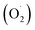Open Access Article
Open Access ArticleEffects of ozone treatment on SOD activity and genes in postharvest cantaloupe
Huijie Zhangab,
Xiaojun Zhangc,
Chenghu Dongb,
Na Zhangb,
Zhaojun Band,
Li Li e,
Jinze Yub,
Yunfeng Hu*a and
Cunkun Chen
e,
Jinze Yub,
Yunfeng Hu*a and
Cunkun Chen *b
*b
aCollege of Food Science and Engineering, Tianjin University of Science and Technology, Tianjin, China. E-mail: hu-yf@163.com
bNational Engineering Technology Research Center for Preservation of Agricultural Products, Key Laboratory of Postharvest Physiology and Storage of Agricultural Products, Ministry of Agriculture of China, Tianjin Key Laboratory of Postharvest Physiology and Storage of Agricultural Products, Tianjin, China. E-mail: chencunkun@126.com
cCollege of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
dZhejiang Provincial Key Laboratory of Chemical and Biological Processing Technology of Farm Products, Zhejiang University of Science and Technology, Hangzhou, China
eKey Laboratory for Agro-Products Postharvest Handling of Ministry of Agriculture, Zhejiang Key Laboratory for Agro-Food Processing, College of Biosystems Engineering and Food Science, Zhejiang University, Hangzhou, China
First published on 5th May 2020
Abstract
Ozone has been shown to play a positive role in the storage and preservation of agricultural products. However, there is little research on the cantaloupe preservation mechanism of ozone treatment (OT), especially the effect on superoxide dismutase (SOD) and the mechanism of scavenging superoxide anion  In this study, xizhoumi 25 was used as a typical cantaloupe material to detect
In this study, xizhoumi 25 was used as a typical cantaloupe material to detect 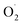 content, hydrogen peroxide (H2O2) and SOD enzyme activity in the pericarp and pulp, respectively, and transcriptomics and qRT-RCR were used for cantaloupe SOD family gene expression. The results showed that the rate of
content, hydrogen peroxide (H2O2) and SOD enzyme activity in the pericarp and pulp, respectively, and transcriptomics and qRT-RCR were used for cantaloupe SOD family gene expression. The results showed that the rate of 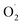 and H2O2 content were inhibited and SOD activity was higher in the treatment group compared with the control (CK) group in the pericarp and pulp; SOD was more active in the pericarp and was higher than that in the pulp. The transcription level of Cu/Zn-SOD, identified as the most abundant component of the cantaloupe SOD gene family, was promoted in the OT group, especially the key gene Cu/Zn-SOD-1. The expression level of the Fe-SOD gene was promoted in the pericarp but regulated in the pulp, while the expression of the Mn-SOD gene was down-regulated in the OT group in both pericarp and pulp. In addition, the results of qRT-PCR were consistent with the transcriptome results. Correlation analysis showed that OT not only enhanced the positive correlation between
and H2O2 content were inhibited and SOD activity was higher in the treatment group compared with the control (CK) group in the pericarp and pulp; SOD was more active in the pericarp and was higher than that in the pulp. The transcription level of Cu/Zn-SOD, identified as the most abundant component of the cantaloupe SOD gene family, was promoted in the OT group, especially the key gene Cu/Zn-SOD-1. The expression level of the Fe-SOD gene was promoted in the pericarp but regulated in the pulp, while the expression of the Mn-SOD gene was down-regulated in the OT group in both pericarp and pulp. In addition, the results of qRT-PCR were consistent with the transcriptome results. Correlation analysis showed that OT not only enhanced the positive correlation between 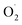 and H2O2 in the whole cantaloupe and the negative correlation between
and H2O2 in the whole cantaloupe and the negative correlation between 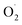 and SOD activity in the pericarp but also altered the correlation between SOD genes and
and SOD activity in the pericarp but also altered the correlation between SOD genes and 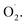 The mechanism of
The mechanism of 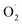 regulation in postharvest cantaloupe treated with ozone may be through stimulating the SOD activity and altering the expression of related genes in the pericarp and pulp.
regulation in postharvest cantaloupe treated with ozone may be through stimulating the SOD activity and altering the expression of related genes in the pericarp and pulp.
1. Introduction
Cantaloupe is a widely grown and typical climacteric fruit with is inclined to decay during ripening.1 Studies have shown that the senescence of cantaloupe is induced by producing a large number of reactive oxygen species (ROS).2,3Ozone has the characteristics of rapid decomposition and oxidation.4 In 2001, it was approved by the US Food and Drug Administration as an antimicrobial additive for direct contact with food.5 As an effective storage condition for fresh fruits and vegetables, ozone can activate the antioxidant defense mechanism in plant cells to metabolize ROS,6,7 which may be an important regulator of plant cells to scavenge the superoxide anion 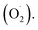 Recent research indicates that the content of
Recent research indicates that the content of 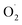 was lower in the ozone treatment group than in the control group, such as coriander,8 raspberries.9 Similarly, the activity of antioxidant system-related enzymes, especially superoxide dismutase (SOD), was also induced by ozone in tangerine10 and fresh-cut green peppers.11 Ozone has begun to receive widespread attention in activating the antioxidant enzyme system in postharvest fruits and vegetables.
was lower in the ozone treatment group than in the control group, such as coriander,8 raspberries.9 Similarly, the activity of antioxidant system-related enzymes, especially superoxide dismutase (SOD), was also induced by ozone in tangerine10 and fresh-cut green peppers.11 Ozone has begun to receive widespread attention in activating the antioxidant enzyme system in postharvest fruits and vegetables.
SOD is the first antioxidant defense line of cellular enzymes and a specific antioxidant enzyme that scavenges 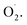 12 With the development of omics, the antioxidant scavenging mechanism of SOD has been further studied. Qian et al. found that antioxidative enzyme genes of cucumber fruit, included the SOD genes, play an important role in regulating ROS formation and clearance by transcriptomics analysis.13 Antioxidant capacity is enhanced by overexpression of Sedum alfredii Cu/Zn-SOD-related genes in transgenic Arabidopsis under Cd stress.14 Moreover, nitric oxide treatment delayed the growth rate of
12 With the development of omics, the antioxidant scavenging mechanism of SOD has been further studied. Qian et al. found that antioxidative enzyme genes of cucumber fruit, included the SOD genes, play an important role in regulating ROS formation and clearance by transcriptomics analysis.13 Antioxidant capacity is enhanced by overexpression of Sedum alfredii Cu/Zn-SOD-related genes in transgenic Arabidopsis under Cd stress.14 Moreover, nitric oxide treatment delayed the growth rate of 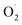 and H2O2 in banana, while SOD activity and SOD gene expression were significantly higher than the control.15 However, the mechanism of scavenging
and H2O2 in banana, while SOD activity and SOD gene expression were significantly higher than the control.15 However, the mechanism of scavenging 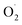 in cantaloupe treated with ozone was hardly investigated, especially the mechanism of the SOD genes in cantaloupe.
in cantaloupe treated with ozone was hardly investigated, especially the mechanism of the SOD genes in cantaloupe.
Therefore, in order to explore the scavenging mechanism of 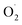 in ozone-treated cantaloupe, xizhoumi 25 was used as the experimental material and the changes in
in ozone-treated cantaloupe, xizhoumi 25 was used as the experimental material and the changes in 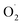 generation rate, SOD activity, and H2O2 content were studied as physical traits. And SOD genes were studied by comparing the transcriptome differences between the CK and OT groups in different storage time. At the same time, the differential expression of the cantaloupe SOD gene expression was verified by qRT-PCR. That study may lay the foundation for the future study of the potential impact of ozone on the SOD of the postharvest fruits and vegetables.
generation rate, SOD activity, and H2O2 content were studied as physical traits. And SOD genes were studied by comparing the transcriptome differences between the CK and OT groups in different storage time. At the same time, the differential expression of the cantaloupe SOD gene expression was verified by qRT-PCR. That study may lay the foundation for the future study of the potential impact of ozone on the SOD of the postharvest fruits and vegetables.
2. Materials and methods
2.1 Fruit materials
Cantaloupes (xizhoumi 25) were hand-harvested at the commercially mature stage from a cantaloupe production base located in Tulufan, Xinjiang Province, China on November 12, 2017. Undamaged cantaloupes of uniform size and firmness were selected and pre-cooled at 4 °C for 10 h. Every four cantaloupes were packed randomly into a special wrapping paper box with uniform holes and then 36 boxes of cantaloupes were transported to the laboratory by plane within 6 h.2.2 Storage condition and treatments
Cantaloupe was randomly divided into two groups of 18 boxes each, one group of fruits was a control (CK) group and the other group was ozone-treated (OT) group. After the cantaloupe was pre-cooled, the OT group was transported to an ozone refrigeration fumigation device (2 m × 1.5 m × 0.8 m, storage capacity 1200 L) for 30 d at 4 ± 1 °C with 75–80% relative humidity (RH), which was developed by the National Agricultural Products Preservation Engineering Research Center.16 The CK group was stored at the same fumigation device for 20 d (decayed before the 30th day). Each group was treated with different concentrations of ozone (0, 15 mg m−3) for 2 h at 0, 10, and 20 d. The specific method of processing as described in our previous article.16,17On the day of sampling, the whole cantaloupe was separated into two parts, the pericarp and the pulp, and the pericarp sample was obtained 1 cm from the outermost layer to the inner, the rest was pulp sample except seeds. Both the pericarp and pulp samples were cut into small pieces, frozen in liquid nitrogen, and stored at −80 °C. 6 cantaloupes were prepared at each time each group and every experiment had three times replicates.
2.3 Determinations of superoxide anion 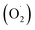 generation rate
generation rate
A 0.9 mL of pre-chilled phosphate buffer (0.01 M, pH 7.4) was added to 0.1 g of fresh sample, ground on icebox, and the homogenate was centrifuged for 20 minutes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C. The supernatant obtained by filtration was assayed for the rate of
000 × g at 4 °C. The supernatant obtained by filtration was assayed for the rate of 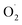 generation according to the Plant ELISA Kit (Shanghai Enzyme-linked Biotechnology Co., Ltd., China). The absorbance was read at a wavelength of 530 nm, and the generation rate of
generation according to the Plant ELISA Kit (Shanghai Enzyme-linked Biotechnology Co., Ltd., China). The absorbance was read at a wavelength of 530 nm, and the generation rate of 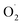 was calculated using a standard curve of the standard. The rate of
was calculated using a standard curve of the standard. The rate of 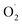 generation rate was expressed in U g−1.
generation rate was expressed in U g−1.
2.4 Determinations of hydrogen peroxide (H2O2) content
After grinding with liquid nitrogen, a 0.1 g fresh sample was accurately weighed with a balance and added 0.9 mL of physiological saline to prepare a homogenate under ice bath, and then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 minutes at 4 °C. The content of H2O2 in the sample was determined using the H2O2 kit (Nanjing Jiancheng Bioengineering Institute). According to the instructions, the absorbance was read at 405 nm by spectrophotometry to calculate the H2O2 content. On the basis of fresh weight, the content of H2O2 is expressed in mmol kg−1.
000 × g for 10 minutes at 4 °C. The content of H2O2 in the sample was determined using the H2O2 kit (Nanjing Jiancheng Bioengineering Institute). According to the instructions, the absorbance was read at 405 nm by spectrophotometry to calculate the H2O2 content. On the basis of fresh weight, the content of H2O2 is expressed in mmol kg−1.
2.5 Determinations of superoxide dismutase (SOD) activity
The activity of total SOD (EC 1.15.1.1) was determined using a SOD assay kit (WST-1 method) purchased from the Nanjing Jiancheng Bioengineering Institute (WST-1 method). A 0.1 g of fresh sample was ground in liquid nitrogen, homogenized in a 0.9 mL ice bath by adding 10 mM phosphate buffer (pH 7.4), and then the mixture was centrifuged at 8000 × g for 10 minutes at 4 °C. SOD activity was expressed as U g−1, where U = 0.01ΔOD450 min−1. And one unit (U) of SOD activity is defined as the amount of enzyme corresponding to a SOD inhibition rate of 50%.2.6 RNA extraction and mRNA library preparation
Total RNA was extracted from ball-milled samples with RNAiso™ Plus (Takara, 9109) combined with Fruit-mate™ for RNA Purification (Takara, 9192) according to the manufacturer's instructions. Then 500 ng total RNA was used for mRNA isolation using NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, 7490) and mRNA library was constructed using NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, E7530) according to the instruction manual. Briefly, library preparation orderly includes RNA fragmentation, reverse transcription using random primers, second-strand cDNA synthesis, end repair, dA-tailing, adapter ligation, U excision and PCR enrichment. The mRNA libraries were sequenced on Illumina HiSeq X Ten platform with 150 bp pair-end reads.2.7 MicroRNA library preparation
A 1 μg of total RNA was used for miRNA library construction using NEBNext Small RNA Library Prep Set for Illumina (NEB, E7330) according to the instruction manual. Briefly, 3′ SR adaptor was first ligated followed by the hybridization of the reverse transcription primer. The reverse transcription was performed after the ligation of 5′ SR adaptor and then followed by 12 cycles of PCR enrichments (94 °C 30 s; 94 °C 15 s, 62 °C 30 s, 70 °C 15 s for 12 cycles; 70 °C 5 min). After amplification, 140 bp DNA fragments were size-selected and recovered after resolving on the 6% PAGE gel. The miRNA libraries were then sequenced on Illumina HiSeq2500 platform with 50 bp single-end read.2.8 Real-time quantitative reverse transcription PCR (qRT-PCR)
The 8 SOD genes were verified using qRT-PCR. Primer 3.0 was used to design specific primers, which were shown in Table 1. Total RNA in fruits was extracted from three parallel samples using the MiniBEST Plant RNA Extraction Kit (TAKARA). Real-time relative quantitative qRT-PCR detection of candidate genes was performed on the ABI Step One Plus Real-Time PCR system, and GAPDH rRNA was used as an internal control and analyzed by the comparative Ct method. The green two-step qRT-PCR Super Mix (Thermo) was used in the three parallel experiments according to the manufacturer's instructions. Equation 2−ΔΔCt is used to calculate relative transcription levels.| Accession number | Primer F (5′–3′) | Primer R (5′–3′) |
|---|---|---|
| MELO3C004342 | AGGGCCACATTTCAACCC | GACCATTATCACCGGCAACC |
| MELO3C008809 | TTGGGGAACATTTATGCAGGT | GAACAATGGCCAACCGAATC |
| MELO3C014007 | GGTGCAACGCCTGTGAAGAT | GGAGAGGGGGCAAATTTTGT |
| MELO3C015351 | CGGAAGACATTGGAGGTTCAC | CTGGGCAGCATTGTTGAACT |
| MELO3C015374 | TCGACATGCAGCCATTGGT | AACATGTCCGCGTCTGTGTC |
| MELO3C017624 | TGCACGGCACCATATAATTTC | AACATGTCCGCGTCTGTGTC |
| MELO3C020487 | CGCTCTTGAGCCCGTCATCA | GATGGCCTCGTGAAGTTGCT |
| MELO3C026955 | GACGTTGAAGGCGTTGTCAC | TTGATTCAGACCCTCCTCAAGT |
2.9 Statistical analysis
A completely randomized experimental design was used. All statistics were analyzed using IBM SPSS Statistics ver. 22 (SPSS Inc, Armonk, New York, USA) software. Data were analyzed by LSD test for one-way ANOVA with a 5% significance level (*P < 0.05), a 1% significance level (**P < 0.01), and a 0.1% significance level (***P < 0.001). Error bars represent the standard error of three replicate experimental data.3. Results and discussion
3.1 Effect of OT on SOD activity, 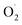 production rate, and H2O2 content
production rate, and H2O2 content
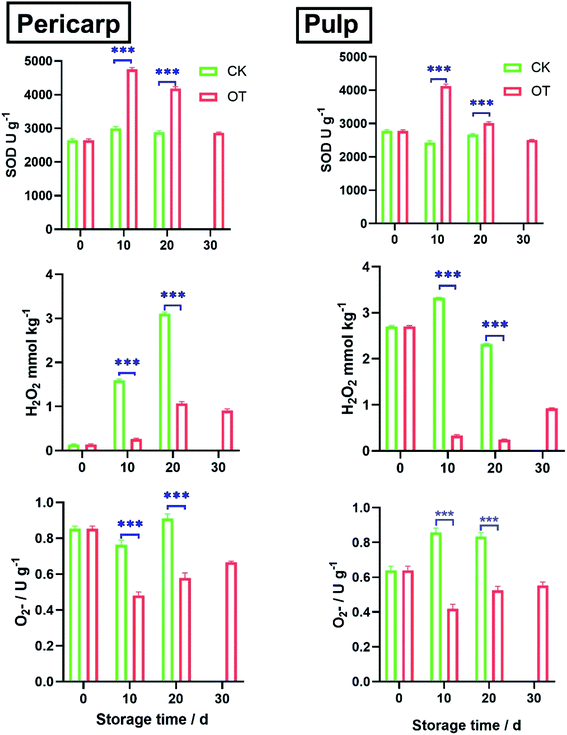
As shown in Fig. 1, the formation of 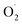 in the OT group decreased rapidly in the first 10 d, especially in the pericarp. After 30 d, the superoxide anion production rate of OT was 40% lower than that of CK in pericarp and 27% lower than that of CK in the pulp. In general, the rate of superoxide anion formation in the CK group was always higher than that in the treatment group during the whole storage period.
in the OT group decreased rapidly in the first 10 d, especially in the pericarp. After 30 d, the superoxide anion production rate of OT was 40% lower than that of CK in pericarp and 27% lower than that of CK in the pulp. In general, the rate of superoxide anion formation in the CK group was always higher than that in the treatment group during the whole storage period.
In addition, it was observed that the trend of SOD activity was opposite to that of 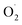 both in the pericarp and pulp. The change of SOD activity of CK group was relatively flat, while it increased first and then decreased in the OT group. The SOD activity of OT group was significantly higher than that of the CK, especially on the 10th day after storage, and the SOD activity of the treatment group was more than 50% higher than that of the CK group, which may due to ozone induces the activity of the SOD in cantaloupe and reduces the formation of
both in the pericarp and pulp. The change of SOD activity of CK group was relatively flat, while it increased first and then decreased in the OT group. The SOD activity of OT group was significantly higher than that of the CK, especially on the 10th day after storage, and the SOD activity of the treatment group was more than 50% higher than that of the CK group, which may due to ozone induces the activity of the SOD in cantaloupe and reduces the formation of 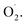 Increasing the activity of SOD can effectively remove superoxide anion radicals from ROS metabolism.18
Increasing the activity of SOD can effectively remove superoxide anion radicals from ROS metabolism.18
H2O2 is converted from superoxide anion catalyzed by SODs.19 The H2O2 content of cantaloupe in the OT group was always lower than that in the CK group, especially decreased sharply on the 10th day of storage in the OT group, which 39% and 20% lower than that of the CK group in pericarp and pulp, respectively.
H2O2 and 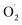 were key substances of active oxygen, which are prone to damage and cell death in plants.20 However, ROS can also act as signal molecules and pass to the entire plant in response to abiotic stresses, allowing plants to produce different antioxidant enzymes.21,22 SOD is a key enzyme that converts superoxide anion to H2O2, while high SOD activity can reduce superoxide radicals that plants overproduce under stress.23 In our study, OT showed a higher free radical scavenging activity of SOD compared to the CK, and a lower rate of
were key substances of active oxygen, which are prone to damage and cell death in plants.20 However, ROS can also act as signal molecules and pass to the entire plant in response to abiotic stresses, allowing plants to produce different antioxidant enzymes.21,22 SOD is a key enzyme that converts superoxide anion to H2O2, while high SOD activity can reduce superoxide radicals that plants overproduce under stress.23 In our study, OT showed a higher free radical scavenging activity of SOD compared to the CK, and a lower rate of 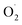 generation and H2O2 content, which was consistent with M. V. Rao's research.6 Ozone also induces an increase in plant SOD activity in oxidative stress.24 In addition, some research and review articles concluded that an appropriate concentration of ozone can effectively induce the production of an antioxidant defense system. Alothman et al. found that the OT group has high antioxidant capacity by comparing the Oxygen Radical Absorption Capacity (ORAC) and Hydroxyl Radical Antioxidant Capacity (HORAC) of Salvia officinalis.25,26
generation and H2O2 content, which was consistent with M. V. Rao's research.6 Ozone also induces an increase in plant SOD activity in oxidative stress.24 In addition, some research and review articles concluded that an appropriate concentration of ozone can effectively induce the production of an antioxidant defense system. Alothman et al. found that the OT group has high antioxidant capacity by comparing the Oxygen Radical Absorption Capacity (ORAC) and Hydroxyl Radical Antioxidant Capacity (HORAC) of Salvia officinalis.25,26
3.2 Identification and sequence analysis of SOD gene family members
SOD in plants can be divided into Cu/Zn-SOD, Mn-SOD and Fe-SOD according to the metal prosthetic content.27,28 Three types of SOD genes were screened from the pericarp and pulp of cantaloupe by genome-wide sequencing. 8 SOD genes were screened out by screening and deleting redundant RNA sequences using cut adapt (v1.10), which were MELO3C015374, MELO3C014007, MELO3C026955, MELO3C004342, MELO3C008809, MELO3C015351, MELO3C017624, MELO3C020487. The type of SOD gene was analyzed using the Cucurbit Genomics Database (http://cucurbitgenomics.org/), five genes were recognized as Cu/Zn-SOD gene, one was Mn-SOD gene and two was Fe-SOD genes, and which were assumed to be named Cu/Zn-SOD-1, Cu/Zn-SOD-2, Cu/ZnSOD-3, Cu/Zn-SOD-4, Cu/Zn-SOD-5, Fe-SOD-1, Fe-SOD-2, Mn-SOD. However, the unknown gene sequence (MELO3C017624, MELO3C020487) was more than 65% similar to the Fe-SOD gene in Arabidopsis. SOD genes of cantaloupe were mainly distributed on seven chromosomes, Cu/Zn-SOD-1 and Fe-SOD-1 on chr2, and the rest of which on chr5, chr6, chr7, chr8, chr11 and chr12. A phylogenetic tree was constructed using the cantaloupe SOD gene sequence (Fig. 2). The phylogenetic relationship of SOD gene indicated that the tree was divided into three main branches, the Fe-SOD-1/2 were clustered into one branch; Mn-SOD and Cu/Zn-SOD-2 were one branch; Cu/Zn-SOD-1/3/4/5 were another branch.3.3 Expression of SOD genes
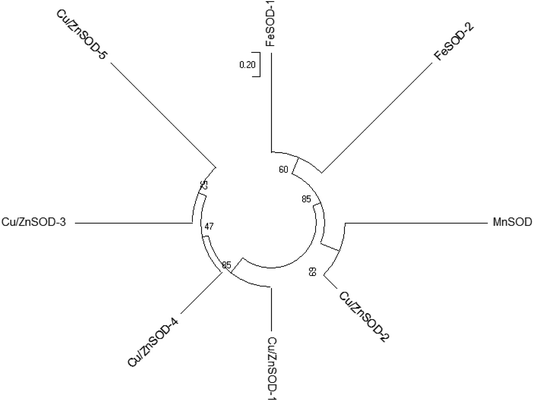
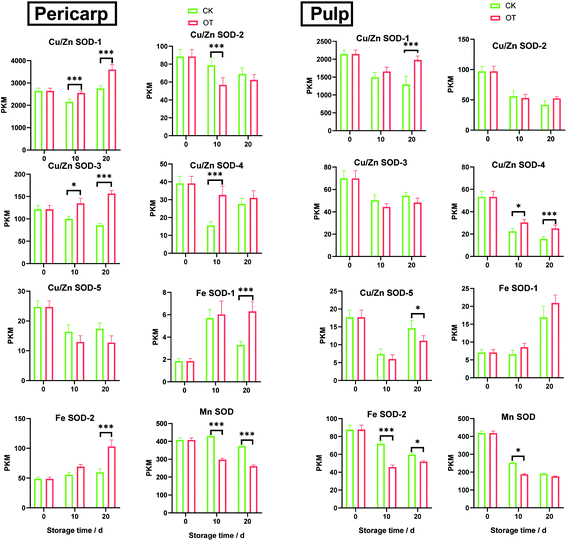
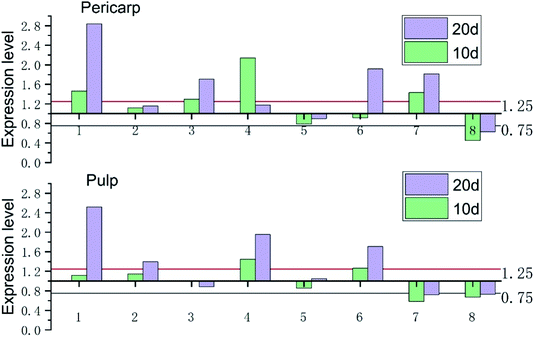
As shown in the Fig. 3, the expression of Cu/Zn-SOD gene in pericarp and pulp accounted for the most of all SOD genes in both OT group and CK group, of which the transcription level of Cu/Zn-SOD-1 was the highest, accounting for 78% and 76% of the pericarp and pulp of the CK group, and 82% and 79% of the pericarp and pulp in OT group, respectively. Cu/Zn-SOD may be the most important and abundant SOD gene in cantaloupe caused by the number and expression of Cu/Zn-SOD gene accounted for the largest proportion. Huan et al. also found that PpaCu/Zn-SODs were the most abundant member of the PpaSOD gene family and expressed stably during fruit development and maturation.29 Wang's construction of a rootless phylogenetic tree indicates that the SODs of Pyrus bretschneideri are mainly composed of Cu/Zn-SODs.30The SOD genes of xizhoumi 25 were detected by transcriptome sequencing in CK and OT groups. The change of SOD genes expression in the pericarp and pulp of cantaloupe during storage was shown in Fig. 4. The SOD genes of the whole cantaloupe without ozone-treated group were down-regulated except for Fe-SOD-1/2 in the pericarp and Fe-SOD-1 in the pulp. Simultaneously, the abundance of SOD genes expression in the pulp of the OT group during storage time was also less than the initial time except for Fe-SOD-1. However, the expression trend of SOD genes in the pericarp of the OT group was significantly different from that of the CK group, the up-regulated genes in the OT group included the same up-regulated genes as those in the CK group as well as Cu/Zn-SOD-1/3. Interestingly, the transcription level of SOD genes in pericarp during storage was higher than that in the pulp, except for Cu/Zn-SOD-4 and Fe-SOD-1/2 in CK group, Fe-SOD-1 in OT group. In addition, it was observed that the gene expression of Cu/Zn-SOD-1/3/4, Fe-SOD-1/2 in the pericarp of the OT group and the expression of Cu/Zn-SOD-1/4 and Fe-SOD-1 in the pulp were higher than that of the CK group. But expression abundance of the Mn-SOD gene in the CK group was higher than that in the OT group from beginning to end, especially in the pericarp. The expression level of Mn-SOD in the CK group was 1.4 times higher than that in the OT group. SOD genes with different metal cofactors have different expression levels in stress response.
The reason for these results may be that different types of SOD genes have different functions according to their subcellular localization.31,32 And Huan et al. concluded the functions of different SOD genes were based on the types and location of cells.29 Under dark conditions, mitochondria are the main source of synthetic ATP, which promotes energy metabolism in plants.33 And the expression of the Mn-SOD gene is related to mitochondria respiratory activity,34 which can inhibit the excessive accumulation of 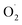 and protect mitochondria from damage.35 Although the expression of the Mn-SOD gene in the CK group was higher than that in the OT group, the trend of Mn-SOD gene expression was of down-regulation in the CK group, which may be caused by inhibition of respiratory rate and low
and protect mitochondria from damage.35 Although the expression of the Mn-SOD gene in the CK group was higher than that in the OT group, the trend of Mn-SOD gene expression was of down-regulation in the CK group, which may be caused by inhibition of respiratory rate and low 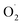 content under low temperature.29 And many studies have shown that the respiration rate of many fruits was inhibited by ozone treatment to reduce mitochondrial energy metabolism and maintain fruit nutrition.11,36,37 Ozone and low temperature may have synergistic effects to inhibit the gene expression of Mn-SOD.
content under low temperature.29 And many studies have shown that the respiration rate of many fruits was inhibited by ozone treatment to reduce mitochondrial energy metabolism and maintain fruit nutrition.11,36,37 Ozone and low temperature may have synergistic effects to inhibit the gene expression of Mn-SOD.
Most of the Fe-SOD genes were up-regulated in both CK and OT groups. Some studies reported that low temperature stimulated the up-regulation of the Fe-SOD gene, which is consistent with our results.38 Fe-SOD, located in the chloroplast mostly, is closely related to photosynthesis,39 which protects the chloroplast from oxidative damage.40 Since the pericarp contains more chloroplasts than the pulp,41 the Fe-SOD expression of the pericarp is higher than that of the pulp in cantaloupe. Guo et al. found that ferric reducing/antioxidant power (FRAP) of the pericarp was higher than that of the pulp by measuring 28 kinds of fruits,42 which was the same as our research results. Interestingly, OT resulted in higher levels of Cu/Zn-SOD-4 and Fe-SOD-2 gene expression in the pericarp than in the pulp, indicating that OT also increased the expression of the Fe-SOD gene in the pericarp.
The gene for Cu/Zn-SOD is present in the cytoplasm and is strongly expressed in most plants, but there were few studies related to the cellular processes involved in Cu/Zn-SOD.43 Cu/Zn-SOD was considered to be the most active SOD substance in tobacco extracts.44 In this study, as the most abundant member of the cantaloupe SOD family, Cu/Zn-SOD is also considered to be key genes for anti-oxidation of cantaloupe, especially Cu/Zn-SOD-1, which contributed more than 70% of all SOD genes. The expression level of Cu/Zn-SOD-1 in the OT group was significantly higher than that of the CK group, especially in the later stage of storage, which was 1.3 times higher than that of the CK group. This indicated that the expression of the Cu/Zn-SOD-1 gene was stimulated treated with ozone in cantaloupe. Y. K. Sharma et al. found that ozone can increase the expression level of Cu/Zn-SOD mRNA in Arabidopsis thaliana.45 In addition, the high expression of Cu/Zn-SOD gene was induced by glutathione molecules,46 and GSH interacts with cell extracts of Arabidopsis can activate Cu/Zn-SOD.47 The content of glutathione was stimulated in Passiflora edulis Sims when exposed to ozone,48 and the accumulation of total glutathione was promoted in pea plants of Little Marvel in the ozone treatment group.49 Ozone may induce the SOD activity and the expression of Cu/Zn-SOD gene by stimulating the synthesis of glutathione to improve the efficiency of scavenging superoxide anion, which requires further verification.
3.4 qRT-PCR verification of the SOD gene
As shown in Fig. 5, the expression level of the SOD genes was further verified using qRT-PCR. The results were consistent with the patterns of change measured by transcriptomics and the folds of Cu/Zn-SOD-1 and Mn-SOD expression were more pronounced. This suggested that Cu/Zn-SOD-1 and Mn-SOD may be key genes for OT to regulate SOD expression in cantaloupe.3.5 Correlation between physicochemical quality and SOD gene
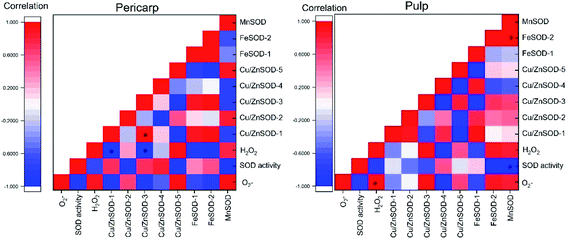
A visual correlation coefficient matrix is used to find correlations between variables. The correlation matrix is made based on the past 3 software, and the correlation coefficients of each index are displayed by a thermodynamic chart using origin software. Correlation analysis was performed on the difference between the OT group and the CK group by linear correlation analysis. As shown in Fig. 6, the range of correlation coefficients between transcription levels and biochemical parameters ranged from 1.0 (maximum positive correlation) to −1.0 (maximum negative correlation), with 0 indicating no correlation. In the whole cantaloupe, the superoxide anion is positively correlated with hydrogen peroxide. However,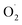 showed a strong negative correlation with SOD activity in the pericarp but was not strong in the pulp. There was a significant negative correlation between SOD activity and Mn-SOD in the pericarp, which may due to the expression of the Mn-SOD gene in the pericarp of the OT group was lower than that of the CK group. The correlation between SOD activity and Fe-SOD-1/2 was significantly negative in the pulp. There was a positive correlation between Fe-SOD-1 and Fe-SOD-2 in the pericarp and no correlation in the pulp, which may be because the chloroplast activity in the pericarp is higher than that in the pulp. As a potential key gene, Cu/Zn-SOD-1 was positively correlated with superoxide anion, Cu/Zn-SOD-3 and Fe-SOD-1/2 in the pericarp, and negatively correlated with Cu/Zn-SOD-5. However, Cu/Zn-SOD-1 was only positively correlated with Cu/Zn-SOD-2 and Fe-SOD-1 in the pulp. In summary, from the difference between the indicators of the OT group and the CK group, the correlation between each indicator of the pericarp was stronger than that of the pulp, and SOD activity and gene expression in cantaloupe were induced by ozone to inhibit superoxide anion.
showed a strong negative correlation with SOD activity in the pericarp but was not strong in the pulp. There was a significant negative correlation between SOD activity and Mn-SOD in the pericarp, which may due to the expression of the Mn-SOD gene in the pericarp of the OT group was lower than that of the CK group. The correlation between SOD activity and Fe-SOD-1/2 was significantly negative in the pulp. There was a positive correlation between Fe-SOD-1 and Fe-SOD-2 in the pericarp and no correlation in the pulp, which may be because the chloroplast activity in the pericarp is higher than that in the pulp. As a potential key gene, Cu/Zn-SOD-1 was positively correlated with superoxide anion, Cu/Zn-SOD-3 and Fe-SOD-1/2 in the pericarp, and negatively correlated with Cu/Zn-SOD-5. However, Cu/Zn-SOD-1 was only positively correlated with Cu/Zn-SOD-2 and Fe-SOD-1 in the pulp. In summary, from the difference between the indicators of the OT group and the CK group, the correlation between each indicator of the pericarp was stronger than that of the pulp, and SOD activity and gene expression in cantaloupe were induced by ozone to inhibit superoxide anion.
4. Conclusion
The study of the scavenging mechanism of superoxide anion by SOD in cantaloupe treated with ozone was little. In this study, the treatment of intermittent ozone was beneficial to remove superoxide anions from cantaloupe after harvest. Ozone-induced oxidative stress directly acted on the pericarp, which may be the reason for the higher SOD activity in the peel than in the pulp. The results of ozone effect on the SOD transcriptome showed that ozone promoted the transcription level of Cu/Zn-SOD, and which was consisted of the results of qRT-PCR. And Cu/Zn-SOD-1 may be the most important member of SOD. The expression level of Fe-SOD is promoted in the pericarp but is inhibited in the pulp, which may be related to the localization of Fe-SOD in the cell. The expression of the Mn-SOD gene located in mitochondria tended to be down-regulated in both the ozone-treated group and CK. The visual correlation matrix indicates that the SOD clearance mechanism in the ozone-treated pericarp has a higher correlation than in the pulp.Conflicts of interest
The authors declare there is no competing financial interest.Acknowledgements
This work was supported by Special Fund for National Natural Science Foundation of China (31501547); Innovation Team of Tianjin Forestry & Pomology Research System (ITTFPRS2018009, ITTFPRS2018010); Innovative research and experimental projects for young researchers (202009, 17011); National Key R&D Program of China (2016YFD0400903-05, 2018YFF0213605-2) and Tianjin Application Basics and Frontier Technology Research Program (15JCYBJC51300).References
- A. L. Amaro, N. D. Spadafora, M. J. Pereira, R. Dhorajiwala, R. J. Herbert, C. T. Müller, H. J. Rogers and M. Pintado, Food Chem., 2018, 241, 222–231 CrossRef CAS PubMed.
- S. Tian, G. Qin and B. Li, Plant Mol. Biol., 2013, 82, 593–602 CrossRef CAS PubMed.
- K. Mondal, N. Sharma, S. Malhotra, K. Dhawan and R. Singh, Biol. Plant., 2004, 48, 49–53 CrossRef CAS.
- R. Pandiselvam, S. Subhashini, E. Banuu Priya, A. Kothakota, S. Ramesh and S. Shahir, Ozone: Sci. Eng., 2019, 41, 17–34 CrossRef CAS.
- US FDA, Fed. Reg., 2001, 66, 33829–33830 Search PubMed.
- M. V. Rao, G. Paliyath and D. P. Ormrod, Plant Physiol., 1996, 110, 125–136 CrossRef CAS PubMed.
- R. G. Alscher, N. Erturk and L. S. Heath, J. Exp. Bot., 2002, 53, 1331–1341 CrossRef CAS PubMed.
- D. Xu, M. Shi, B. Jia, Z. Yan, L. Gao, W. Guan, Q. Wang and J. Zuo, J. Food Process. Preserv., 2019, e14020 Search PubMed.
- T. Piechowiak and M. Balawejder, Food Chem., 2019, 298, 125093 CrossRef CAS PubMed.
- P. Boonkorn, H. Gemma, S. Sugaya, S. Setha, J. Uthaibutra and K. Whangchai, Postharvest Biol. Technol., 2012, 67, 25–28 CrossRef CAS.
- J. Chen, Y. Hu, J. Wang, H. Hu and H. Cui, J. Food Process. Preserv., 2016, 40, 1145–1150 CrossRef CAS.
- O. Ighodaro and O. Akinloye, Alexandria J. Med., 2018, 54, 287–293 CrossRef.
- C.-L. Qian, Y.-Y. Zhao, H.-B. Mi, X.-H. Chen, L.-J. Guo and L.-C. Mao, Biol. Plant., 2012, 56, 793–797 CrossRef CAS.
- Z. Li, X. Han, X. Song, Y. Zhang, J. Jiang, Q. Han, M. Liu, G. Qiao and R. Zhuo, Front. Plant Sci, 2017, 8, 1010 CrossRef PubMed.
- B. Wu, Q. Guo, Q. Li, Y. Ha, X. Li and W. Chen, Postharvest Biol. Technol., 2014, 92, 157–163 CrossRef CAS.
- C. Chen, X. Zhang, H. Zhang, Z. Ban, L. Li, C. Dong, H. Ji and W. Xue, RSC Adv., 2019, 9, 676–689 RSC.
- C. Chen, H. Zhang, X. Zhang, C. Dong, W. Xue and W. Xu, Postharvest Biol. Technol., 2020, 163, 111124 CrossRef CAS.
- Y. Luo, Y. Zhou and K. Zeng, Crop Prot., 2013, 53, 96–102 CrossRef.
- J. M. Tepperman and P. Dunsmuir, Plant Mol. Biol., 1990, 14, 501–511 CrossRef CAS PubMed.
- J. Dat, S. Vandenabeele, E. Vranová, M. Van Montagu, D. Inzé and F. Van Breusegem, Cell. Mol. Life Sci., 2000, 57, 779–795 CrossRef CAS PubMed.
- R. Mittler, S. Vanderauwera, N. Suzuki, G. Miller, V. B. Tognetti, K. Vandepoele, M. Gollery, V. Shulaev and F. Van Breusegem, Trends Plant Sci., 2011, 16, 300–309 CrossRef CAS PubMed.
- X.-J. Xia, Y.-H. Zhou, K. Shi, J. Zhou, C. H. Foyer and J.-Q. Yu, J. Exp. Bot., 2015, 66, 2839–2856 CrossRef CAS PubMed.
- C. Bowler, M. v. Montagu and D. Inze, Annu. Rev. Plant Biol., 1992, 43, 83–116 CrossRef CAS.
- V. Calatayud, F. Marco, J. Cerveró, G. Sánchez-Peña and M. J. Sanz, Environ. Pollut., 2010, 158, 3580–3587 CrossRef CAS PubMed.
- A. Marchica, G. Lorenzini, R. Papini, R. Bernardi, C. Nali and E. Pellegrini, Sci. Total Environ., 2019, 657, 568–576 CrossRef CAS PubMed.
- M. Alothman, B. Kaur, A. Fazilah, R. Bhat and A. A. Karim, Innovative Food Sci. Emerging Technol., 2010, 11, 666–671 CrossRef CAS.
- R. Mittler, Trends Plant Sci., 2002, 7, 405–410 CrossRef CAS PubMed.
- W. Wang, M. Xia, J. Chen, R. Yuan, F. Deng and F. Shen, Biochemistry (Moscow), 2016, 81, 465–480 CrossRef CAS PubMed.
- C. Huan, L. Jiang, X. An, M. Yu, Y. Xu, R. Ma and Z. Yu, Plant Physiol. Biochem., 2016, 104, 294–303 CrossRef CAS PubMed.
- L. Wang, L. Wang, Z. Zhang, M. Ma, R. Wang, M. Qian and S. Zhang, Postharvest Biol. Technol., 2018, 143, 68–77 CrossRef CAS.
- F. J. Corpas, A. Fernandez-Ocana, A. Carreras, R. Valderrama, F. Luque, F. J. Esteban, M. Rodríguez-Serrano, M. Chaki, J. R. Pedrajas and L. M. Sandalio, Plant Cell Physiol., 2006, 47, 984–994 CrossRef CAS PubMed.
- X. Feng, Z. Lai, Y. Lin, G. Lai and C. Lian, BMC Genomics, 2015, 16, 823 CrossRef PubMed.
- A. C. Asensio, M. Gil-Monreal, L. Pires, Y. Gogorcena, P. M. Aparicio-Tejo and J. F. Moran, J. Plant Physiol., 2012, 169, 1253–1260 CrossRef CAS PubMed.
- W. Van Camp, D. Hérouart, H. Willekens, H. Takahashi, K. Saito, M. Van Montagu and D. Inzé, Plant Physiol., 1996, 112, 525–535 CrossRef CAS PubMed.
- Y. Wang, Y. Ying, J. Chen and X. Wang, Plant Sci., 2004, 167, 671–677 CrossRef CAS.
- Q. Han, H. Gao, H. Chen, X. Fang and W. Wu, Food Chem., 2017, 221, 1947–1953 CrossRef CAS PubMed.
- M. Z. Islam, M. A. Mele, K. A. Hussein and H.-M. Kang, J. Biotech, 2018, 13, 51–55 CAS.
- F. Van Breusegem, M. Van Montagu and D. Inzé, Outlook Agric., 1998, 27, 115–124 CrossRef.
- A. Ribera-Fonseca, C. Inostroza-Blancheteau, P. Cartes, Z. Rengel and M. Mora, Plant Physiol. Biochem., 2013, 73, 77–82 CrossRef CAS PubMed.
- W. Van Camp, K. Capiau, M. Van Montagu, D. Inze and L. Slooten, Plant Physiol., 1996, 112, 1703–1714 CrossRef CAS PubMed.
- M. Roca and M. I. Mínguez-Mosquera, J. Agric. Food Chem., 2001, 49, 832–839 CrossRef CAS PubMed.
- C. Guo, J. Yang, J. Wei, Y. Li, J. Xu and Y. Jiang, Nutr. Res., 2003, 23, 1719–1726 CrossRef CAS.
- W. Van Camp, D. Inzé and M. Van Montagu, Free Radicals Biol. Med., 1997, 23, 515–520 CrossRef CAS PubMed.
- L. Slooten, K. Capiau, W. Van Camp, M. Van Montagu, C. Sybesma and D. Inze, Plant Physiol., 1995, 107, 737–750 CrossRef CAS PubMed.
- Y. K. Sharma and K. R. Davis, Plant Physiol., 1994, 105, 1089–1096 CrossRef CAS PubMed.
- D. Herouart, M. Van Montagu and D. Inze, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3108–3112 CrossRef CAS PubMed.
- C.-H. Huang, W.-Y. Kuo, C. Weiss and T.-L. Jinn, Plant Physiol., 2012, 158, 737–746 CrossRef CAS PubMed.
- F. F. Fernandes, M. P. Esposito, M. R. G. da Silva Engela, P. Cardoso-Gustavson, C. M. Furlan, Y. Hoshika, E. Carrari, G. Magni, M. Domingos and E. Paoletti, Sci. Total Environ., 2019, 656, 1091–1101 CrossRef CAS PubMed.
- I. Hassan, N. Haiba, R. Badr, J. Basahi, T. Almeelbi, I. Ismail and W. Taia, Pak. J. Bot., 2017, 49, 47–55 CAS.
| This journal is © The Royal Society of Chemistry 2020 |