Open Access Article
Open Access ArticleRaman profile alterations of irradiated human nasopharyngeal cancer cells detected with laser tweezer Raman spectroscopy
Sufang Qiu†
*ab,
Youliang Weng†c,
Ying Li†a,
Yang Chen d,
Yuhui Panc,
Jun Liuef,
Wanzun Lina,
Xiaochuan Chena,
Miaomiao Lia,
Ting Lina,
Wei Liua,
Lurong Zhangg and
Duo Lin
d,
Yuhui Panc,
Jun Liuef,
Wanzun Lina,
Xiaochuan Chena,
Miaomiao Lia,
Ting Lina,
Wei Liua,
Lurong Zhangg and
Duo Lin h
h
aFujian Medical University Cancer Hospital, Fujian Cancer Hospital, Fuzhou 350014, China. E-mail: sfqiu@126.com
bFujian Provincial Key Laboratory of Translational Cancer Medicine, Fuzhou 350014, China
cDepartment of Radiation Oncology, Fujian Cancer Hospital, Fujian Medical University Cancer Hospital, Fuzhou 350014, China
dDepartment of Laboratory Medicine, Fujian Medical University, Fuzhou 350004, China
eCancer Bio-immunotherapy Center, Fujian Medical University Cancer Hospital, Fujian Cancer Hospital, Fuzhou 350014, China
fDepartment of Medical Oncology, Fujian Medical University Cancer Hospital, Fujian Cancer Hospital, Fuzhou 350014, China
gLaboratory of Radiation Oncology and Radiobiology, Fujian Cancer Hospital, Fujian Medical University Cancer Hospital, Fuzhou 350014, China
hKey Laboratory of Optoelectronic Science and Technology for Medicine, Ministry of Education, Fujian Provincial Key Laboratory for Photonics Technology, Fujian Normal University, Fuzhou 350007, China
First published on 8th April 2020
Abstract
Radiotherapy has been widely used for nasopharyngeal carcinoma (NPC) treatment, which causes DNA damage and alterations of macromolecules of cancer cells. However, the Raman profile alterations of irradiated NPC cells remain unclear. In the present study, we used laser tweezers Raman spectroscopy (LTRS) to monitor internal structural changes and chemical modifications in NPC cells after exposure at a clinical dose (2.3 Gy) to X-ray irradiation (IR) at a single-cell level. Two types of NPC cell lines, CNE2 (EBV-negative cell line) and C666-1 (EBV-positive cell line), were used. The Raman spectra of cells before and after radiation treatment were recorded by LTRS. The analysis of spectral differences indicated that the IR caused Raman profile alterations of intracellular proteins, DNA base and lipids. Moreover, by using the multivariate statistical analysis including principal component analysis (PCA) and linear discriminant analysis (LDA) algorithm, an accuracy of 90.0% for classification between CNE2 cells before and after IR could be achieved, which was 10% better than that of C666-1 cells. The results demonstrated that CNE2 cells were more sensitive to IR in comparison to C666-1 cells, providing useful information for creating a treatment strategy in clinical practice. This exploratory study suggested that LTRS combined with multivariate statistical analysis would be a novel and effective tool for evaluating the radiotherapeutic effect on tumor cells, and for detection of the corresponding alterations at the molecular level.
Introduction
Nasopharyngeal carcinoma (NPC) is a prevalent malignant tumor with an extremely high incidence in Southeast Asia.1 Due to its anatomic localization and extremely radiosensitive nature, radiation therapy (RT) is the mainstay of the treatment for NPC.1–4Various imaging technologies are applied for evaluating the curative effect and revealing the molecular mechanisms of RT.5 Raman spectroscopy (RS) is a rapid, accurate and convenient technology providing spectral fingerprint information of the cellular biochemical changes and identifying radiation-induced alterations at the molecular level, including changes of the content and conformation of key macromolecules (nucleic acids, lipids, proteins and carbohydrates).6,7 The radioresistant lung and breast cells showed radiation-induced RS spectral features as glycogen accumulation at low RT doses, whereas the radiosensitive prostate cells showed some membrane phospholipid-related alterations.8,9 Roman et al. analyzed cytoplasmic and nuclear regions of irradiated prostate cancer cells (PC-3) and suggested various DNA damages located in those two regions.10 Lasalvia et al. observed that DNA was damaged in breast cells (MCF10A) irradiated by protons, in particular the O–P–O stretching mode at about 784 cm−1.11 However, there is barely attention toward the Raman technique study on radiation-induced cellular response of NPC cells.
There are still some limitations associated with conventional RS technique, such as the interference from chemical agent for cell fixation, making it different to actually reflect cellular biological activity after radiation.12 The advent of laser tweezers Raman spectroscopy (LTRS) technology provides an alternative strategy to circumvent the limitations by optically capturing alive single cells in suspension without fixation and simultaneously measuring the Raman signals of the trapped cells.13 Recently, LTRS has been widely applied in red blood cell detection,14 diagnosis of cancer, and detection of cell apoptosis.15
This study is to explore the feasibility of applying the LTRS technique for studying NPC cellular biochemical changes induced by IR and to address the following questions: (1) could any alterations of Raman profile reflecting the cellular macromolecules (protein, nucleic acid and lipid) after NPC cells exposed to the clinical relevant 2.3 Gy IR dose? (2) Does alteration of Raman profile differ between EBV negative CNE2 cells and EBV positive C666-1 cells? Multivariate statistics methods, including principal component analysis and linear discriminant analysis (PCA–LDA) algorithms, were utilized to analyze and classify different cellular macromolecules based on spectral data. To our best acknowledge, this is the first time to investigate the spectral characterization of human NPC cells after radiation using LTRS, and the results would aid in understanding the NPC cellular responses to the radiotherapy at the molecular level.
Materials and methods
Cell lines and irradiations
Human NPC cell lines CNE2 and C666-1 were provided from Cancer Research Institute, Fujian Provincial Tumor Hospital (Fuzhou, China) and were routinely cultured in medium RPMI-1640 containing 10% fetal bovine serum (FBS), 100 U mL−1 penicillin and streptomycin in a humidified incubator with 5% CO2 at 37 °C. The usage of these cell lines and protocols in this study was approved by the Ethics Committee of Fujian Provincial Tumor Hospital. The CNE2 and C666-1 cells in logarithmic growth phase were divided into a blank group and radiated group. The irradiation was performed using a 6 MV electron beam produced by an Elekta Precise linear accelerator equipped with the Precise Treatment System™ (Elekta AB, Stockholm, Sweden). Cells exposed to commonly clinical doses of radiation 2.3 Gy with a dose rate of 400 μ min−1 were studied together with control group (0 Gy). 48 hours post irradiation, the adherent NPC cells were digested by 2 mL 0.25% trypsin solution, suspension in culture medium, centrifuged at 1000 rpm for 5 min to collect cells, and resuspended in phosphate-buffered saline (PBS) solution for analysis with LTRS.The LTRS system and Raman spectra measurements
Thirty living cells were collected randomly from each group in suspension via laser tweezers and the Raman spectrum of each living cell was recorded using the same laser beam by our homemade LTRS system.16 Briefly, a 785 nm laser (LE-LS-XXRaman, StarBright Laser AB, Sweden) was used to trap single cell through beam expansion and shaping (Fig. 1), as well as a 100× oil-immersion objective (N.A. = 1.3, IX71; Olympus, Center Valley, PA, USA). Then, the Raman scattering signals of this cell were collected by the same objective and finally recorded by a combination of a spectrograph (Holospec-f/2.2-NIR) and a CCD camera (Princeton Instruments), with the acquisition time of 40 s and the laser power of ∼6 mW in the region of 400–1800 cm−1. All the spectral data preprocessing was under the same condition.Spectral processing and data analysis
Due to that the raw Raman spectra of single cell containing strong autofluorescence and background noises, a Vancouver Raman algorithm was applied as a data preprocessing for background and autofluorescence removal to extract pure Raman spectra signals by performing a fifth-order polynomial fitting procedure.17 The spectra were subsequently normalized according to the area under the curve to minimize the potential variations caused by the system and measurement environment. Finally, we used PCA–LDA method to explore statistical significance and perform classifications between the control and radiated groups using the SPSS software package (version 20.0, IBM, USA).Results and discussions
Assessment of radiation dose in NPC cells
NPC is an extremely radiosensitive disease. Assessment of radiation dose is especially important in the treatment of NPC. The personalized RT aims to achieve maximal tumor control while minimum toxicity. However, the irradiated response monitoring better treatment doses and fractionation schemes remains a major impediment.18 Conventional clinical doses for curative RT are on the order of 2–10 Gy. It has been confirmed that RS is capable of identifying radiation-induced biochemical changes in tumor cells at these doses. But there is no such a study on NPC cells.9 The 2.3 Gy is a commonly used clinical dose for NPC treatment.19,20 Hence, two different radiation doses of 0 Gy (control) and 2.3 Gy were chosen in this study.Raman spectroscopy of NPC cells after 2.3 Gy irradiation and the cellular effect of radiation
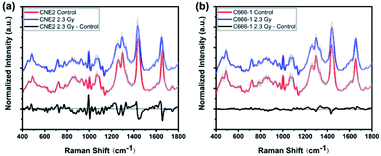
To investigate radiation-induced cellular responses of single-living NPC cell, we recorded the Raman spectra using label-free LTRS technique. Fig. 2 shows the averaged Raman spectrum and its differences between 0 Gy (control) and 2.3 Gy in both CNE2 (Fig. 2(a)) and C666-1 (Fig. 2(b)) cells, demonstrating the significant differences of Raman signals after irradiation, especially for CNE2 groups. The shaded areas (grey) represent the standard deviations of the means, indicating that the spectral results of LTRS is highly reproducible and that the spectral variances among 30 cell samples in each group are relatively small.As showed in Fig. 2 black line at the bottom, some changes in some Raman peaks appeared in both NPC cell lines after irradiation, suggesting that the Raman profile could reflect alteration of cellular macromolecules (protein, nucleic acid and lipid) after NPC cells exposed to the clinical relevant 2.3 Gy IR dose. Table 1 lists the tentative assignments for several observed prominent Raman bands, according to previous studies, which can be assigned to nucleic acids, carbohydrates, proteins or lipids and provide rich information to assess cellular molecular structure and content changes via spectral characteristics.21–25
| Peak position (cm−1) | Assignment |
|---|---|
| 492 | Tyrosine |
| 724 | A (ring breathing mode of DNA/RNA bases) |
| 752 | T (ring breathing mode of DNA/RNA bases) |
| 785 | U, T, C (ring breathing modes in the DNA/RNA bases), backbone O–P–O |
| 853 | Ring breathing mode of tyrosine & C–C stretch of proline ring |
| 875 | Tryptophan |
| 970 | Lipid: chain C–C |
| 1003 | Symmetric ring breathing of phenylalanine |
| 1033 | Phenylalanine, C–N stretching of proteins, C–H in-plane phenylalanine (proteins) |
| 1065 | Protein: C–N stretch, lipid: chain C–C stretch |
| 1156 | C–C, C–N stretching (protein) |
| 1264 | Amide III, T, A (ring breathing modes of the DNA/RNA bases), ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H bend (protein) C–H bend (protein) |
| 1297 | CH2 twisting (lipids) |
| 1335 | A, G (ring breathing modes in the DNA bases), C–H deformation (protein) |
| 1440 | CH2 deformation (lipids and proteins) |
| 1552 | Tryptophan |
| 1612 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching mode of tyrosine & tryptophan C stretching mode of tyrosine & tryptophan |
| 1655 | Amide I (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching mode of proteins, α-helix conformation), C O stretching mode of proteins, α-helix conformation), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C lipid stretch C lipid stretch |
| 1743 | Lipids: C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester O ester |
Comparing the mean spectra and their difference spectra between control and irradiated cells, the significant decreased signals were detected at 1297 and 1440 cm−1, corresponding to CH2 bending mode of protein and lipid components, and 1655 cm−1 reflected amide I and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C lipid stretch (Fig. 2(a) and (b)) after two NPC cell lines irradiated. The cellular Raman spectral features of the three significant bands were assigned to bending modes of proteins and lipids as reported by Movasaghi et al.,22 which are main components in cells, indicating the damage of cellular components after radiation. In addition, the radiation-induced cellular responses observed in CNE2 cells were generally more notable than C666-1 cells. For instance, the differences of spectra at 1264 cm−1 (came from protein), 970 cm−1 and 1065 cm−1 (attributed to C–C stretching mode of lipids) showed less significant (Fig. 2(a)). It indicates that the relative content of cellular components with corresponding bond to the spectrum decreased after radiation, which can be attributed to special structure scissions and conformation aggregation. However, the band at 1003 cm−1 (from symmetric ring breathing of phenylalanine) in CNE2 cells showed an increased signal as compared to the control group. The intensity of certain Raman peaks associated with DNA damage increased in irradiated group. In particular, the intensity of a Raman peak at 752 cm−1 (constituted of the ribose bands of thymine) and 1335 cm−1 (from the ribose bands of adenine and guanine) were related to the fragmentation of nucleic acids bases, as a result of radiation-induced DNA damage and bases unstacked. All other bands showed relatively little change after irradiation.
C lipid stretch (Fig. 2(a) and (b)) after two NPC cell lines irradiated. The cellular Raman spectral features of the three significant bands were assigned to bending modes of proteins and lipids as reported by Movasaghi et al.,22 which are main components in cells, indicating the damage of cellular components after radiation. In addition, the radiation-induced cellular responses observed in CNE2 cells were generally more notable than C666-1 cells. For instance, the differences of spectra at 1264 cm−1 (came from protein), 970 cm−1 and 1065 cm−1 (attributed to C–C stretching mode of lipids) showed less significant (Fig. 2(a)). It indicates that the relative content of cellular components with corresponding bond to the spectrum decreased after radiation, which can be attributed to special structure scissions and conformation aggregation. However, the band at 1003 cm−1 (from symmetric ring breathing of phenylalanine) in CNE2 cells showed an increased signal as compared to the control group. The intensity of certain Raman peaks associated with DNA damage increased in irradiated group. In particular, the intensity of a Raman peak at 752 cm−1 (constituted of the ribose bands of thymine) and 1335 cm−1 (from the ribose bands of adenine and guanine) were related to the fragmentation of nucleic acids bases, as a result of radiation-induced DNA damage and bases unstacked. All other bands showed relatively little change after irradiation.
Above results indicate that Raman spectroscopy is extremely sensitive to identify the information about IR-induced proteins- and lipids-rearrangements and DNA damage at the clinical dose. DNA is considered to be the most important target for radiation, mediating the lethal cellular effects and delivering benefit after treatment. Radiotherapy is effective in inducing the apoptosis of tumor cells through breaking chemical bonds,26 single- and double-strand nucleic acid breaks, and modifying bases of DNA and RNA. The interesting findings from Raman peak analysis and simplistic peak intensity analysis described above only used limited peak information. There are other significant variations and overlapping intensities of Raman spectra between control and treated cells. Hence, multivariate statistical analyses were conducted by employing whole spectral data for further analysis.
Multivariate analysis of Raman spectra from NPC cells
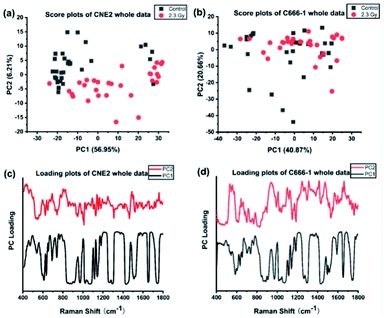
We first performed PCA on the whole Raman data with the factor analysis to demonstrate significant spectral differences between control and radiated group. The first two principal components (PCs) describe the most variance in original spectral information and contain primary effects in the PCA process, while the explained variance gradually decline by PCs > 3. In addition, the significance of first two PCs scores were determined by independent sample T test (p < 0.05). Therefore we generated the scatter plot depicted by scores of the first 2 PCs, which can keep the near original spectral information shown in the Fig. 3. In Fig. 3(a), we found the radiated and control CNE2 were distributed in obvious separate regions with some degree of overlap. However, most of radiated C666-1 cells samples overlapped with control cells shown in Fig. 3(b). These results suggested greater variation of cellular components appear in CNE2 cells, compared with C666-1 after radiation exposure. These phenomena suggest that the damage of radiated-induced stress is common to both cell lines, but the degree of the damage between cell lines is not the same, which results in different distribution based on PC score, and is consistent with above spectral peak analysis.Next, the most significant PCs defined by the independent T-test were loaded into the LDA model to explore effective classification algorithms for different NPC cell groups. The obtained classification sensitivity and specificity using PCA–LDA with the leave-one-out, cross-validation method for comparison are shown in Table 2. The accuracy of 90.0% for classification between CNE2 cells before and after radiation could be achieved, which was 10% improvement than that for C666-1 cells. It further confirms that sensitivity of CNE2 cells to IR was significantly higher than that of C666-1 cells. A previous in vitro study also found that the survival rate of C666-1 cells was significantly higher than that of CNE2 cells with photodynamic therapy, which indicating the biological characteristics of NPC cells can affect the susceptibility of therapy.27 The reason might be attributed to different EBV transcript expression. In this study, the CNE2 cells do not express EBV transcripts in long-term culture, whereas the C666-1 is the only EBV+ human NPC cell line in long-term culture. EBV may promote development and progression of NPC by regulating special RNA expressions, such as miR-BART4, consequently inhibiting its radiosensitivity,28 which confirmed the radiosensitivity of EBV-CNE2 cells is higher than EBV+ C666-1 cells.
| Model | Cell | Predicted | ||
|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | ||
| Control vs. 2.3 Gy | CNE2 | 90.00% | 86.70% | 93.30% |
| C666-1 | 78.35% | 76.70% | 80.00% | |
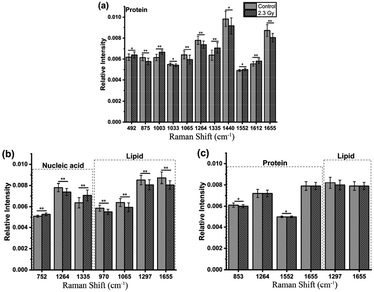
Furthermore, loadings of the first two components (PCs) contributed for the classification are shown in Fig. 3(c) and (d), respectively, where we can explore the important Raman bands contributing to discriminant analysis. Fig. 4 displays the comparison of the mean intensities of the selected peaks with the most significant differences between control and IR treated cell samples for CNE2 group and C666-1 group. The normality and significate variance of these spectral intensities were assessed by Shapiro–Wilk test and F-test followed by Mann–Whitney U test and T-test. We found that the difference in proteins was prominent, compared to other cellular components between the control and irradiated group at the cell level, suggesting that the radiation effect was concentrated in the majority of some amino acids molecules or deformation mode.9 For amino acids in CNE2 cells (Fig. 4(a)), the relative contents of protein bands such as tryptophan (the peaks at 875, 1552 and 1612 cm−1) and tyrosine (the peaks at 492 and 1612 cm−1) were significantly different between the control and irradiated group (p < 0.05). For example, the intensity at 1612 cm−1 was significantly increased in the irradiated group, demonstrating that the internal C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching mode of tryptophan and tyrosine were affected by radiation. It also showed that the band of 853 cm−1 (related to ring breathing mode of tyrosine) (F = 5.973, p < 0.05) and 1552 cm−1 (related to tryptophan) (F = 5.236, p < 0.05) were significantly increased in C666-1 cells (Fig. 4(c)). Such changes also suggested the molecular structural vibration of proteins was sensitive to radiation. With the help of KEGG online database, characteristic metabolites with significant changes from Raman bands when compared with normal cells could be rationally used to derive the potential disordered metabolic networks of NPC cell lines after radiation. The phenylalanine together with tyrosine contributed to the common pathways such as TCA cycle and several amino acid metabolisms in NPC cells with RT.29 Moreover, the bands related to amide (1264 and 1655 cm−1 from amide I and amide III respectively), C–N stretching (i.e. 1065 cm−1) and C–H stretching (i.e. 1033, 1335 and 1440 cm−1) of proteins were deviated obviously in irradiated group in CNE2 cells (p < 0.05). The change of amides may suggest that a desamidization was triggered and resulted in modification of the spatial structure and eventually leaded to cellular protein function lost.10 However, we failed to find the significant difference in changes of amide in C666-1 cells.
C stretching mode of tryptophan and tyrosine were affected by radiation. It also showed that the band of 853 cm−1 (related to ring breathing mode of tyrosine) (F = 5.973, p < 0.05) and 1552 cm−1 (related to tryptophan) (F = 5.236, p < 0.05) were significantly increased in C666-1 cells (Fig. 4(c)). Such changes also suggested the molecular structural vibration of proteins was sensitive to radiation. With the help of KEGG online database, characteristic metabolites with significant changes from Raman bands when compared with normal cells could be rationally used to derive the potential disordered metabolic networks of NPC cell lines after radiation. The phenylalanine together with tyrosine contributed to the common pathways such as TCA cycle and several amino acid metabolisms in NPC cells with RT.29 Moreover, the bands related to amide (1264 and 1655 cm−1 from amide I and amide III respectively), C–N stretching (i.e. 1065 cm−1) and C–H stretching (i.e. 1033, 1335 and 1440 cm−1) of proteins were deviated obviously in irradiated group in CNE2 cells (p < 0.05). The change of amides may suggest that a desamidization was triggered and resulted in modification of the spatial structure and eventually leaded to cellular protein function lost.10 However, we failed to find the significant difference in changes of amide in C666-1 cells.
In addition, the significant changes occurring in DNA, as the most important target, due to the radiation can be seen at a clinical dose of 2.3 Gy in CNE2 cells, confirming that it was very sensitive to radiation. Such phenomenon was particularly shown in the peaks of 752, 1264 and 1335 cm−1 in CNE2 cells, for which a statistically significant difference (p < 0.001) were found between control cells and radiation exposed cells. According to the PC loading shown in Fig. 3, the PC1 score of the loading of the 1335 cm−1 (from A and G) and 752 cm−1 (from T) also have a positive value for the irradiated samples, thus meaning that the contribution of the positive feature is increased in the spectra from irradiated cells. DNA damage induced by IR activates the DNA damage responses of a complex network of proteins that involves in cell cycle coordination, DNA repair and apoptosis.30 It is essentially step to protein-mediated modifications and activating downstream signaling pathways.31 It also suggests that RT-induced damage can be corrected by various DNA repair pathways.30 The Raman spectroscopy provides a new method to explore the intricacy of DNA damage response in cancer cells and normal cells. For lipid as the main component of cell membrane, we also analyzed the intensity of some lipid peaks. Several relative contents from lipid also altered significantly, compared to the control group, in CNE2 cells, but not in C666-1 cells (Fig. 4(b) and (c)). The failure of radiation to play an aggressive role in DNA and lipid damage suggested its damage in C666-1 cells was limited.
So according to above results, we have reciprocally authenticated that Raman spectra of the NPC cells have significant changes after the radiation exposure. Together with several changes of Raman band intensity by combining the above metabolic pathway analysis with KEGG database, we confirmed that several radiated-induced pathways were involved, including several amino acid metabolisms, energy metabolism and purine metabolism. Such a phenomenon was resulted from the biological effect of radiation interacting with tumor cells.32 The detailed molecular mechanism is remained to be further elucidated. We further confirmed that the dose of 2.3 Gy was capable of triggering the radiation-induced cellular response. In addition, we found CNE2 cells is more sensitivity than C666-1 cells, which may be due to the biological characteristics of NPC cell lines, including different EBV transcript expression.
Conclusions
We provide evidences about the potential utilization of LTRS in the observation of IR-induced NPC cell damages. A multitude of changes of cellular biochemistry within NPC cells were observed after radiation treatment. In addition, the PCA–LDA methods were applied to explore statistical analysis on significant modifications and provide classification results. The Raman spectrum highlight variations were found in specific protein and lipids rearrangements and DNA damage, such as the relative content of specific Raman bands related to the C–H deformation and DNA bases. This study demonstrates that the LTRS technology can identify unique biochemical signatures occurring in NPC cells after response to radiation at the level of single living cells. The results may contribute to further understanding interacting mechanism between tumor cells and radiation and the RS would be a complementary technique for assessing cellular response to radiotherapy.Conflicts of interest
There is no conflict to declare.Acknowledgements
Authors would like to acknowledge the financial support from Fujian Provincial Health Technology Project (2018-CX-13) and Joint Funds for the Innovation of Science and Technology, Fujian Province (2018Y9105). Authors are thankful to United Fujian Provincial Health and Education Project for Tackling the Key Research, China (2019-WJ-03) for their support during this research work. Authors also like to acknowledge National Natural Science Foundation of China (NSFC) (U1605253, 11974077), Fujian Provincial Key Laboratory of Translational Cancer Medicine and Science and Technology Program of Fujian Province, China (2018Y2003) for providing the financial support for the fellows working in the project.References
- Y. P. Chen, A. T. C. Chan, Q. T. Le, P. Blanchard, Y. Sun and J. Ma, Lancet, 2019, 394, 64–80 CrossRef.
- F. Perri, G. D. V. Scarpati, F. Caponigro, F. Ionna, F. Longo, S. Buonopane, P. Muto, M. Di Marzo, S. Pisconti and R. Solla, OncoTargets Ther., 2019, 12, 1583–1591 CrossRef CAS PubMed.
- J. Lang, C. Hu, T. Lu, J. Pan and T. Lin, Cancer Manage. Res., 2019, 11, 6365–6376 CrossRef CAS PubMed.
- W. Liao, M. Tian and N. Chen, Cancer Manage. Res., 2019, 11, 8431–8442 CrossRef PubMed.
- M. Lyu, D. Zhu, Y. Duo, Y. Li and H. Quan, Biomaterials, 2019, 119656 Search PubMed.
- S. Bakhtiaridoost, H. Habibiyan, S. Muhammadnejad, M. Haddadi, H. Ghafoorifard, H. Arabalibeik and S. Amanpour, RSC Adv., 2016, 6, 50027–50033 RSC.
- N. Kuhar, S. Sil, T. Verma and S. Umapathy, RSC Adv., 2018, 8, 25888–25908 RSC.
- Q. Matthews, A. Jirasek, J. J. Lum and A. G. Brolo, Phys. Med. Biol., 2011, 56, 6839–6855 CrossRef CAS PubMed.
- S. J. Harder, Q. Matthews, M. Isabelle, A. G. Brolo, J. J. Lum and A. Jirasek, Appl. Spectrosc., 2015, 69, 193–204 CrossRef CAS PubMed.
- M. Roman, T. P. Wrobel, A. Panek, E. Efeoglu, J. Wiltowska-Zuber, C. Paluszkiewicz, H. J. Byrne and W. M. Kwiatek, Sci. Rep., 2019, 9, 8715 CrossRef PubMed.
- M. Lasalvia, G. Perna, L. Manti, J. Rasero, S. Stramaglia and V. Capozzi, Int. J. Radiat. Biol., 2019, 95, 207–214 CrossRef CAS PubMed.
- K. Eberhardt, C. Stiebing, C. Matthaus, M. Schmitt and J. Popp, Expert Rev. Mol. Diagn., 2015, 15, 773–787 CrossRef CAS PubMed.
- S. Casabella, P. Scully, N. Goddard and P. Gardner, Analyst, 2016, 141, 689–696 RSC.
- J. Lukose, N. Mithun, G. Mohan, S. Shastry and S. Chidangil, RSC Adv., 2019, 9, 7878–7884 RSC.
- C. Krafft, I. W. Schie, T. Meyer, M. Schmitt and J. Popp, Chem. Soc. Rev., 2016, 45, 1819–1849 RSC.
- D. Lin, Z. Zheng, Q. Wang, H. Huang, Z. Huang, Y. Yu, S. Qiu, C. Wen, M. Cheng and S. Feng, Opt. Express, 2016, 24, 24750–24759 CrossRef CAS PubMed.
- S. Zhu, X. Cui, W. Xu, S. Chen and W. Qian, RSC Adv., 2019, 9, 9500–9508 RSC.
- S. K. Paidi, P. M. Diaz, S. Dadgar, S. V. Jenkins, C. M. Quick, R. J. Griffin, R. P. M. Dings, N. Rajaram and I. Barman, Cancer Res., 2019, 79, 2054–2064 CrossRef CAS PubMed.
- J. Bernier and S. M. Bentzen, Eur. J. Cancer, 2003, 39, 560–571 CrossRef CAS PubMed.
- E. B. Butler, B. S. Teh, W. H. Grant, 3rd, B. M. Uhl, R. B. Kuppersmith, J. K. Chiu, D. T. Donovan and S. Y. Woo, Int. J. Radiat. Oncol., Biol., Phys., 1999, 45, 21–32 CrossRef CAS.
- J. De Gelder, K. De Gussem, P. Vandenabeele and L. Moens, J. Raman Spectrosc., 2007, 38, 1133–1147 CrossRef CAS.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- F. Zheng, Y. Qin and K. Chen, J. Biomed. Opt., 2007, 12, 034002 CrossRef PubMed.
- Q. Matthews, A. Jirasek, J. Lum, X. Duan and A. G. Brolo, Appl. Spectrosc., 2010, 64, 871–887 CrossRef CAS PubMed.
- Q. Matthews, A. Brolo, J. Lum, X. Duan and A. Jirasek, Phys. Med. Biol., 2011, 56, 19–38 CrossRef CAS PubMed.
- G. Borrego-Soto, R. Ortiz-Lopez and A. Rojas-Martinez, Genet. Mol. Biol., 2015, 38, 420–432 CrossRef CAS PubMed.
- B. Li, Z. Chen, L. Liu, Z. Huang, Z. Huang and S. Xie, Photodiagn. Photodyn. Ther., 2010, 7, 204–209 CrossRef PubMed.
- Q. Wu, T. Han, X. Sheng, N. Zhang and P. Wang, Biomed. Pharmacother., 2018, 108, 741–751 CrossRef CAS PubMed.
- Y. Chen, Z. Chen, Y. Su, D. Lin, M. Chen, S. Feng and C. Zhou, Cancer Cell Int., 2019, 19, 37 CrossRef PubMed.
- J. Biau, E. Chautard, P. Verrelle and M. Dutreix, Front. Oncol., 2019, 9, 1009 CrossRef PubMed.
- S. P. Jackson and J. Bartek, Nature, 2009, 461, 1071–1078 CrossRef CAS PubMed.
- J. M. Surmacki, Anal. Methods, 2020, 12, 383–391 RSC.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |