Open Access Article
Open Access ArticleQuantification of EGFR and EGFR-overexpressed cancer cells based on carbon dots@bimetallic CuCo Prussian blue analogue†
Yingpan Song ,
Lina He,
Kun Chen,
Minghua Wang,
Longyu Yang,
Linghao He,
Chuanpan Guo,
Qiaojuan Jia and
Zhihong Zhang
,
Lina He,
Kun Chen,
Minghua Wang,
Longyu Yang,
Linghao He,
Chuanpan Guo,
Qiaojuan Jia and
Zhihong Zhang *
*
Henan Provincial Key Laboratory of Surface and Interface Science, Zhengzhou University of Light Industry, No. 136, Science Avenue, Zhengzhou 450001, P. R. China. E-mail: mainzhh@163.com
First published on 29th July 2020
Abstract
A new bimetallic CuCo Prussian blue analogue (CuCo PBA) loaded with carbon dots (CDs) was prepared (represented by CD@CuCoPBA) and developed as a scaffold for anchoring the epidermal growth factor receptor (EGFR) aptamer to detect EGFR and living EGFR-overexpressed cancer cells. The basic characterizations revealed CuCo PBA exhibited nanocube shape and still remained its nanostructure and physical/chemical properties after coupling with large amounts of CDs. As compared with the pristine CuCo PBA, the CD@CuCoPBA displayed good electrochemical activity, strong binding interaction toward aptamer, and high stability of aptamer-EGFR G-quadruplex in aqueous solution. As such, the results of electrochemical impedance spectroscopy measurements indicated that the CD@CuCoPBA-based aptasensor displayed an ultra-low detection limit toward EGFR (0.42 fg mL−1) and living EGFR-overexpressed MCF-7 cancer cells (80 cell per mL), as well as high selectivity, good reproducibility, high stability, repeatability, and acceptable applicability. Consequently, the constructed CD@CuCoPBA-based aptasensor can be extended to be a promising universal method for early diagnosis of cancers.
1. Introduction
As a transmembrane glycoprotein,1 epidermal growth factor receptor (EGFR) is vital in regulating normal cell signaling. The mutation of EGFR is closely related to cell proliferation, invasion, angiogenesis, metastasis, and inhibition of apoptosis, thus representing the pathogenesis and progression of tumor cells.2 The overexpression of EGFR is closely associated with some cancer such as breast, lung, bladder, prostate, pancreatic, colorectal, and ovarian cancer.3 Therefore, the in situ analysis of the EGFR level in cell membranes, for which ligand binding kinetics and affinity is essential in the early diagnosis of cancer and cancer treatment. At moment, the radiolabeling assay is the most widely utilized method to receptor density measurement,4 for which synthesizing radio-ligand often needs special training and strict safety protection. The integration of metallic nanoparticles with EGFR,5 fluorescent polystyrene nanoparticles conjugated with an anti-EGFR monoclonal antibody,6 and a nano-biochip to analyze the EGFR level7 are some recently reported materials used in EGFR detection. EGFR was imaged on a single living mammalian cell and investigated by scanning electrochemical optical microscopy.8 A facile and efficient multifunctional (anti-EGFR antibody and EGFR aptamer conjugated) gold NPs-based resonance Rayleigh scattering assay for sensitively determining the trace EGFR.9 Consequently, the common approaches for determine EGFR or EGFR-overexpressed cancer cells often need complex instrumentation, special operations, the isolation of biomarkers, time consuming, and clinically high cost.10 To develop advanced biosensors with high sensitivity, simplification, fast response, and low-cost is promising for the early diagnosis EGFR in human serum.Aptamers, one kind of artificial nucleic acid molecules, which exhibit antibody-like characteristics, are capable for recognizing diverse targets with high affinity and specificity. Differentiating from antibodies, aptamer strands are relatively small and lacking in immunogenicity. For cellular research, aptamers have been applied to distinguish native protein targets on the cellule surface and identify different types of cell lines.11 Therefore, aptamers may have great potential for therapeutic, analytical, and diagnostic applications. Developing electrochemical aptamer sensors (aptasensors) has aroused attentions owning to their good feasibility, cheapness, and high sensitivity for detecting cancer biomarkers. For instance, a zirconium metal framework embedded with a nanocomposite of silver nanoclusters was provided and applied as a scaffold to fabricate electrochemical and SPR aptasensors for determining carcinoembryonic antigens.12 The protein tyrosine kinase-7-targeted aptamer was strong anchored onto the bimetallic ZnZr metal organic framework (MOF) architecture to develop PTK-7 sensor, giving a low detection limit (LOD) of 0.66 pg mL−1.13 In comparison with the other approaches, electrochemical techniques offer short analytical time, high detection limit, and wide linear detection range. It is vital to efficiently anchor aptamer strands on to construct an efficient electrochemical aptasensor. Usually, aptamer strands have to be modified with some functional groups such as thiol group, biotin, amino, or azide group to link with the electrode materials via the formation of different covalent bonds.14 Additionally, aptamers also can be labelled with electroactive labels (methylene blue, ferrocene derivatives, and ruthenium complexes) to modify the electrochemical signals.15 Undoubtedly, these strategies can heighten the cost of the aptasensor fabrication and make the fussy construction procedure increase. Thereby, developing advanced electrode materials is essential to fabricate advanced electrochemical aptasensors.16 Various types of materials are being actively investigated, and they include gold nanoparticles (AuNPs),17 carbon nanomaterials (nanotubes, grapheme, and carbon dots (CDs)),18 conducting polymers,19 metal oxides, and porous organic frameworks.20 Good electrical conductivity, thermal property, and high electrocatalytic activity enable carbon nanomaterials to be favorable carriers for aptamers or targets.18 Nevertheless, carbon nanomaterials can easily aggregate through the π–π* stacking interaction, thus further reducing sensing performance. Therefore, combining different nanomaterials into the same electrochemical biosensor can enhance the aptamer immobilization, signal transduction, and amplification. Among the different carbon nanomaterials, CDs exhibit low toxicity, high biocompatibility, and good stability and electrochemical activity,21 all of which have been explored for biosensing and bioimaging applications.22 Consequently, CD-based aptasensors can be developed to detect cancer markers and living cancer cells.
Prussian blue analogues (PBAs) are well-studied materials and have been extensively explored in the aspects of electrocatalytic,23 electrochemical,24 magnetic,25 and photophysical properties,26 as well as their potential analytical applications.27 Bimetallic MnFe PBA coupled with AuNP (MnFePBA@AuNP) was used as biosensing platform to immobilize aptamers for detecting trace biomarkers (human epidermal growth factor receptor-2 (HER2) and living MCF-7 cells).28 A novel three-dimensional silver nanocube@Prussian blue core–shell material was provided to develop an ultrasensitive aptasensor.29 Core–shell heterostructured PBA nanospheres were composited with silver nanoclusters and further applied as aptasensors for detecting bleomycin.30 Especially, PBA nanocube precursors containing different metal ion centers such as nickel, cobalt, and iron were used to keep a general nanocuboidal structure while optimizing the composition of metal centers to efficiently anchor the probe biomolecules. Nevertheless, PBA-based biosensors for detecting EGFR have not been reported.
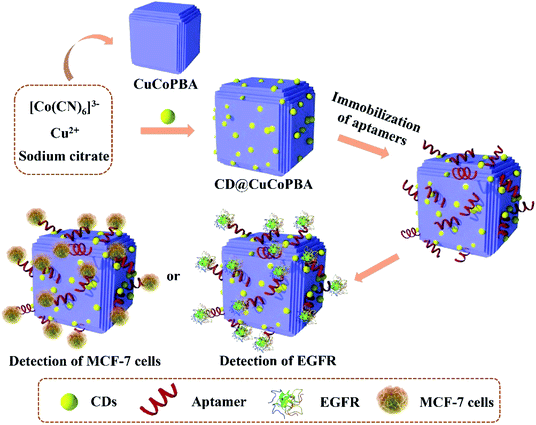
Considering the merits of PBA and CDs, we designed and synthesized a novel bimetallic CuCo PBA coupled with large amounts of CDs (denoted as CD@CoCuPBA) and explored it as a sensing platform for anchoring EGFR aptamers to detect EGFR and EGFR-overexpressed MCF-7 cells (Scheme 1). The presence of CDs in CuCo PBA could facilitate aptamer immobilization and enhance electrochemical activities. In comparison with other routine nanomaterials, the CD@CuCoPBA displayed outstanding advantages, such as (i) large specific active surface area, high porosity, as well as robust structure of PBAs,31 (ii) active nitrogen species of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N of PBAs, (iii) tunable electronic structure by integrating active transition metals showing different numbers of outer electrons,32 and (iv) excellent electrical conductivity and biocompatibility of CDs.33 As a result, the proposed CD@CuCoPBA-based aptasensor exhibits a very low LOD of 0.42 fg mL−1, high selectivity, and good stability. This present approach can provide a new approach for constructing promising aptasensors by using PBA materials as platforms for detecting other analytes.
N of PBAs, (iii) tunable electronic structure by integrating active transition metals showing different numbers of outer electrons,32 and (iv) excellent electrical conductivity and biocompatibility of CDs.33 As a result, the proposed CD@CuCoPBA-based aptasensor exhibits a very low LOD of 0.42 fg mL−1, high selectivity, and good stability. This present approach can provide a new approach for constructing promising aptasensors by using PBA materials as platforms for detecting other analytes.
2. Experimental section
The parts of chemicals and regents, preparations of all solutions, CDs and CuCo PBA, basic characterizations, electrochemical measurements, cell culture, cytotoxicity in vitro, and cell imaging were provided in the ESI S1.†2.1 Synthesis of CuCo PBA and CD@CuCoPBA composite
The preparation of CD@CuCoPBA was performed in according to the similar procedure with that of CuCo PBA, besides the precursor were dissolved in CDs solution (15 mL) instead of Milli-Q water.2.2 Development of the CD@CuCoPBA-aptasensor
The CD@CuCoPBA powder (1.0 mg) was decentralized in 1.0 mL of Milli-Q water and ultrasonically treated for 30 min to obtain the homogeneous CD@CuCoPBA suspension with the concentration of 1.0 mg mL−1. The CD@CuCoPBA dispersion (5 μL) was coated on the pre-treated Au electrode (AE) surface, following by dried at room temperature with ultrapure N2 (CD@CuCoPBA/AE). Then, the CD@CuCoPBA/AE was incubated with the aptamer solution (100 nM) for 30 min. As such, the EGFR-targeted aptamer can be tightly anchored on the electrode surface (Apt/CD@CuCoPBA/AE). Finally, the Apt/CD@CuCoPBA/AE was incubated with the EGFR or MCF-7 cells solution with a series of concentrations for analyzing the sensing performance (EGFR/Apt/CD@CuCoPBA/AE). For comparison, the CoCu PBA dispersion and construction of CuCoPBA-based sensor were developed by the same method.2.3 Electrochemical measurements
All the electrochemical measurements, including EIS and cyclic voltammetry (CV), were carried out on the CHI660D electrochemical workstation (Shanghai Chenhua, China). The traditional three-electrode system includes a modified electrode as the working electrode, an Ag/AgCl (saturated KCl) electrode as the reference electrode, and a platinum wire as counter electrode. CV curves were measured in the mixture of 5.0 mM K3[Fe(CN)6]/K4[Fe(CN)6] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as a redox probe dissolved in 0.10 M PBS at potentials ranging from −0.2 to 0.8 V vs. Ag/AgCl (saturated KCl) at the scan rate of 100 mV s−1. EIS spectra were recorded with 5 mV amplitude in the frequency ranging from 0.01 Hz to 100 kHz under open circuit potential (Fig. S1†). EIS data were analyzed using Zview2 software, of which EIS spectra were simulated using an equivalent circuit consisted of solution resistance (Rs), charge-transfer resistance (Rct), constant-phase element (CPE), and Warburg impedance (Wo) (the inset in Fig. S1†). Each measurement was repeated at least three times. All EIS data obtained in the present work were analyzed using Zview2 software. A nonlinear least-square method was used to fit and determine the parameters of the elements in an equivalent circuit (Fig. S1†).
1) as a redox probe dissolved in 0.10 M PBS at potentials ranging from −0.2 to 0.8 V vs. Ag/AgCl (saturated KCl) at the scan rate of 100 mV s−1. EIS spectra were recorded with 5 mV amplitude in the frequency ranging from 0.01 Hz to 100 kHz under open circuit potential (Fig. S1†). EIS data were analyzed using Zview2 software, of which EIS spectra were simulated using an equivalent circuit consisted of solution resistance (Rs), charge-transfer resistance (Rct), constant-phase element (CPE), and Warburg impedance (Wo) (the inset in Fig. S1†). Each measurement was repeated at least three times. All EIS data obtained in the present work were analyzed using Zview2 software. A nonlinear least-square method was used to fit and determine the parameters of the elements in an equivalent circuit (Fig. S1†).
The constructed CD@CuCoPBA-based sensor was immersed in the EGFR solution with different concentrations to monitor the limit of detection (LOD) against EGFR and tested using electrochemical measurements. The selectivity of the developed sensor for detecting EGFR was investigated by incubating with the possibly coexisted interferences (0.5 pg mL−1) with the 100-folds of the concentration of EGFR, including HER2, mouse immunoglobulin G (IgG), immunoglobulin E (IgE), prostate specific antigen (PSA), bovine serum albumin (BSA), CEA, and their mixed solution with EGFR. The reproducibility was simultaneously assessed using five developed sensors independently, while the stability was tested by continuously storing the sensor in a refrigerator (4 °C) for 15 days and measured by EIS every day. Additionally, to verify the applicability, the EGFR solution with the same concentration for analyzing the LOD were diluted in the human serum, which was purchased from Solarbio Bioengineering Ltd. Company (Beijing, China).
To investigate the sensing performance of the developed biosensor for determining the living MCF-7 cells in the biological solution such as the LOD toward MCF-7 cells, selectivity, and stability, the similarly electrochemical operations were carried out, which were described in the following parts.
3. Results and discussion
3.1 Working mechanism of CD@CuCoPBA-based aptasensor for detecting EGFR and MCF-7 cells
Here, a CD@CuCoPBA composite was chosen as the sensitive platform for constructing electrochemical aptasensor, which was further exploited to determine trace EGFR or MCF-7 cells (Scheme 1). After dried at room temperature with ultrapure N2, the CD@CuCoPBA was uniformly covered onto the AE surface due to the surface effect and quantum size effect. This combination was stable and the CD@CuCoPBA did not release from the AE surface during the whole detection process. Next, when the CD@CuCoPBA/AE was incubated in the aptamer solution, the aptamer strands tended to approach to the surface of the CD@CuCoPBA composite. As aforementioned, CD@CuCoPBA exhibited outstanding properties, including large specific active surface area, high porosity, robust structure of PBAs, excellent electrical conductivity and biocompatibility of CDs, which were favorable for the immobilization of aptamer strands, leading to large amounts of aptamer strands adsorbed onto the surface of the nanocomposite. Then, the Apt/CD@CuCoPBA/AE was used to sensitively detect EGFR or MCF-7 cells. The aptamer strands generated specific recognition with EGFR and formed Apt–EGFR complex, thus causing the conformational change of aptamers. Each step during the construction of CD@CuCoPBA-based aptasensor, including the electrode modification with CD@CuCoPBA, aptamers adsorption, and detection of targets, caused the change of electrochemical signals, such as the peak current density of cyclic voltammetry (CV) curve and/or the electron transfer resistance at the electrode/electrolyte interface, which were measured by CV and electrochemical impedance spectroscopy (EIS) techniques.343.2 Chemical component and nanostructure of CuCo PBA and CD@CuCoPBA
The chemical components and structure of CD@CuCoPBA were analyzed by Fourier transform infrared spectroscopy (FT-IR) and X-ray diffraction measurements (XRD), as well as CuCo PBA. The XRD patterns (Fig. S2a†) of CuCo PBA (curve i) and CD@CuCoPBA (curve ii) show main diffraction peaks located in the range of 10–60°, of which all diffraction peaks correspond to pure CuCo PBA and matched well with the simulation results.35 FT-IR spectra (Fig. S2b†) of the CuCo PBA and CD@CuCoPBA illustrate a clear peak at the band of 2185 cm−1, which is attributed the stretching vibration peak of –C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N.36 Meanwhile, the bands at 3420 and 1610 cm−1 are respectively due to the presence of stretching and bending vibration of O–H containing in the crystal water in the PBA framework,37 respectively.
N.36 Meanwhile, the bands at 3420 and 1610 cm−1 are respectively due to the presence of stretching and bending vibration of O–H containing in the crystal water in the PBA framework,37 respectively.
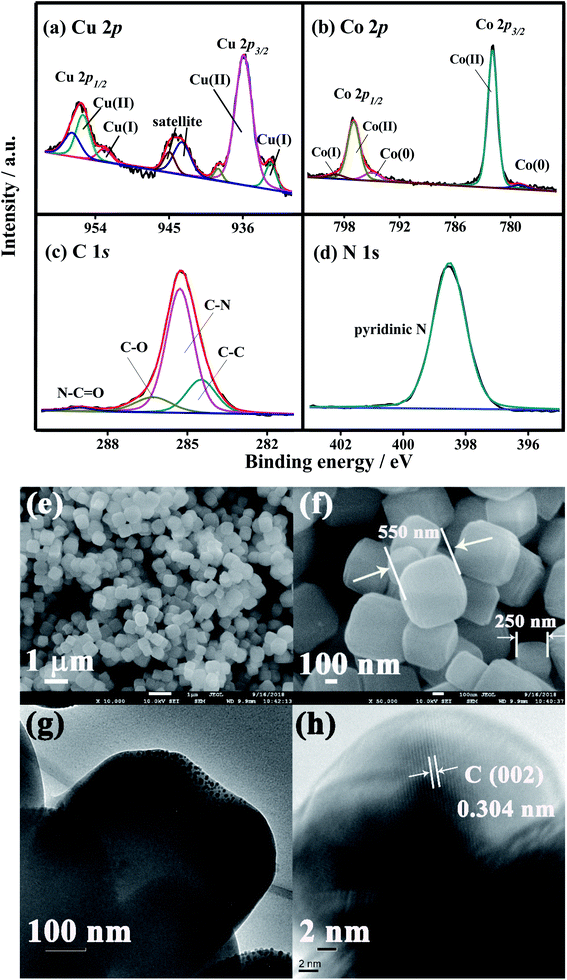
To probe the variation in the chemical structures and environments before and after the composite of CDs with CoCu PBA, their X-ray photoelectron spectroscopy (XPS) characterizations were performed, as depicted in Fig. S2c.† The clear signals of Cu 2p, Co 2p, C 1s, N 1s, and O 1s signals are present in both CuCo PBA and CD@CuCoPBA. As demonstrated in Fig. 1a–d, the high-resolution Cu 2p, Co 2p, C 1s, and N 1s XPS spectra of the CD@CuCoPBA nanospheres were deconvoluted by the XPSPEAK1 software and analyzed. The high-resolution Cu 2p XPS spectrum was decomposed to the peaks at the binding energies (BEs) of 932.5 and 952.9 eV (Fig. 1a), due to the Cu 2p3/2 and Cu 2p1/2 of Cu(I), respectively, while the peaks at the 935.9 and 955.6 eV are corresponded to Cu 2p3/2 and Cu 2p1/2 of Cu(II), respectively. Additionally, the characteristic satellite peaks at 963–962 and 943–942 eV are also obtained. The high-resolution Co 2p XPS spectrum (Fig. 1b) can be separated into the peaks at the BEs of 778.8 and 781.9 eV, which are respectively due to Co(0) and Co(II) of Co 2p3/2, whereas the peaks at the BEs of 795 and 797 eV are respectively assigned with the Co(0) and Co(II) of Co 2p1/2. The other peak at the BE of 798.9 eV is attributed to the satellite peak of Co(I). The high-resolution C 1s XPS spectrum (Fig. 1c) is simulated to three main peaks at the BEs of 284.5, 285.3, and 286.3 eV, respectively corresponding to C–C, C–N, and C–O, while the peak at the BE of 289 eV is assigned with the N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group. Only one main peak at 398.6 eV is appeared in the high-resolution N 1s XPS spectrum (Fig. 1d), corresponding to the pyridine N binding. Concurrently, the high-resolution Cu 2p, Co 2p, C 1s, and N 1s XPS spectra of the pristine CuCo PBA were displayed in Fig. S3.† Evidently, there is no substantial difference between them, suggesting that the addition of CDs into CoCu PBA has no much effect on the chemical structure and environment of each element.
O group. Only one main peak at 398.6 eV is appeared in the high-resolution N 1s XPS spectrum (Fig. 1d), corresponding to the pyridine N binding. Concurrently, the high-resolution Cu 2p, Co 2p, C 1s, and N 1s XPS spectra of the pristine CuCo PBA were displayed in Fig. S3.† Evidently, there is no substantial difference between them, suggesting that the addition of CDs into CoCu PBA has no much effect on the chemical structure and environment of each element.
3.3 Surface morphologies of CD@CuCo PBA nanospheres
FE-SEM and TEM characterizations were used to estimate the surface morphologies of CuCo PBA and CD@CuCoPBA. The FE-SEM images of CuCo PBA nanocubes (Fig. S4†) show uniform shape with a size of 700 nm to 1.0 μm and smooth surface. Meanwhile, every face is composed of several layers, which can be further proved by its TEM image (Fig. S4c and d†). For the CD@CuCoPBA nanocomposite (Fig. 1e and f), the CuCo PBA retains its nanocube shape, giving an average size of 50 nm. From its TEM images, however, the CDs are clearly distributed within the CuCo PBA interior (Fig. 1g). A clear lattice spacing (0.304 nm) of carbon is observed in the HR-TEM image (Fig. 1h). All results suggest the successful combination CDs with CuCo PBA nanocubes.3.4 Biocompatibility of CD@CuCoPBA against MCF-7 cells and in vitro cell uptake
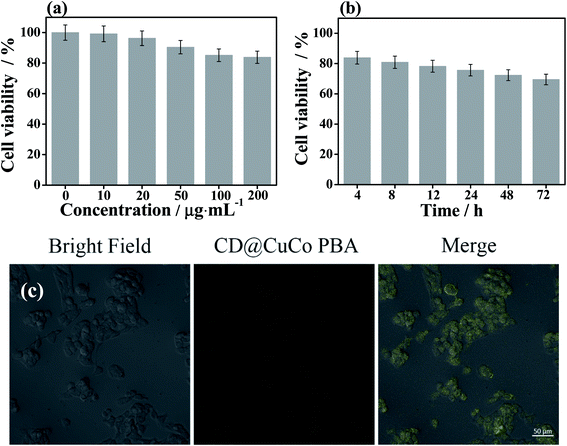
The cytotoxicity and cell uptake behavior of the CD@CuCoPBA composite were characterized and depicted in Fig. 2. In vitro cytotoxicity of the CD@CuCoPBA nanocomposite to MCF-7 cells was evaluated at various concentrations (10, 20, 50, 100, and 200 μg mL−1) by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). As displayed in Fig. 2a, the CD@CuCoPBA composite exhibits slight toxicity on MCF-7 cells at a dose of 50 μg mL−1 and above. Approximately 90.4% cells are alive, which further confirmed that CD@CuCoPBA have good biocompatibility. Besides, the influence of different culture time intervals with MTT (4, 8, 12, 24, 48, and 72 h) on the in vitro cytotoxicity of CD@CuCoPBA composite (200 μg mL−1) was also studied. As shown in Fig. 2b, the CD@CuCoPBA composite exhibits acceptable toxicity on MCF-7 cells at the culture time of 12 h, and approximately 78.3% cells are alive. The cell cytotoxicity increases along with the culture time, when the culture time prolongs to 72 h, 69.5% cells are alive. Therefore, the CD@CuCoPBA-based platform can be explored to anchor aptamer strands, further directly detecting living cancer cells in which EGFR are overexpressed. The culture time with MTT was set as 4 h for better performance. The cell uptake behavior of the CD@CuCoPBA composite was measured by confocal laser scanning microscopy (CLSM). As displayed in Fig. 2b, the cell uptake of CD@CuCoPBA is clearly observed after cells were gestated with 50 μg mL−1 of CD@CuCoPBA for 1 h. After irradiation of MCF-7 cells by a 488 nm laser, weak yellow fluorescence is found, which is originated from the fluorescent property of CD@CuCoPBA. The merged photo proves that the CD@CuCoPBA composite was accumulated in MCF-7 cells and directly generated fluorescence without requiring the use of other dyes. This result indicates the CD@CuCoPBA can be applied for live cell imaging.3.5 Sensing performance of the CD@CuCoPBA-based aptasensor for detecting EGFR
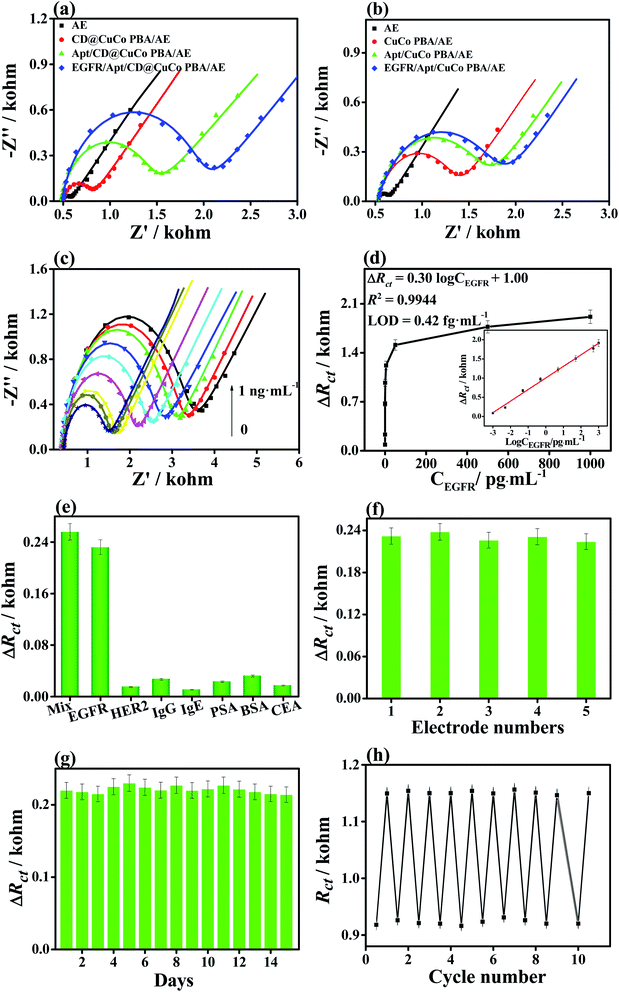
EIS and CV results of each step during the detection of EGFR using the CuCoPBA- and CD@CuCoPBA-based aptasensor are shown in Fig. 3 and S5,† respectively. All electrochemical tests were performed on the electron transfer kinetics of [Fe(CN)6]3−/4− redox indicator on the electrode surface. In terms of the CD@CuCoPBA-based aptasensor, EIS Nyquist plots for all measurement step for detecting EGFR are demonstrated in Fig. 3a. After fitting, the Rct values for each step were obtained. The Rct values of the bare AE, CD@CuCoPBA/AE, and Apt/CD@CuCoPBA/AE are 0.103, 0.321, and 1.058 kohm, respectively. After detecting EGFR, the obtained EGFR/Apt/CD@CuCoPBA/AE exhibits a Rct value of 1.601 kohm. The increase of the Rct value for the CD@CuCoPBA/AE indicates that the CD@CuCoPBA nanocomposite displays relatively poor conductivity.38 After the EGFR aptamer immobilization, the Rct value of the modified electrode increases from 0.321 to 1.058 kohm suggests that the EGFR aptamer was successfully immobilized onto the CD@CuCoPBA/AE, further hampering the charge transfer.39 When detecting EGFR, the Rct value further increases because the specifically binding between EGFR and the aptamer to form a tight complex.40 It further hinders the electron transfer and the diffusion of redox materials. As shown in Fig. 3b, the similar discovery is obtained for the CuCoPBA-based sensor for the detection of EGFR. The achieved Rct value caused by each step also raised successively. The Rct values are 0.142, 0.872, 1.732, and 1.887 kohm for the bare AE, CuCoPBA/AE, Apt/CuCoPBA/AE, and EGFR/Apt/CuCoPBA/AE, respectively. The corresponding CV curves show clear redox peak of [Fe(CN)6]3−/4− (Fig. S5†). The highest peak currents of the bare AE surface are found. The oxidation and reduction peaks decrease slightly due to the blocking effect of the CD@CuCoPBA nanospheres. After immobilization of the EGFR aptamer on to the CD@CuCoPBA/AE surface, a decrease is appeared in reversible oxidation and reduction peaks, which is attributed to the charge repulsion between [Fe(CN)6]3−/4− and the phosphate groups containing in aptamer strands. When the Apt/CD@CuCoPBA/AE surface is bound with EGFR, the oxidation and reduction peak current densities are reduced, suggesting that EGFR is successfully interacted on the Apt/CD@CuCoPBA/AE surface and acted as a barrier, thus hindering the electron transfer at the solid/liquid interface.The detection sensitivity of the EGFR aptasensor was also checked by EIS (Fig. 3c). The EIS results reveal that EIS responses gradually increase with increasing the EGFR concentrations (0.001, 0.005, 0.05, 0.5, 5, 50, 500, and 1000 pg mL−1). Similarly, the ΔRct values increase with increasing the EGFR concentration (Fig. 3d), of which ΔRct values vary linearly with the logarithm of EGFR concentration within the range from 1.0 fg mL−1 to 1.0 ng mL−1 (the inset of Fig. 3d). The regression equation can be expressed as ΔRct (kohm) = 0.30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ConEGFR (pg mL−1) + 1.00 with a coefficient of determination (R2) of 0.9944, where ConEGFR is the target EGFR concentration, showing an ultralow LOD value of 0.42 fg mL−1 (S/N = 3). Compared with other EGFR biosensors, the present CD@CuCoPBA detection system exhibits superior sensing performance toward EGFR (Table 1).
ConEGFR (pg mL−1) + 1.00 with a coefficient of determination (R2) of 0.9944, where ConEGFR is the target EGFR concentration, showing an ultralow LOD value of 0.42 fg mL−1 (S/N = 3). Compared with other EGFR biosensors, the present CD@CuCoPBA detection system exhibits superior sensing performance toward EGFR (Table 1).
| Materials | Methods | Detection range (pg mL−1) | LOD (pg mL−1) | Ref. |
|---|---|---|---|---|
| Dithiobissuccinimidyl propionate | EIS | 1–1 × 105 | 1 | 41 |
| AuNPs | Differential pulse voltammetry (DPV) | 1 × 103 to 4 × 104 | 50 | 10 |
| AuNPs | Resonance Rayleigh scattering | 2 × 104 to 1 × 105 | 100 | 9 |
| CMK-3/poly-(AC-co-MDHLA) | CV | 10−5 × 104 | 3.03 | 42 |
| Ferrocene moiety-peptide ligands | DPV | 100–1 × 106 | 37 | 43 |
| AuNPs-protein G | EIS | 1–1 × 106 | 0.34 | 44 |
| CD@CuCoPBA | EIS | 1 × 10−3 to 1 × 103 | 4.2 × 10−4 | This work |
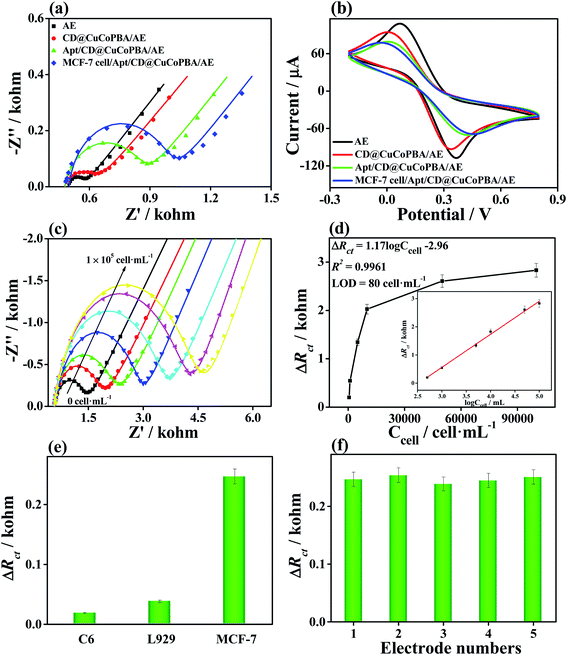
After analyzing the LOD of the CD@CuCoPBA-based aptasensor for detecting EGFR, we further investigated its selectivity, reproducibility, and stability of the proposed sensor. The proteins or biomarkers probably coexisted with EGFR in the human serum, including HER2, IgG, IgE, PSA, BSA, CEA, and their mixed solution were investigated under the same conditions to assess the selectivity of the proposed sensor. As illustrated in Fig. 3e, when the CD@CuCoPBA-based aptasensor was applied to separately analyze different interferents with a concentration of 0.5 pg mL−1, EIS responses exhibit nearly no difference compared with the background test. Howbeit, a large EIS response is found once the aptasensor is incubated with the mixed solution of EGFR and interferents, which is comparable with the case of EGFR only. All results indicate high selectivity of the present aptasensor. In terms of the reproducibility, five different CD@CuCoPBA-based sensors with 5.0 fg mL−1 EGFR were examined under the same measurement conditions (Fig. 3f). The results demonstrate that similar ΔRct values are obtained, giving a low relative standard deviation (RSD) of 2.4%. To assess the stability, the long-term stability experiment was tested (Fig. 3g). After 15 days, ΔRct value retains 97.3% of the initial value, suggesting good stability of the constructed aptasensor. To probe the repeatability, the CD@CuCoPBA-based aptasensor bound with EGFR was dispersed in 0.5 M KOH solution for 5 min. After incubating in the EGFR solution, good electrochemical EIS responses for 10 cycles are observed (Fig. 3h). Consequently, this investigation can broaden the PBA applications in sensing fields, such as environmental monitoring and early determination of cancer markers.
3.6 Practicability of the developed aptasensor toward EGFR
The analytical reliability of the present constructed sensor for detecting EGFR was tested by recovery experiments. The CD@CuCoPBA-based sensor was used to determine the healthy human serum samples containing EGFR with various concentrations. Corresponding to the results (Fig. 3d), the concentration of EGFR was calculated by using the constructed CD@CuCoPBA-based aptasensor (Table S1†). As for the healthy human serum samples, the recovery varied from 94.4% to 104.0%, together with RSDs within a range of 1.9% to 4.5%. Hence, it is promising for the utilization of the developed aptasensor to monitor EGFR concentration in serum samples.3.7 Electrochemical performance of the CD@CuCoPBA-based sensor for determining living MCF-7 cells
Considering the sensing performance, low cell cytotoxicity, and good cell image of the CD@CuCoPBA, we also applied it as the scaffold nanomaterial for EGFR aptasensor to detect the cancer cells which overexpress EGFR, MCF-7 cells. The CD@CuCoPBA-based sensor was applied to monitor MCF-7 cells using the EIS technique. The stepwise fabrication process is displayed in Fig. 4a, including the CD@CuCoPBA/AE, Apt/CD@CuCoPBA/AE, and MCF-7/Apt/CD@CuCoPBA/AE, which leads to the gradually increases in the Rct value. This result indicates that recognition interaction took place between aptamer and the MCF-7 cell surface. As can be seen from Fig. 4b, there is a decrease in peak current, along with the step-by-step fabrication of the aptasensor, which is consistent with the EIS results. Thereby, the constructed CD@CuCoPBA-based sensor can be applied to directly analyze the living cancer cells by the electrochemical technique.The sensitivity of CD@CuCoPBA-based sensor for determining MCF-7 cells was investigated. MCF-7 cells were added into PBS to form the cell solution with different concentrations of 5 × 102, 1 × 103, 5 × 103, 1 × 104, 5 × 104, and 1 × 105 cell per mL before use. As shown in Fig. 4c, the impedance of the aptasensor increase with increasing the concentration of MCF-7 cells with the concentration ranging from 5 × 102 to 1 × 105 cell per mL owning to the recognition interaction between the EGFR-targeted aptamer and EGFR expressed in MCF-7 cells. By analyzing the Rct value with the concentrations of MCF-7 cell (Fig. 4d), the good linear relationship was achieved between the obtained ΔRct value and the logarithm of MCF-7 cell concentration (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ConMCF-7) (the inset of Fig. 4d). The linear equation is presented as ΔRct (kohm) = 1.17
ConMCF-7) (the inset of Fig. 4d). The linear equation is presented as ΔRct (kohm) = 1.17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ConMCF-7 (cell per mL) − 2.96 with a R2 of 0.9961. The LOD was deduced to be 80 cell per mL at an S/N ratio of 3. The analytical performance of the aptasensor is also compared with previously reported biosensors for MCF-7 cells (Table S2†). The result hints that the excellent sensing performance of the CD@CuCoPBA-based sensor is contributed to high electrochemical activity and strong aptamer binding interaction.
ConMCF-7 (cell per mL) − 2.96 with a R2 of 0.9961. The LOD was deduced to be 80 cell per mL at an S/N ratio of 3. The analytical performance of the aptasensor is also compared with previously reported biosensors for MCF-7 cells (Table S2†). The result hints that the excellent sensing performance of the CD@CuCoPBA-based sensor is contributed to high electrochemical activity and strong aptamer binding interaction.
To test the selectivity of CD@CuCoPBA-based biosensor as a sensitive layer for detecting living cells, cancer cell lines (C6) and normal cell lines (L929) were also measured by EIS. As depicted in Fig. 4e, there is no substantial change in the ΔRct value after the modified electrode is incubated in C6 and L929 cell solutions (500 cell per mL). In terms of the reproducibility of the CD@CuCoPBA-based biosensor sensor, parallel tests of five aptasensors were tested under same experiment conditions (Fig. 4f). The RSD of five measurements is calculated to be 3.2%. This founding further proves that the constructed CD@CuCoPBA-based sensor can not only be utilized in detecting living cancer markers but also be applied in clinical analysis.
4. Conclusion
In summary, a novel nanocomposite of CuCo PBA loaded with abundant CDs was synthesized and applied it as a bioplatform for immobilizing EGFR aptamer to construct the electrochemical aptasensor to detect EGFR and MCF-7 cells. The combined valences of Cu (Cu(I) and Cu(II)) and Co (Co(0), Co(I), and Co(II)) species were coexisted, while rich carbon-related functional groups were observed in the CD@CuCoPBA. From the TEM analysis, it is clear that the CDs were distributed within the interior of the CuCo PBA networks, which can facilitate the electrochemical conductivity, fluorescence performance, and good biocompatibility. Therefore, compared with the CoCuPBA-based sensor, the CD@CuCoPBA-based one demonstrated a superior sensing performance toward EGFR and EGFR-overexpressed cancer cells. The electrochemical results demonstrated that the CDs@CuCoPBA-based sensor gave low LODs for the detection of EGFR (0.42 fg mL−1) and MCF-7 cancer cells (80 cell per mL), together with high selectivity, good reproducibility, high stability, repeatability, and acceptable applicability. We anticipate that this work can benefit researchers on biosensing and clinical assay because of the outstanding sensing performance of CuCo PBA.Conflicts of interest
The authors declare no competing conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. U1604127), Doctoral research foundation of Zhengzhou University of Light Industry (No. 2016BSJJ031).References
- P. Wee and Z. Wang, Cancers, 2017, 9, 52 CrossRef PubMed.
- S. Huang, J. Li and P. M. Harari, Mol. Cancer Ther., 2002, 1, 507–514 CAS.
- H. Lo and M. Hung, Br. J. Cancer, 2006, 94, 184–188 CrossRef CAS PubMed.
- E. Koumarianou, T. A. Slastnikova, M. Pruszynski, A. A. Rosenkranz, G. Vaidyanathan, A. S. Sobolev and M. R. Zalutsky, Nucl. Med. Biol., 2014, 41, 441–449 CrossRef CAS PubMed.
- J. Aaron, K. Travis, N. Harrison and K. Sokolov, Nano Lett., 2009, 9, 3612–3618 CrossRef CAS PubMed.
- L. W. Chan, Y. Wang, L. Y. Lin, M. P. Upton, J. H. Hwang and S. H. Pun, Bioconjugate Chem., 2013, 24, 167–175 CrossRef CAS PubMed.
- S. E. Weigum, P. N. Floriano, S. W. Redding, C. Yeh, S. D. Westbrook, H. S. McGuff, A. Lin, F. R. Miller, F. Villarreal, S. D. Rowan, N. Vigneswaran, M. D. Williams and J. T. McDevitt, Cancer Prev. Res., 2010, 1940–6207 Search PubMed.
- Y. Takahashi, H. Shiku, T. Murata, T. Yasukawa and T. Matsue, Anal. Chem., 2009, 81, 9674–9681 CrossRef CAS PubMed.
- J. Li, J. Wang, X. Zhang, H. Chang and W. Wei, Sens. Actuators, B, 2018, 273, 1300–1306 CrossRef CAS.
- H. Ilkhani, M. Sarparast, A. Noori, S. Zahra Bathaie and M. F. Mousavi, Biosens. Bioelectron., 2015, 74, 491–497 CrossRef CAS PubMed.
- S. Yazdanparast, A. Benvidi, M. Banaei, H. Nikukar, M. D. Tezerjani and M. Azimzadeh, Microchim. Acta, 2018, 185, 405 CrossRef PubMed.
- C. Guo, F. Su, Y. Song, B. Hu, M. Wang, L. He, D. Peng and Z. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 41188–41199 CrossRef CAS PubMed.
- N. Zhou, F. Su, C. Guo, L. He, Z. Jia, M. Wang, Q. Jia, Z. Zhang and S. Lu, Biosens. Bioelectron., 2019, 123, 51–58 CrossRef CAS PubMed.
- S. Song, L. Wang, J. Li, C. Fan and J. Zhao, TrAC, Trends Anal. Chem., 2008, 27, 108–117 CrossRef CAS.
- E. G. Hvastkovs and D. A. Buttry, Analyst, 2010, 135, 1817–1829 RSC.
- Y. Qi, F. Xiu, M. Zheng and B. Li, Biosens. Bioelectron., 2016, 83, 243–249 CrossRef CAS PubMed.
- Y. Huo, L. Qi, X. Lv, T. Lai, J. Zhang and Z. Zhang, Biosens. Bioelectron., 2016, 78, 315–320 CrossRef CAS PubMed.
- Z. Wang, J. Yu, R. Gui, H. Jin and Y. Xia, Biosens. Bioelectron., 2016, 79, 136–149 CrossRef CAS PubMed.
- W. Wang, W. Wang, J. J. Davis and X. Luo, Microchim. Acta, 2015, 182, 1123–1129 CrossRef CAS.
- V. Urbanová, K. Jayaramulu, A. Schneemann, Š. Kment, R. A. Fischer and R. Zbořil, ACS Appl. Mater. Interfaces, 2018, 10, 41089–41097 CrossRef PubMed.
- X. T. Zheng, A. Ananthanarayanan, K. Q. Luo and P. Chen, Small, 2015, 11, 1620–1636 CrossRef CAS PubMed.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- J. Lee, G. Nam, J. Sun, S. Higashi, H. Lee, S. Lee, W. Chen, Y. Cui and J. Cho, Adv. Energy Mater., 2016, 6, 1601052 CrossRef.
- A. A. Karyakin, O. V. Gitelmacher and E. E. Karyakina, Anal. Chem., 1995, 67, 2419–2423 CrossRef CAS.
- D. Liu, R. Qiang, Y. Du, Y. Wang, C. Tian and X. Han, J. Colloid Interface Sci., 2018, 514, 10–20 CrossRef CAS PubMed.
- X. Li, J. Liu, A. I. Rykov, H. Han, C. Jin, X. Liu and J. Wang, Appl. Catal., B, 2015, 179, 196–205 CrossRef CAS.
- L. Chao, X. Xiong, J. Liu, A. Xu, T. Huang, F. He and Q. Xie, Sens. Actuators, B, 2017, 253, 603–611 CrossRef CAS.
- N. Zhou, F. Su, Z. Li, X. Yan, C. Zhang, B. Hu, L. He, M. Wang and Z. Zhang, Microchim. Acta, 2019, 186, 75 CrossRef PubMed.
- P. Yang, J. Pang, F. Hu, J. Peng, D. Jiang, Z. Chu and W. Jin, Sens. Actuators, B, 2018, 276, 31–41 CrossRef CAS.
- N. Zhou, L. Yang, B. Hu, Y. Song, L. He, W. Chen, Z. Zhang, Z. Liu and S. Lu, Anal. Chem., 2018, 90, 13624–13631 CrossRef CAS PubMed.
- X. Cai, W. Gao, M. Ma, M. Wu, L. Zhang, Y. Zheng, H. Chen and J. Shi, Adv. Mater., 2015, 27, 6382–6389 CrossRef CAS PubMed.
- W. Ahn, M. G. Park, D. U. Lee, M. H. Seo, G. Jiang, Z. P. Cano, F. M. Hassan and Z. Chen, Adv. Funct. Mater., 2018, 28, 1802129 CrossRef.
- G. Zhang, M. Yan, X. Teng, H. Bi, Y. Han, M. Tian and M. Wang, Chem. Commun., 2014, 50, 10244–10247 RSC.
- Y. Song, M. Xu, Z. Li, L. He, M. Hu, L. He, Z. Zhang and M. Du, Sens. Actuators, B, 2020, 311, 127927 CrossRef CAS.
- L. Han, T. Yu, W. Lei, W. Liu, K. Feng, Y. Ding, G. Jiang, P. Xu and Z. Chen, J. Mater. Chem. A, 2017, 5, 16568–16572 RSC.
- G. Zhou, L. Veron, A. Elaissari, T. Delair and C. Pichot, Polym. Int., 2004, 53, 603–608 CrossRef CAS.
- M. Majdan, O. Maryuk, A. Gładysz-Płaska, S. Pikus and R. Kwiatkowski, J. Mol. Struct., 2008, 874, 101–107 CrossRef CAS.
- Y. Song, F. Duan, S. Zhang, J. Tian, Z. Zhang, Z. Wang, C. Liu, W. Xu and M. Du, J. Mater. Chem. A, 2017, 5, 19378–19389 RSC.
- M. Wang, L. Yang, B. Hu, J. Liu, L. He, Q. Jia, Y. Song and Z. Zhang, Biosens. Bioelectron., 2018, 113, 16–24 CrossRef CAS PubMed.
- X. Yan, Y. Song, J. Liu, N. Zhou, C. Zhang, L. He, Z. Zhang and Z. Liu, Biosens. Bioelectron., 2019, 126, 734–742 CrossRef CAS PubMed.
- A. Vasudev, A. Kaushik and S. Bhansali, Biosens. Bioelectron., 2013, 39, 300–305 CrossRef CAS PubMed.
- M. Regiart, M. A. Fernández-Baldo, J. Villarroel-Rocha, G. A. Messina, F. A. Bertolino, K. Sapag, A. T. Timperman and J. Raba, Anal. Chim. Acta, 2017, 963, 83–92 CrossRef CAS PubMed.
- R. Li, H. Huang, L. Huang, Z. Lin, L. Guo, B. Qiu and G. Chen, Electrochim. Acta, 2013, 109, 233–237 CrossRef CAS.
- R. Elshafey, A. C. Tavares, M. Siaj and M. Zourob, Biosens. Bioelectron., 2013, 50, 143–149 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra01439g |
| This journal is © The Royal Society of Chemistry 2020 |