the fcc Ag phase.
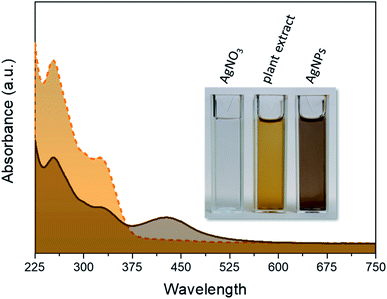
Typical UV-Vis spectra of the pure plant extract and AgNPs obtained from the optimized synthesis are shown in Fig. 2. The two broad bands in the region of 225–375 nm in this spectrum are mainly due to the polyphenolic and quinone compounds present in the plant extract.22
The reddish-brown color of the suspension shown in the inset of Fig. 4 arises owing to the excitation of surface plasmon resonance in the metallic nanoparticles dispersed in solution and corresponds to the broad absorbance (λmax ∼ 420) nm in the spectrum of AgNPs sample, indicating the successful formation of nanosized structures.23 In this spectrum, the bands owing to the plant extract are also present, indicating a successful capping of AgNPs with natural molecules.
A detailed discussion on FTIR analysis of the AgNPs obtained from optimized synthesis is shown in ESI.† The analysis of FTIR spectrum (Fig. S3†) revealed a series of moieties that strongly suggests functionalization of AgNPs with the plant antioxidant extract molecules. Park et al. showed major compounds with biological activity in this plant include polyphenolic species, furan-naphthoquinones, quinones, naphthoquinones, carboxylic acids, benzaldehyde derivatives, cyclopentene dialdehyde and flavonoids.24
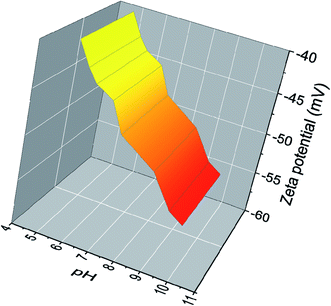
In Fig. 3 the zeta potential of AgNPs dispersion obtained from optimized synthesis is depicted at different pH values. The zeta potential of AgNPs, which is strongly pH-sensitive, varies from ∼−40 mV to ∼−55 mV in the range of the investigated pH. One correlates this with the presence of adsorbed natural molecules which have functional groups capable of deprotonation under pH-dependent conditions inducing the observed surface charges. This can be easily realized in the case of the –COOH/–COO− equilibrium of the very common carboxyl moieties present in these biomolecules.25 With increasing pH, these functional groups reach a higher degree of deprotonation, thus the amount of negative surface charge increases simultaneously. Therefore, the adsorption of these polyfunctional molecules enhances the electrostatic repulsion between particles and can cause a screening of the unbalanced attractive van der Waals forces, in an electrosteric model of repulsion to increase the kinetic stability of colloids.26
The stability in time of AgNPs sols (pH ∼ 7) was monitored by accompanying the variation of the hydrodynamic size and zeta potential during 120 days. During this period, the hydrodynamic diameter (Fig. S4a†) slightly increased but ranged between 130 nm and 145 nm (no agglomeration tendency), while the zeta potential (Fig. S4b†) also varied but remained ∼–50 mV during the period of observation. The high value of zeta potential normally implies increased kinetic stability – it is generally assumed zeta potentials higher than |30 mV| to achieve this condition in charged colloids.27 In addition, no sedimentation was observed and pH variation was ±0.2 during the 120 days of evaluation, also indicating the high colloidal stability of our samples.
Biological tests
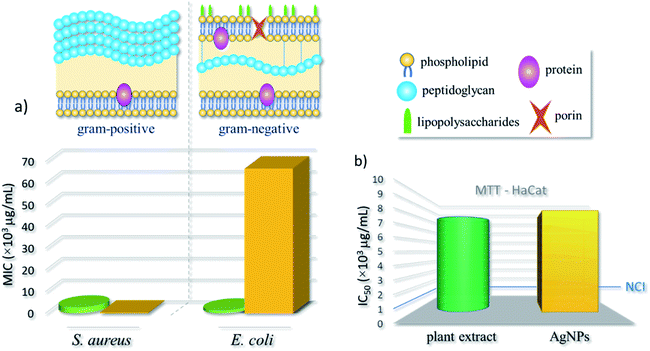
Biological tests for the AgNPs obtained from our optimized green synthesis were performed and compared to the outcomes of pure underbark extract of Handroanthus impetiginous. More specifically, the half maximal inhibitory concentration (IC50) was obtained from MTT assays with HaCat keratinocyte lineage of human origin. Besides, the minimum inhibitory concentration (MIC), and thus the antibacterial effect of AgNPs and plant extract, was evaluated against Staphylococcus aureus (Gram-positive) and Escherichia coli (Gram-negative) bacteria. These strains are frequently used as models to evaluate the antibacterial action of AgNPs, including those obtained by plant extract-mediated synthesis.28The estimated MIC values, shown in Table 1 and schematized in the bar chart of Fig. 4a, demonstrate a distinct behavior of strains against the nanoformulation. The AgNPs induced a growth inhibition against the microorganism S. aureus, with a low MIC value (3.1 × 102 μg mL−1), otherwise with E. coli strain, the MIC was higher (6.7 × 104 μg mL−1). For pure extract, the MIC concentrations were relatively low, equals 2.7 × 103 μg mL−1 and 1.2 × 103 μg mL−1 for S. aureus and E. coli, respectively. Thus, a better antibacterial activity was found for S. aureus – a Gram-positive species – when compared to E. coli (Gram-negative). As expected, the pure extract presented efficient antibacterial activity against both strains.
| IC50 (μg mL−1) | MIC (μg mL−1) | ||||
|---|---|---|---|---|---|
| HaCat | S. aureus | E. coli | |||
| Plant extract | AgNPs | Plant extract | AgNPs | Plant extract | AgNPs |
| a The confidence interval values are between parenthesis. | |||||
| 7.8 | 8.3 | 2.7 | 3.1 | 1.2 | 6.7 |
| (4.3–9.3)a [×103] | (5.9–10.2)a [×103] | (1.6–3.4)a [×103] | (1.9–4.9)a [×102] | (0.1–65.6)a [×103] | (3.3–28.8)a [×104] |
Although the mechanism of action of AgNPs against bacteria is not yet well known, most acceptable theories are that the interaction of AgNPs with membrane proteins of bacteria increases their permeability, leading to the disruption and extravasation of intracellular content; and the production of reactive oxygen species (ROS) by the interaction with AgNPs leads to the inhibition of respiratory enzymes and, consequently, death.1 Positively charged AgNPs normally present stronger antibacterial effect than negatively charged nanoparticles against both Gram-negative and Gram-positive strains.29 This is often associated to the electrostatic attraction between positively charged nanoparticles and negatively charged bacterial outer shells.30 Although, their different membrane design, most Gram-positive and Gram-negative bacteria have a negative superficial charge due to the presence of ionizable moieties. The outer surface of Gram-positive bacteria is mainly composed of a thick layer of peptidoglycan. Otherwise, the Gram-negative bacteria have a functional lipid bilayer outer membrane composed mainly of phospholipids in the inner leaflet and lipopolysaccharides in the outer layer. In addition, the outer membrane features porin proteins that encloses a central aqueous channel which can regulate the passage of hydrophilic species (see Fig. 4).31 Moreover, AgNPs are more effective against Gram-negative strains, this normally associated to the difference in the composition and cell wall thickness (Gram-positive > Gram-negative).32
In this work, the plant extract acted as a reducing and capping agent, thus adding new chemical moieties on AgNPs surface – which becomes negatively charged – as schematized in Fig. S3.† Some researches that use AgNPs obtained from plant extract mediated-synthesis (with similar results against S. aureus) ascribe this improved antibacterial activity to the presence of molecules, such as flavonoids and quinones, on AgNPs surface and originated from the natural extract.33,34
As also mentioned, the AgNPs can interact with the cell wall producing pits, or can penetrate the bacterial cell, acting internally, interfering in the oxidative phosphorylation or producing complexes with nucleic acids. Besides, it may be possible that specific bacterial enzymes promote the production of ROS due to the reduction of existing moieties, such as in the quinolinic species of natural molecules.31,35
Moreover, from the MTT assay, the IC50 values of the plant extract and AgNPs were evaluated, respectively as 7.8 × 103 μg mL−1 and of 8.3 × 103 μg mL−1. According to cytotoxicity criteria for preliminary tests of new compounds – as established by the United States' National Cancer Institute (NCI) – the IC50 values of plant extract and AgNPs are superior to103 μg mL−1, and thus could be considered harmless to humans (see bar chart in Fig. 4b).36
Therefore, although negatively charged, our AgNPs presented a high efficiency against Gram-positive bacteria (S. aureus) and low toxicity – the antibacterial effectiveness of a given material is usually related to its activity to eradicate or reduce the growth of a bacteria strain, without provoking general toxicity to the tissues.37 This can be associated to the existence of a capping layer containing compounds with antimicrobial properties, and considered an alternative to commonly obtained AgNPs from conventional routes that present antibacterial effectiveness preferentially against Gram-negative strains.
However, further investigation must be performed to elucidate the mechanism of nanoparticles formation, the composition of organic capping and the biochemical mechanism that explains the specific action of these nanoparticles and the extent of the synergistic effect of natural capping molecules with the AgNP silver cores against the studied bacteria.
Conclusion
Microwave-assisted synthesis using a natural plant extract has proven to be a very fast green methodology for the obtention of well crystallized negatively charged AgNPs in a stable colloidal dispersion, with low toxicity against human cells and effective bactericidal activity, possibly because of the synergistic combination of the properties of metallic silver cores and the organic capping. Therefore, the AgNPs synthesized using the route described in this work have potential for biomedical and therapeutic applications.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
The authors gratefully acknowledge financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Apoio à Pesquisa do Distrito Federal (FAPDF), Decanato de Pesquisa e Inovação (DPI-UnB) and Fundação de Empreendimentos Científicos e Tecnológicos (FINATEC). The authors also thank LabMic-UFG for TEM measurements.Notes and references
- N. Durán, M. Durán, M. B. de Jesus, A. B. Seabra, W. J. Fávaro and G. Nakazato, Nanomed. Nanotechnol. Biol. Med., 2016, 12, 789–799 CrossRef PubMed.
- D. Hebbalalu, J. Lalley, M. N. Nadagouda and R. S. Varma, ACS Sustain. Chem. Eng., 2013, 1, 703–712 CrossRef CAS.
- R. S. Varma, Curr. Opin. Chem. Eng., 2012, 1, 123–128 CrossRef CAS.
- M. N. Nadagouda, N. Iyanna, J. Lalley, C. Han, D. D. Dionysiou and R. S. Varma, ACS Sustain. Chem. Eng., 2014, 2, 1717–1723 CrossRef CAS.
- J. Kou and R. S. Varma, RSC Adv., 2012, 2, 10283–10290 RSC.
- J. Kou and R. S. Varma, ChemSusChem, 2012, 5, 2435–2441 CrossRef CAS PubMed.
- B. Baruwati, V. Polshettiwar and R. S. Varma, Green Chem., 2009, 11, 926–930 RSC.
- B. Baruwati and R. S. Varma, ChemSusChem, 2009, 2, 1041–1044 CrossRef CAS PubMed.
- M. N. Nadagouda and R. S. Varma, Green Chem., 2008, 10, 859–862 RSC.
- M. N. Nadagouda and R. S. Varma, Green Chem., 2006, 8, 516–518 RSC.
- N. Tarannum, Divya and Y. K. Gautam, RSC Adv., 2019, 9, 34926–34948 RSC.
- J. Kou, C. Bennett-Stamper and R. S. Varma, ACS Sustain. Chem. Eng., 2013, 1, 810–816 CrossRef CAS.
- J. Kou and R. S. Varma, Chem. Commun., 2013, 49, 692–694 RSC.
- M. N. Nadagouda, T. F. Speth and R. S. Varma, Acc. Chem. Res., 2011, 44, 469–478 CrossRef CAS PubMed.
- M. B. Gawande, S. N. Shelke, R. Zboril and R. S. Varma, Acc. Chem. Res., 2014, 47, 1338–1348 CrossRef CAS PubMed.
- M. Suo, H. Isao, H. Kato, F. Takano and T. Ohta, Fitoterapia, 2012, 83, 1484–1488 CrossRef CAS PubMed.
- J. B. Harborne, Phytochemical Methods A Guide To Modern Tecniques Of Plant Analysis, 3rd edn, 1973 Search PubMed.
- S. Bhattacharjee, J. Control. Release, 2016, 235, 337–351 CrossRef CAS PubMed.
- M. P. Weinstein; J. B. Patel and C.-A. Burnham, M07 - Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Clinical and Laboratory Standards Institute, 2018 Search PubMed.
- K. V. Jardim, A. F. Palomec-Garfias, B. Y. G. Andrade, J. A. Chaker, S. N. Báo, C. Márquez-Beltrán, S. E. Moya, A. L. Parize and M. H. Sousa, Mater. Sci. Eng. C, 2018, 92, 184–195 CrossRef CAS PubMed.
- P. Rauwel, S. Küünal, S. Ferdov and E. Rauwel, Adv. Mater. Sci. Eng., 2015, 2015, 1–9 Search PubMed.
- E. Segoloni and F. Di Maria, Eur. J. Wood Wood Prod., 2018, 76, 1547–1561 CrossRef CAS.
- G. Moreno-Martin, M. E. León-González and Y. Madrid, Talanta, 2018, 188, 393–403 CrossRef CAS PubMed.
- B. Park, K. Lee, T. Shibamoto, S. Lee and G. R. Takeoka, J. Agric. Food Chem., 2003, 51, 295–300 CrossRef CAS PubMed.
- K. A. Huynh and K. L. Chen, Environ. Sci. Technol., 2011, 45, 5564–5571 CrossRef CAS PubMed.
- D. V. Goia and E. Matijević, New J. Chem., 1998, 22, 1203–1215 RSC.
- S. Mourdikoudis, R. M. Pallares and N. T. K. Thanh, Nanoscale, 2018, 10, 12871–12934 RSC.
- N. Gogoi, P. J. Babu, C. Mahanta and U. Bora, Mater. Sci. Eng. C, 2015, 46, 463–469 CrossRef CAS PubMed.
- L. F. Wicks, J. Nanomater., 2014, 2015, 1–8 Search PubMed.
- L. C. Yun’an Qing, R. Li, G. Liu, Y. Zhang, X. Tang, J. Wang, H. Liu and Y. Qin, Int. J. Nanomed., 2018, 13, 3311–3327 CrossRef PubMed.
- K. B. Holt and A. J. Bard, Biochemistry, 2005, 44, 13214–13223 CrossRef CAS PubMed.
- S. Shrivastava, T. Bera, A. Roy, G. Singh, P. Ramachandrarao and D. Dash, Nanotechnology, 2007, 18, 1–9 CrossRef.
- C. Dipankar and S. Murugan, Colloids Surf. B Biointerfaces, 2012, 98, 112–119 CrossRef CAS PubMed.
- M. R. Bindhu, M. Umadevi, G. A. Esmail, N. A. Al-Dhabi and M. V. Arasu, J. Photochem. Photobiol. B Biol., 2020, 205, 111836 CrossRef CAS PubMed.
- I. Sondi and B. Salopek-Sondi, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef CAS PubMed.
- E. Hameed, Eur. J. Med. Plants, 2012, 2, 93–112 CrossRef.
- M. J. Hajipour, K. M. Fromm, A. Akbar Ashkarran, D. Jimenez de Aberasturi, I. R. de Larramendi, T. Rojo, V. Serpooshan, W. J. Parak and M. Mahmoudi, Trends Biotechnol., 2012, 30, 499–511 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03240a |
| This journal is © The Royal Society of Chemistry 2020 |