Open Access Article
Open Access ArticleAntibacterial activity evaluation and mode of action study of novel thiazole-quinolinium derivatives†
Ying Li‡
a,
Ning Sun‡ *abc,
Hooi-Leng Serad,
Wei Longa,
Yanan Lie,
Cuicui Chena,
Boxin Zhenga,
Xuanhe Huanga,
Zhihua Liu*b and
Yu-Jing Lu*a
*abc,
Hooi-Leng Serad,
Wei Longa,
Yanan Lie,
Cuicui Chena,
Boxin Zhenga,
Xuanhe Huanga,
Zhihua Liu*b and
Yu-Jing Lu*a
aSchool of Biomedical and Pharmaceutical Sciences, Guangdong University of Technology, Guangzhou 510006, PR China. E-mail: luyj@gdut.edu.cn
bThe State Key Laboratory of Chemical Biology and Drug Discovery, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hung Hom, Kowloon, Hong Kong, China. E-mail: ning.sun@connect.polyu.hk; liuzhihualzh@hotmail.com
cThe Fifth Affiliated Hospital of Guangzhou Medical University, Guangzhou 510700, PR China
dNovel Bacteria and Drug Discovery (NBDD) Research Group, Microbiome and Bioresource Research Strength, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, 47500 Bandar Sunway, Selangor Darul Ehsan, Malaysia
eDepartment of Pharmacy, The Fifth Affiliated Hospital of Sun Yat-sen University, Zhuhai, 519000, P. R. China
First published on 16th April 2020
Abstract
New antimicrobial agents are urgently needed to address the emergence of multi-drug resistant organisms, especially those active compounds with new mechanisms of action. Based on the molecular structures of the FtsZ inhibitors reported, a variety of thiazole-quinolinium derivatives with aliphatic amino and/or styrene substituents were synthesized from benzothiazolidine derivatives. In the present study, to further explore the antibacterial potential of thiazole-quinolinium derivatives, several Gram-positive and Gram-negative bacteria were treated with the newly modified compounds and the biological effects were studied in detail in order to understand the bactericidal action of the compounds. Our findings reveal that some of these derivatives possess good potent bactericidal activity as they can inhibit Gram-positive methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus and also some Gram-negative organisms and NDM-1 Escherichia coli. Furthermore, compounds 4a1–4a4 and 4b1–4b4 altered the morphology of bacterial cells and the cells displayed a more-elongated shape compared to the untreated cells. Biochemical assays showed that 4a4 and 4b4 stimulate FtsZ polymerization in bacterial cells, which eventually disrupts its dynamic assembly and Z-ring formation. The inhibition of this crucial step in bacterial cell division could potentially represent their main mechanism of antibacterial activity. Cytotoxicity assay and hemolysis assay suggested that 4a4 and 4b4 possess low cytotoxicity. In summary, these results further highlight the importance of 4a4 and 4b4 that could be developed as potent and effective bacteriostatic agents against multi-drug resistant bacteria.
1. Introduction
Since the discovery of penicillin in the 1930s, antibiotics have been the most common choice and effective practice to treat bacterial infection and related diseases. Even so, the overuse and mismanagement of antibiotics have resulted in the antimicrobial resistance crisis. For instance, methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE) and NDM-1 Escherichia coli pose great threat to human health as these strains are resistant to most of the conventional antibiotics. In fact, most of them carry antibiotic resistance genes that make them survive against antibiotics (e.g. enzymatic degradation).1,2 Nowadays, the emergence of “superbugs” is considered as one of the greatest scientific challenges worldwide and urges the discovery of new antibacterial agents with novel mechanisms of actions.3–5One of the classical target pathways for antibacterial agents is cell division process.6 Bacterial cell division is mediated by a highly dynamic, large protein complex divisome, which involves a time-dependent assembly of specific cell division proteins. Divisome formation is orchestrated by the essential tubulin homolog FtsZ (filamenting temperature sensitive mutant).7 At the initiation of cell division, FtsZ migrates to the division site and assembles into protofilaments in a GTP dependent manner. After that, a ring-like structure, which was called the Z-ring, forms at the prospective division site. Towards the end of the cell division process, Z-ring constricts to induce septum formation before the completion of cell division.8 FtsZ shows high conservation, structural similarity and functional importance among bacteria and thus it presents as one of the imperative targets for the development of new antibiotic agents.
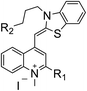
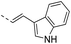
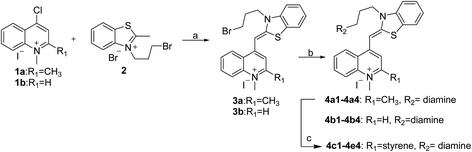
In recent years, some compounds with inhibitory action on bacterial cell division are potential candidates to be developed as new antibiotics and they have also gained much attention from the scientific community and drug companies.8 FtsZ inhibitors are capable of disturbing the dynamic polymerization of FtsZ and Z-ring formation, which ultimately impede bacterial cell division.6,9–12 Among the known FtsZ inhibitors, taxane derivative7 and SRI-3072 (ref. 13) were known to inhibit FtsZ polymerization effectively, particularly against drug-resistant Mycobacterium tuberculosis. Besides, Zantrins inhibited GTPase activity of FtsZ and perturbed cytokinetic ring assembly.6 PC190723 displayed potent bactericidal activity with good selectivity against Staphylococci, as shown in the in vitro screening assay using multi-drug resistant Staphylococcus aureus and a rodent model with MRSA infection.11 Recently, we have reported some FtsZ inhibitors with good antibacterial activity including berberine derivatives,14 tiplaxtinin,15 and quinolinium derivative.4 In addition, some macrolide derivatives containing quinoline or benzothiazole scaffolds,16 quinoline and quinolone derivatives17 were found exhibiting good antibacterial effects. We analyzed the molecular structures of these active compounds and found that the scaffold of benzothiazole or quinoline is generally effective in preventing bacterial cell division by interrupting the normal function of FtsZ protein. Our recent study targeting FtsZ inhibition by applying thiazole orange4 and benzofuroquinolinium derivatives18 (Fig. 1) prompted us to further investigate the antibacterial activity and mode of action for a combined scaffold of thiazole-quinolinium. In the present study, we synthesized 17 new thiazole-quinolinium derivatives (Table 1) and conducted a series of bioassays to investigate their antibacterial ability and elucidate the possible mode of action.
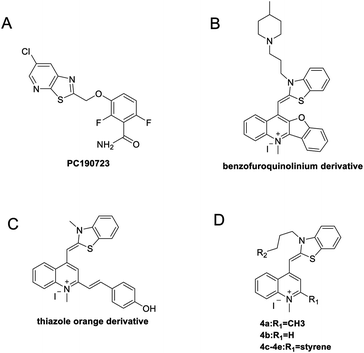 | ||
| Fig. 1 Chemical structures of PC190723 (A), benzofuroquinolinium derivative (B), thiazole orange derivative (C), and thiazole-quinolinium derivatives 4a–4e (D). | ||
2. Results and discussion
2.1. Design and synthesis of the thiazole-quinolinium derivatives
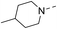
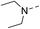
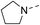
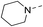
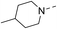
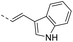
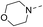
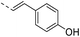
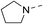
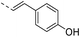
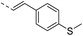
Our results showed that some molecules contain benzothiazole or quinoline scaffold may have good antibacterial activity by interrupting the function of FtsZ protein.4,19 Thiazole orange derivative (Fig. 1C) possesses strong bacterial cell growth inhibitory effect. This compound enhances FtsZ polymerization in vitro. The prediction model also showed that it binds to the interdomain cleft delimited by H7-helix and T7-loop of FtsZ.4 Benzofuroquinolinium derivative18 (Fig. 1B) has also been demonstrated targeting FtsZ and showing good antibacterial activity. We therefore retained the benzothiazolidine or quinone core for designing new antibacterial compounds. In the present study, we introduced a fatty amino substituent at 3-position of benzothiazole of the compound. In addition, a styrene substituent was introduced at 2-position of quinoline in compounds 4c1–4e4. The antibacterial activity and mode of action were used to investigate whether these new thiazole-quinolinium compounds possess antibacterial activity targeting FtsZ.The new thiazole-quinolinium derivatives (4a–e) shown in Table 1 were prepared in good yields by following the reported procedures20,21 (Scheme 1). Intermediates 3a and 3b were obtained by the reaction of 1,2-dimethyl-4-chloroquinolinium iodide or N-methyl-4-chloroquinolinium with N-(3-bromopropyl)-2-methylbenzo[d]thiazolium iodide in the presence of sodium bicarbonate. The reaction of 3a or 3b with a secondary amine including morpholine, diethylamine, piperidine and pyrrolidine afforded compounds 4a and 4b in good yields. Further reaction of 4a or 4b with different aromatic aldehydes including 4-hydroxybenzaldehyde, 4-(methylthio)benzaldehyde, 4-(dimethylamino)benzaldehyde, and indole-2-carboxaldehyde to afford the targeted compounds 4c–e. All the compounds were purified by flash silica gel column chromatography and were confirmed with 1H NMR, 13C NMR, ESI-MS or HRMS (Fig. S11–S24†). The synthesis of intermediates 1a, 1b and 2 was followed the reported methods.21 Compounds 4a2, 4b1 and 4b2 were reported as the fluorescent bio-molecular sensors.21
2.2. Antibacterial activity of the thiazole-quinolinium derivatives
The antimicrobial test showed that these thiazole-quinolinium derivatives possess broad-spectrum, potent antibacterial activity against a majority of selected Gram-positive and some Gram-negative strains, including four MRSA strains and drug resistant E. coli strain as shown in Tables 2 and 3. In general, all compounds reduced significantly the growth of antibiotic-susceptible and antibiotic-resistant S. aureus strains with MIC values in a range of 1–32 μg mL−1. Some analogues against MRSA strains were found 100 times more potent than methicillin. In addition, S. epidermidis was inhibited by the compounds with MIC values ranged from 0.25 to 16 μg mL−1. These thiazole-quinolinium derivatives were able to inhibit the growth of vancomycin-resistant E. faecium with MIC values of 2–32 μg mL−1 and also indicated that these compounds were much more effective than vancomycin. Interestingly, while all positive controls used in the study including methicillin, vancomycin, ampicillin and rifampin were not able to reduce the growth of the antibiotics resistant E. coli mutant NDM-1 strain (MIC > 64 μg mL−1), the new compounds inhibited both antibiotic susceptible strain and NDM-1 strain with MIC values of 0.5–64 μg mL−1.| Bacterial strains | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus ATCC 29213 | S. aureus ATCC 43300a | S. aureus ATCC BAA-41a | S. aureus ATCC BAA-44 | S. aureus ATCC BAA-1747a | S. aureus ATCC BAA-1720a | E. faecalis ATCC 29212 | E. faecium ATCC 49624 | E. faecium ATCC 700221b | S. epidermidis ATCC 12228 | B. subtilis 168 | |
| a These strains are MRSA.b Vancomycin-resistant strain, MIC of vancomycin is 1024 μg mL−1.c amp: ampicillin; van: vancomycin; meth: methicillin; rif: rifampin. | |||||||||||
| 4a1 | 2 | 4 | 2 | 4 | 8 | 8 | 32 | 16 | 16 | 1 | 8 |
| 4a2 | 32 | 32 | 8 | 8 | 32 | 32 | 64 | 32 | 32 | 4 | 8 |
| 4a3 | 32 | 32 | 8 | 8 | 32 | 32 | 64 | 32 | 32 | 8 | 8 |
| 4a4 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 0.5 | 1 |
| 4b1 | 2 | 8 | 2 | 2 | 16 | 16 | 32 | 8 | 2 | 1 | 8 |
| 4b2 | 32 | 8 | 4 | 4 | 16 | 16 | 32 | 16 | 32 | 4 | 8 |
| 4b3 | 32 | 8 | 4 | 4 | 8 | 16 | >64 | 32 | 32 | 2 | 16 |
| 4b4 | 2 | 8 | 1 | 2 | 4 | 8 | 16 | 8 | 16 | 0.25 | 4 |
| 4c1 | 16 | 16 | 16 | 8 | 16 | 16 | 16 | 16 | 8 | 2 | 8 |
| 4c2 | 16 | 16 | 8 | 8 | 16 | 32 | 16 | 8 | 4 | 4 | 4 |
| 4d1 | 16 | 8 | 4 | 4 | 8 | 8 | 32 | 8 | 16 | 2 | 2 |
| 4d2 | 32 | 32 | 32 | 32 | 32 | 32 | 64 | 32 | 32 | 16 | 16 |
| 4d3 | 16 | 32 | 32 | 32 | 32 | 32 | 64 | 64 | 32 | 16 | 16 |
| 4e1 | 16 | 16 | 16 | 16 | 32 | 16 | >64 | 32 | 8 | 16 | 8 |
| 4e2 | 16 | 8 | 16 | 16 | 32 | 16 | >64 | 16 | 8 | 8 | 4 |
| 4e3 | 8 | 8 | 16 | 8 | 16 | 16 | >64 | 16 | 8 | 4 | 8 |
| 4e4 | 16 | 8 | 16 | 8 | 16 | 8 | 32 | 32 | 4 | 2 | 8 |
| ampc | >64 | >64 | >64 | 16 | >64 | >64 | 0.5 | 0.25 | >64 | >64 | 32 |
| vanc | 2 | 2 | 2 | 1 | 1 | 0.5 | 2 | 0.5 | 1024 | >64 | 0.25 |
| methc | 0.5 | 512 | 1024 | 1024 | 256 | 1024 | 1 | 4 | 1.5 | 0.5 | 0.625 |
| rifc | 0.5 | <0.125 | <0.125 | 2 | <0.125 | <0.125 | 0.5 | <0.125 | <0.125 | 1 | 4 |
| Bacterial strains | |||||||
|---|---|---|---|---|---|---|---|
| E. coli ATCC 25922 | E. coli ATCC 8739 | E. coli ATCC BAA-2469a | K. pneumoniae ATCC BAA-2470a | A. baumannii ATCC 19606b | P. aeruginosa ATCC BAA-2108b | E. cloacae ATCC BAA-1143c | |
| a NDM-1 expressing strain, which is resistant to most of the antibiotics, except tigecycline and colistin.b Multi-drug resistant strain.c AmpC beta-lactamase expressing strain.d amp: ampicillin; van: vancomycin; meth: methicillin; rif: rifampin. | |||||||
| 4a1 | 2 | 4 | 8 | 32 | 64 | 64 | >64 |
| 4a2 | 4 | 8 | 4 | 64 | 64 | 64 | 64 |
| 4a3 | 4 | 4 | 4 | 32 | 64 | 64 | >64 |
| 4a4 | 0.5 | 1 | 1 | 8 | 16 | 64 | 64 |
| 4b1 | 2 | 8 | 1 | 32 | 64 | 64 | >64 |
| 4b2 | 4 | 4 | 2 | 32 | 64 | 64 | >64 |
| 4b3 | 2 | 4 | 4 | 32 | 64 | 64 | >64 |
| 4b4 | 2 | 2 | 0.5 | 8 | 16 | 16 | >64 |
| 4c1 | 8 | 64 | 64 | >64 | >64 | >64 | >64 |
| 4c2 | 4 | 32 | 8 | >64 | >64 | >64 | >64 |
| 4d1 | 4 | 16 | 4 | 16 | 32 | 32 | 64 |
| 4d2 | 32 | 32 | 16 | >64 | >64 | >64 | >64 |
| 4d3 | 16 | 32 | 32 | 64 | 64 | 64 | >64 |
| 4e1 | 16 | 32 | 16 | 64 | >64 | >64 | >64 |
| 4e2 | 4 | 16 | 8 | 64 | >64 | 64 | >64 |
| 4e3 | 16 | 32 | 16 | 64 | 64 | 32 | 64 |
| 4e4 | 16 | 16 | 8 | 32 | >64 | 64 | 64 |
| ampd | 1 | 2 | >64 | >64 | 64 | >64 | >64 |
| vand | >64 | >64 | >64 | >64 | 64 | >64 | >64 |
| methd | 2 | >64 | >2048 | >2048 | >256 | >64 | >96 |
| rifd | 4 | 8 | >64 | >64 | 1 | >64 | 64 |
By analyzing the MIC results obtained from the antimicrobial screening with respect to the molecular structure of the compounds, the antibacterial activity was found obviously influenced by the two substituent groups, R1 on the quinoline fragment and R2 on the benzothiazole fragment. It seems that R1 group gives a more determining effect on the antibacterial activity. We found that the size of the R1 group was also critical, in which CH3 group was the best as it is larger than H and is much less bulky than p-substituted styryl group and indolyl group. The effect of R1 group in antibacterial activity may roughly show in the following order: CH3 > H ≫ indolyl ≈ styryl. This observation may also suggest that the binding pocket of the FtsZ protein was steric sensitivity. For R2 group substituted N-benzothiazole fragments, there were five amino groups including diethyl amine, pyrrolidine, piperidine, 4-methylpiperidine, and morpholine, which were attached to N-benzothiazole moiety through an ethyl bridge. When R1 group was either CH3 or H, both diethyl amine and 4-methylpiperidine gave the best antibacterial activity, whereas pyrrolidine and piperidine gave much less inhibitory potency. When R1 was a bulky group, the substituent of R2 seems no significant influence on the inhibitory potency. However, for the compounds with the substituent R1 was an indolyl (4c1, 4d1, 4e1), the pyrrolidine substituent gave slightly better antibacterial activity than other analogues. In general, the antibacterial results indicated that the amino terminal group of benzothiazole fragment and a small group substituted at the quinoline 2-position were essential in conferring the potent antibacterial activity of the thiazole-quinolinium derivatives.
In the present study, we screened out four compounds (4a1, 4a4, 4b1, 4b4) that showed greater antibacterial activity than their analogues (Tables 2 and 3). For example, 4a4 inhibited the growth of antibiotic-susceptible and antibiotic-resistant S. aureus strains with MIC values in a very low range of 1–2 μg mL−1. The antibacterial activity of 4a4 against MRSA strains was lower than rifampin but comparable to that of vancomycin. In addition, the growth of E. faecium, S. epidermidis and E. coli could be inhibited by 4a4 with MIC values of 0.5–2 μg mL−1. Moreover, 4a4 also displayed moderate antibacterial activity against drug-resistant K. pneumoniae, A. baumannii, P. aeruginosa and E. cloacae, which were resistant to most of the positive controls of commonly prescribed antibacterial agents such as methicillin, vancomycin and ampicillin (Tables 2 and 3).
In order to further investigate the antibacterial activity of these compounds, we evaluated the minimum bactericidal concentrations (MBCs) of four selected compounds (4a1, 4a4, 4b1 and 4b4) against S. aureus ATCC 29213 and E. faecium ATCC 700221. The MBC values were summarized in Table 4. As indicated by the MBC/MIC ratio > 4, the results suggested that the compounds may exert bacteriostatic action (Table 5).22,23
| MBC (μg mL−1) | MIC (μg mL−1) | MBC/MIC | |
|---|---|---|---|
| E. faecium ATCC 700221 | |||
| 4a1 | 128 | 16 | 8 |
| 4a4 | >64 | 2 | >32 |
| 4b1 | 16 | 2 | 8 |
| 4b4 | 128 | 16 | 8 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| S. aureus ATCC 29213 | |||
| 4a1 | 64 | 2 | 32 |
| 4a4 | 8 | 1 | 8 |
| 4b1 | 32 | 2 | 16 |
| 4b4 | 16 | 2 | 8 |
| S. aureus ATCC 29213 (MIC, μg mL−1) | IC50, μg mL−1 | |||
|---|---|---|---|---|
| 4a4 | 4b4 | 4a4 | 4b4 | |
| 16HBE | 1 | 2 | 14.3 | 20.2 |
| HK-2 | 1 | 2 | 26.0 | 18.6 |
| L929 | 1 | 2 | 12.6 | 23.4 |
2.3. Thiazole-quinolinium derivatives inhibited bacterial cell division
FtsZ inhibitors can effectively inhibit bacterial cell division, which triggers the elongation of B. subtilis cells.11 We therefore visualized the cell morphology of B. subtilis incubated with or without the thiazole-quinolinium derivatives. The results showed that the compounds bearing a small group, either H or CH3, at the 2-position of quinoline, such as 4a1–4a4 and 4b1–4b4, were able to induce bacterial cells elongation (Fig. 2 and S1†). For example, the cell length of B. subtilis 168 in the control group was 5–10 μm, whereas after incubation with 4a4 at 0.5 μg mL−1 for 4 h, the bacteria were found longer than 10 μm, suggesting a mechanism of antibacterial-induced cell filamentation. However, for other compounds, including all analogues from 4c, 4d, and 4e series, bearing a bulky substituent group (strylyl or indolyl moiety) at the 2-position of quinoline, they only exhibited very little effect on bacterial cell elongation. These results suggested that the addition of bulky substitute group at 2-position of quinoline core may cause significant reduction in cell division targetability and antibacterial activity.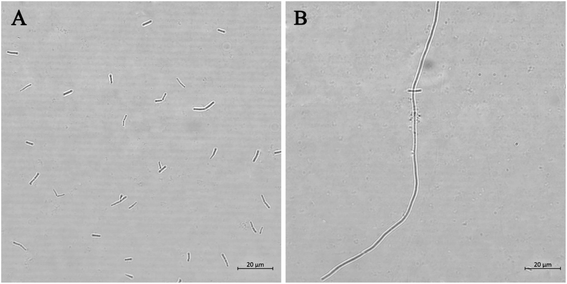 | ||
| Fig. 2 Morphology analysis of B. subtilis 168. Cells were grown in the absence (A) and presence of 4a4 (B). Scale bar = 20 μm. | ||
2.4. Thiazole-quinolinium derivatives disrupted FtsZ function in vitro
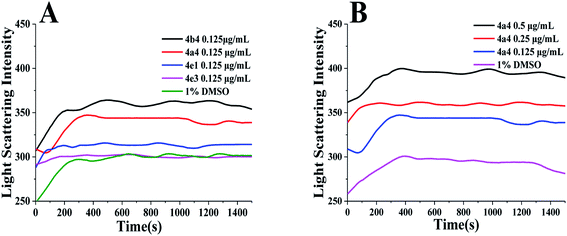
Some inhibitors such as PC190723 (ref. 11) and berberine10 induce cell filamentation by interacting with FtsZ protein. The cell elongation-induced by our compounds may be also via the interactions with FtsZ protein. In order to elucidate the mechanism of antibacterial activity, dynamic polymerization of FtsZ by light scattering test was studied using two groups of selected compounds: (a) compounds 4a4 and 4b4 induced elongation of the bacteria; (b) compounds 4e1 and 4e3 did not promote elongation of bacteria. First, the light scattering signals of 4a4, 4b4, 4e1 and 4e3 with the same concentration were measured under the same test conditions (Fig. 3A). It was found that 4a4 and 4b4 caused a significant increase in light scattering signal compared to the control signal. On the contrary, 4e1 and 4e3 caused almost no signal change. We then further measured the light scattering signals of 4a4, 4b4, 4e1 and 4e3 at a certain concentration gradient. The results showed that 4a4 and 4b4 could enhance FtsZ polymerization in a dose-dependent manner (Fig. 3B and S4†). Similar enhancement effect on polymerization of FtsZ protein could also be observed in some of the FtsZ-targeting compounds, such as thiazole orange derivative (Fig. 1C),4 PC190723 (Fig. 1A)24 and its analogs.25 However, under the same conditions, the light scattering signals of 4e1 and 4e3 did not show significant change (Fig. S4†). These results therefore revealed that the compounds with H or CH3 group at 2-position of quinoline core (4a4 and 4b4) had a stronger stimulation effect on FtsZ polymerization than those with bulky groups (4e1 and 4e3). This indicated that the small-sized substituent at 2-position of quinoline core plays a pivotal role in the inhibitor–FtsZ interaction. Conversely, the bulky substituent group may hamper compound–FtsZ interaction that renders these compounds fail to target FtsZ and lose inhibitory ability to cell division.The dynamic assembly of FtsZ was strictly regulated by its GTPase activity,11,26 the GTPase activity of S. aureus FtsZ (SaFtsZ) was determined using a ATPase/GTPase Activity Assay Kit (Sigma-Aldrich, USA) according to the manufacturer's instructions with minor modifications.27 The results showed that 4a4 and 4b4 did not show any effect on the GTPase activity of SaFtsZ (Fig. S5†).
2.5. Binding study of the compounds with SaFtsZ
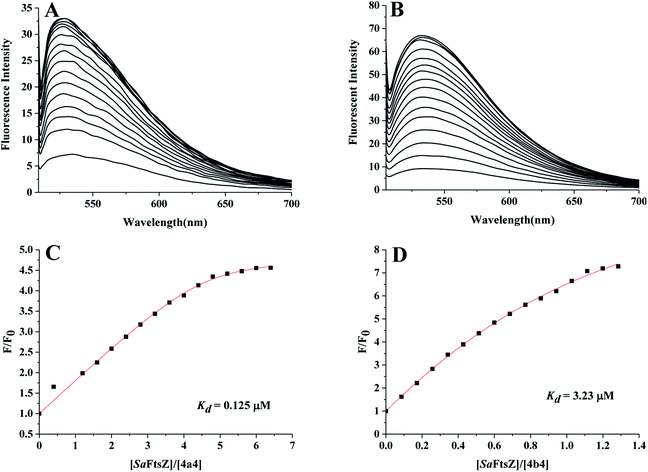
To further determine the ability of a thiazoquinoline derivative to target SaFtsZ. We selected two compounds 4a4 and 4b4 with good antibacterial activity and monitored the intrinsic fluorescence of the compound with the addition of SaFtsZ. From Fig. 4A and B, when increasing the SaFtsZ concentration, the fluorescence intensity of both compounds was enhanced remarkably. The FtsZ-induced fluorescence indicated the compound bound to SaFtsZ. We therefore determined the FtsZ-compound dissociation constant (Kd) of the binding reaction according to the independent site model28 by nonlinear fitting to the equation:29 F/F0 = 1 + (Q − 1)/2 (N + 1 + X − ((X + 1 + N)24X)1/2). In this relationship, F0 and F are the fluorescence emission intensities of the compound in the absence and presence of protein, respectively; N = (KaCcompound)−1, Q = Fmax(F0)−1, X = nCSaFtsZ(Ccompound)−1, Ka = Kd−1, Fmax is the fluorescence intensity upon saturation of SaFtsZ.From Fig. 4C and D, the Kd values of 4a4 and 4b4 were 0.125 μM and 3.23 μM, respectively, by comparing the dissociation constants estimated, 4a4 with a CH3 group at 2-position of quinoline fragment had a greater affinity for SaFtsZ than 4b4. This may indicated that the CH3 group is more likely to enhance the binding affinity of the compound.
2.6. Effects of the synthesized compounds on B. subtilis Z-ring formation in B. subtilis
The perturbation of the Z-ring can also pre-maturely inhibit the bacterial cell division and induce bacterial cell elongation.10,30,31 Therefore, four compounds with strong antibacterial activity and cell inhibitory effects, 4a1, 4a4, 4b1 and 4b4, were used to treat B. subtilis before determining their effects on Z-ring formation (Fig. 5 and S6†). In the absence of compounds, a bright green fluorescent band at the cell midpoint was apparently observed, representing the septation–competent localized Z-rings (cytoskeletal structures) (Fig. 5A). Comparatively, GFP-FtsZ dispensed as discrete foci throughout the elongated cell after the treatment with 4a1, 4a4, 4b1 or 4b4 (Fig. 5B and S6†), suggesting that these compounds promoted the mislocalization of the FtsZ protein.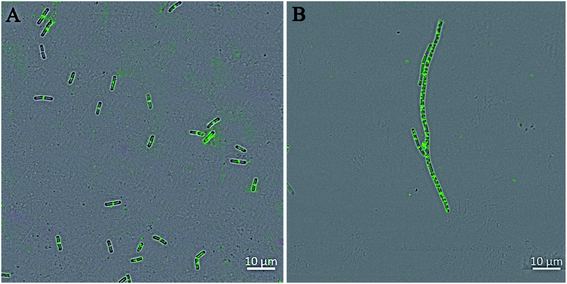 | ||
| Fig. 5 The perturbation of the cytokinetic Z-ring in B. subtilis. Cells of B. subtilis were grown in the absence (A) and presence of 4a4 (B). Scale bar = 10 μm. | ||
2.7. Cytotoxicity of selected synthesized compounds
Cytotoxicity of two representative thiazole-quinolinium derivatives, 4a4 and 4b4, with best antibacterial activity was examined against human bronchial epithelium cells (16HBE), human renal tubular epithelial cells (HK-2), and mouse fibroblast cells (L929). The IC50 values of 4b4 for the 16HBE and L929 cell lines were greater than 4a4, whereas the IC50 values of 4b4 were less than 4a4 for the HK-2 cell line. For these three cell lines, the IC50 values of 4a4 and 4b4 were much greater than MIC values of these compounds against S. aureus ATCC 29213, which indicated that 4a4 and 4b4 may not have significant cytotoxicity against normal human cells. In addition, hemolytic activity test32 was also used to detect the toxicity of 4a4 and 4b4. The result showed that 4a4 and 4b4 did not exhibit hemolytic toxicity to human erythrocytes (Fig. S7†).2.8. In vitro evaluation of potential resistance development
In order to evaluate the potential development of drug resistance of 4a4 and 4b4, three commercial antibiotics (ciprofloxacin, vancomycin and rifampin) were used as positive controls against S. aureus ATCC 29213. As shown in Fig. 6, the MIC values of 4a4 and 4b4 against S. aureus ATCC 29213 only increased by 16-fold and 8-fold, respectively, after 35 passages. Comparatively, the MIC values of ciprofloxacin, vancomycin and rifampin increased 32-fold, 512-fold and 8192-fold, respectively. Similar results were obtained when these compounds were used to treat B. subtilis 168 and E. coli ATCC 25922 (Fig. S8 and S9†). Based on the results of drug resistance study, 4a4 and 4b4 produced low forced induction of resistance on the tested strains.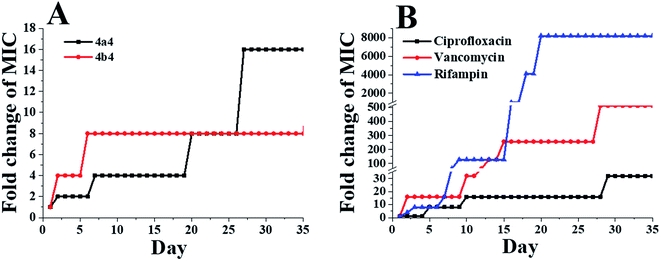 | ||
| Fig. 6 Bacterial resistance study of selected compounds against S. aureus ATCC 29213: 4a4 and 4b4 (A) and ciprofloxacin, vancomycin and rifampin (B). | ||
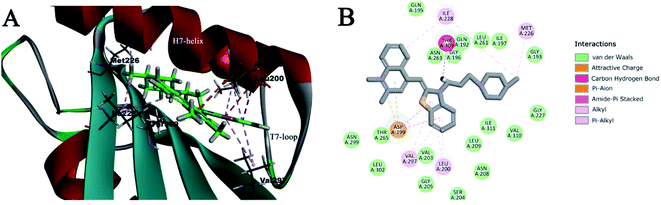
2.9. Predicted binding mode of 4a4 to FtsZ
The biological assay results indicated that 4a4–4b4 may potentially be effective FtsZ targeting inhibitors. Molecular modeling studies were performed to detect potential binding sites of 4a4 and 4b4 to FtsZ protein. One relatively inactive compound 4e1 was also performed in this test for comparison. As a result, the highest docking score for 4a4 and 4b4 binding were obtained near the T7-loop and the H7-helix, and the binding site was a relatively narrow interdomain cleft defined by H7-helix, T7-loop and quadruplex β-sheets (Fig. 7A and S10A†). The whole structure of 4a4 (Fig. S10A,† green) and 4b4 (Fig. S10A,† yellow) was found to locate in the cleft. On the other hand, only part of 4e1 binded into the binding site (Fig. S10A,† purple). A 2D diagram (Fig. 7B) showed the predicted interactions between 4a4 and FtsZ residues. It was predicted that a large number of hydrophobic interactions between 4a4 and Asp 199, Ile228, Met226, Val297 and Leu200 of FtsZ protein. In addition, electrostatic interactions and hydrophobic interactions could be found between benzothiazolidine and Asp199. A carbon–hydrogen bond was discovered between the fatty ammonia carbon chain and Thr309. Furthermore, in the binding pocket, a large number of amino acid residues such as Gln195, Val310, Gly205, Ile311 may have interactions with 4a4 via van der Waals forces. The predicted interactions between 4b4 and FtsZ were similar to that of 4a4. However, as shown in Fig. S10B,† Thr309 and Ile311 cannot establish interaction with 4b4, and this maybe the reason that 4a4 with a CH3 group at 2-position of quinoline fragment had a greater affinity for SaFtsZ than 4b4. In contrast, fewer interactions between 4e1 and FtsZ were predicted (Fig. S10C†) when compared with those of 4a4 and 4b4. For instance, it was found that 4e1 showed a charge attraction with Asp199 only.3. Discussion and conclusions
In summary, a series of new thiazole-quinolinium derivatives were synthesized and their antibacterial activity and possible mode of action were thoroughly investigated. Antimicrobial susceptibility test confirmed that these thiazole-quinolinium derivatives possess broad-spectrum antibacterial activity against Gram-positive and Gram-negative strains, including some highly resistant strains. Among the compounds tested, 4a4 and 4b4 bearing a small group (CH3 or H) at 2-position of quinoline fragment exhibited potent antibacterial activity against MRSA and VRE and low MIC values at 1–8 μg mL−1 and 2–16 μg mL−1 were obtained. The inhibitory effects were comparable to the reported FtsZ inhibitors. 9-Phenoxyalkylberberine derivative 2 inhibited the cell proliferation with MIC values from 2 to 8 μg mL−1 against MRSA and VRE.14 A benzamide prodrug of PC190723 (TXA709) inhibited the cell division with MIC values from 2 to 4 μg mL−1 against MRSA.12 PC190723 inhibited the proliferation of S. aureus strains at 1 μg mL−1.33 In addition, 4a4 and 4b4 inhibited the growth of E. coli and its antibiotics resistant mutant with MIC values of 0.5–2 μg mL−1. To date, a few FtsZ inhibitors have been reported with strong antibacterial activity against Gram-negative bacteria. For example, 9-phenoxyalkylberberine derivative 2 inhibited the growth of E. coli with MIC value of 32 μg mL−1.14 Thiazole orange derivative 1 (Fig. 1C) has MIC values from 0.75 to 48 μg mL−1 against the Gram-negative strains.4 The antibacterial activity of 4a4 and 4b4 against Gram-negative bacteria may be attributed to the ability of the compound to pass through the outer membrane of Gram-negative bacteria. In the morphological study, both compounds significantly increased the cell length of B. subtilis 168 by inhibiting Z-ring formation, without interfering with its membrane integrity, suggesting that compounds interact with Z-ring to inhibit bacterial cytokinesis. 4a4 and 4b4 were also found to bind to FtsZ and thus to promote FtsZ polymerization in a dose-dependent manner without interfering GTPase activity. Similar enhancement effect on polymerization of FtsZ protein could also be observed in some of FtsZ-targeting compounds, such as thiazole orange derivative (Fig. 1C),4 PC190723 (Fig. 1A)24 and its analogs.25 The phenomenon that the compounds did not significantly affect the GTPase activity of SaFtsZ also appears in some reported FtsZ inhibitors, such as 2,6-difluoro-3-aminobenzamide derivative34 and the derivatives of PC190723,35 which were reported to bind to the interdomain cleft of FtsZ. Moreover, both cytotoxic and hemolysis experiments indicated that 4a4 and 4b4 showed no significant cytotoxicity to normal cells. Drug resistance experiments also showed that 4a4 and 4b4 were not easy to induce bacterial resistance. Based on these results obtained, 4a4 and 4b4 may be good candidates for the development of new antibacterial agents targeting drug-resistant bacteria.4. Materials and methods
4.1. Chemistry
All analytical grade reagents and chemicals were used as received without further purification and purchased from Aladdin and Sigma. Mass spectra (MS) were recorded on Bruker amazon SL mass spectrometer with an ESI or ACPI mass selective detector and the high-resolution mass spectra (HRMS) were recorded with Agilent 12606230TOF. 1H and 13C NMR spectra were recorded in DMSO using TMS as the reference with a Bruker BioSpin GmbH spectrometer at 400 MHz and 100 MHz, respectively. Elemental analyses were carried out on an Elementar Vario EL CHNS elemental analyzer.4.2. General synthetic procedure for the intermediates (1a–1b, 2a and 3a–3b)
Intermediates 1a and 1b was obtained by the reaction of 4-chloro-2-methylquinoline (2 g, 11.24 mmol) or 4-chloroquinoline (2 g, 12.27 mmol) with iodomethane (9.6 g) in tetramethylene sulfone (4.5 mL). After stirring at 65 °C for 24 h the reaction mixed was cooled and suction filtration was performed before the addition of ethyl acetate. The solid was washed again with ethyl acetate and dried in vacuum to give of intermediates 1a and 1b, yields were 78% and 75%, respectively. Compound 2 was obtained by the reaction of 2-methylbenzo-[d]-thiazole (4 g, 15.6 mmol) with 1,3-dibromopropane (4 mL, 20 mmol). The reaction mixture was stirred at 100 °C for 24 h and cooled before suction filtration. The solid was washed with ethyl acetate, and vacuum dried to obtain a white solid, yield was 69%. The key intermediate compound 3a or 3b was then prepared by the reaction of 1a (2 g, 6.2 mmol) or 1b (1.89 g, 6.2 mmol) with 2 (2.17 g, 6.2 mmol) and potassium iodide (2.49 g, 15 mmol) in methanol (30 mL) respectively. The reaction mixture was stirred at 45 °C for 3 h. Ethyl acetate was added after the shock, filtered and washed with ethyl acetate before drying with vacuum to obtain red solid. Yields were recorded to be 90.1% and 88.6%, respectively.4.3. General synthetic procedure for thiazole-quinolinium derivatives (4a1–4a4, 4b1–4b4, 4c1–4c2, 4d1–4d3 and 4e1–4e4)
The intermediate 3a (0.2 mmol) or 3b (0.2 mmol) was mixed with secondary amine (1.6 mmol) in 5 mL of DMSO. Then, the reaction mixture was stirred at 45 °C for 24 h, cooled and ethyl acetate was added after the shock, suction filtration, the solid was washed with ethyl acetate, and dried in vacuum to obtain compounds 4a1–4a4 and 4b1–4b4 in good yields. Then, 4a1–4a4 were subjected to a condensation reaction with p-methylthiobenzaldehyde, p-hydroxybenzaldehyde and indole-3-carbaldehyde in n-butanol the reaction mixture was stirred at 130 °C for 3 h. After cooled to room temperature, to the reaction mixture ethyl acetate was added to precipitate the solid products. The residuals were collected by suction filtration and dried in vacuum. The residuals were purified by silica column chromatography (dichloromethane: methanol (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v), from 100
v), from 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 to 100
3 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) to obtain 4c1–4c2, 4d1–4d3 and 4e1–4e4. The characterizations of newly synthesized thiazole-quinolinium derivatives are as follow. For 4a2, 4b1 and 4b2, please refer to previous reports.21
20) to obtain 4c1–4c2, 4d1–4d3 and 4e1–4e4. The characterizations of newly synthesized thiazole-quinolinium derivatives are as follow. For 4a2, 4b1 and 4b2, please refer to previous reports.21
4.4. MIC and MBC assays
MIC tests were conducted using high throughput 96-well microtiter plates method with clinically relevant bacterial strains – (a) Gram-positive: S. aureus ATCC 29213, S. aureus ATCC 43300, S. aureus ATCC BAA-41, S. aureus ATCC BAA-44, S. aureus ATCC BAA-1747, S. aureus ATCC BAA-1720, E. faecalis ATCC 29212, E. faecium ATCC 49624, E. faecium ATCC 700221, S. epidermidis ATCC 12228, B. subtilis 168; (b) Gram-negative: E. coli ATCC 25922, E. coli ATCC 8739, E. coli ATCC BAA-2469, K. pneumoniae ATCC BAA-2470, A. baumannii ATCC 19606, P. aeruginosa ATCC BAA-2108, E. cloacae ATCC BAA-1143. The overnight culture was diluted in Mueller–Hinton broth medium to a final concentration of 5 × 105 CFU mL−1, compounds were serially diluted with DMSO and then added into microtiter plates containing the test bacteria with a final DMSO concentration of 1%. After 18–24 h incubation at 37 °C, the minimum drug concentration without bacterial growth was determined as the MIC value. The MIC was defined as the lowest compound concentration at which 90% of bacteria growth was inhibited. Methicillin, ampicillin, vancomycin and rifampin were applied as a reference compound under the same assay conditions. For the minimal bactericidal concentration (MBC) assays, 100 μL of medium from the MIC microtiter plates assay with no visible turbidity was spread on freshly prepared MH agar plates and incubated at 37 °C for 24 h before colony counting. The MBC of the derivatives was described as the lowest concentration of the derivative with less than five colonies grow on each plate.36 Then, the MBC/MIC ratios were calculated, a compound was considered as bactericidal when the MBC/MIC ratio was ≤4 and bacteriostatic when the MBC/MIC ratio was >4.22,23 Each assay was carried out in triplicates.4.5. Bacterial morphology studies
The B. subtilis strain 168 cells were grown in LB medium. Bacterial culture was prepared at concentration of 1 × 105 CFU mL−1 and inoculated in the same medium containing the test compound at a concentration of 0.5 × MIC. After incubated at 37 °C for 4 h, the cell pellet was collected before resuspension in PBS. 3 μL of the suspension mixture was then loaded on a microscopic slide precoated with 0.1% (w/v) poly-L-lysine and the morphology of the bacterial cells was visualized under a high resolution laser confocal microscope at 40× oil immersion magnification. The images were captured using a ZEISS LSM 800 with Airscan microscope.4.6. Membrane integrity studies
B. subtilis 168 cells were grown in LB medium. The bacterial culture was then diluted to 1 × 105 CFU mL−1 and inoculated in the same medium added with the test compound at a concentration of 0.5 × MIC. After incubation period of 4 h at 37 °C, 1.6 μM of FM 4–64 (a fluorescent dye) was added into the medium and further incubated for an additional 15 min at 37 °C without shaking. The cells were then collected via centrifugation and resuspended in PBS. Then, 3 μL of PBS containing bacterial cells was placed on the microscopy slide precoated with 0.1% (w/v) poly-L-lysine. Changes in morphology of the bacterial cells were monitored under a high resolution laser confocal microscope at 40× oil immersion magnification. The images were captured using a ZEISS LSM 800 with Airscan microscope.4.7. Light-scattering assay
As the polymer mass of FtsZ correlates directly to the magnitude of light scattering signal, the dynamics of FtsZ assembly can be studied in vitro.37 The polymerization of S. aureus FtsZ was monitored by 90° light scattering in a PerkinElmer LS-45 fluorescence spectrometer. Both excitation and emission wavelengths were set at 600 nm with a slit width of 2.5 nm.38 S. aureus FtsZ (4 μM) in 50 mM MOPS buffer (pH 6.5) was incubated with vehicle control (1% DMSO) or varied concentrations of the tested compound for 10 min at 25 °C. Subsequently, baseline was established with 50 mM KCl and 10 mM MgCl2. Ten minutes after the baseline establishment, 1 mM GTP was added at the last fraction and the increase in light scattering was measured for another 25 minutes. The rate and extent of polymerization were calculated with subtraction of background using appropriate blank. Each assay was carried out in triplicates.4.8. SaFtsZ binding affinity assay
Fluorescence spectroscopy is the most rapid and direct means of detecting protein and small molecule interactions. The binding of 4a4 and 4b4 to SaFtsZ was determined by monitoring changes in protein-induced intrinsic fluorescence of the compounds. An aliquot of the SaFtsZ stock solution was sequentially added to a buffer solution (600 μL) containing 4a4 (3.51 μM) or 4b4 (3.68 μM) to increase the protein concentration by a gradient of 0.4 μM. After each addition of the protein, the reaction was equilibrated for 2 minutes and then the emission spectra were obtained from 700 nm to 500 nm in 1 nm increments. Subtract the background spectrum produced by protein titration into a separate buffer. Fluorescence spectra were recorded on a PerkinElmer LS-45 fluorescence spectrometer with an excitation wavelength set at 493 nm and a buffer condition of 50 mM Tris–HCl (pH 7.4), 50 mM KCl and 2 mM magnesium acetate. All measurements were taken at 25 °C.4.9. GTPase activity assay
The GTPase activity of S. aureus FtsZ was determined using a ATPase/GTPase Activity Assay Kit (Sigma-Aldrich, USA) according the manufacturer's instructions with minor modifications.27 S. aureus FtsZ (7.3 μM) was added to assay buffer with vehicle (1% DMSO) or varied concentrations of test compound. Reactions were initiated by the addition of 200 μM GTP and incubated at room temperature. After 30 min of incubation, 200 μL of reagent was added into each well and incubated for another 30 minutes at room temperature to halt the enzymatic reaction and generate the colorimetric product. Inorganic phosphate was quantified spectrometrically at 620 nm with a microplate reader. Each assay was carried out in triplicates.4.10. Visualization of Z-ring in bacterial cells
B. subtilis containing the IPTG-inducible plasmid for the production of green fluorescent protein (GFP)-tagged FtsZ was included in the current study to facilitate visualization of Z-ring. A culture of B. subtilis was grown in LB medium (supplemented with 30 μg mL−1 of chloramphenicol) at 37 °C in shaking incubator. Overnight grown cultures were diluted to 1% with LB medium containing 40 μM of IPTG and selected test compound at a concentration of 0.5 × MIC. After 4 h incubation at 37 °C, B. subtilis cells were fixed, harvested and resuspended in PBS buffer. 3 μL of sample mixtures were added to the pretreated microscopy slide with 0.1% (w/v) poly-L-lysine and visualized using a high resolution laser confocal microscope at 40× oil immersion magnification. The images were captured using a ZEISS LSM 800 with Airscan microscope.4.11. CCK-8 cytotoxicity assay
The cytotoxicity of compounds was explored with human bronchial epithelium cells (16HBE), human renal tubular epithelial cells (HK-2) and mouse fibroblast cells (L929) by CCK-8 (Cell Counting Kit-8) method. For the cytotoxicity assay, 5000 cells were seeded per well in a 96-wells microtiter plate and allowed to adhere overnight at 37 °C before treatment with selected compound. After 48 h of treatment with selected compounds, 10 μL CCK-8 was added to each well and incubated for another 3 h, then the absorbance were measured at 450 nm with a microplate reader. Each assay was carried out in triplicates.4.12. Hemolytic activity assay
Compounds were tested with human erythrocytes at a concentration of 2% as previously described.32 Serial dilution of selected compounds was prepared in PBS and then 150 μL was loaded per well in a 96-well microtiter plate before addition of 150 μL of human blood. After incubated 1 h at 37 °C, plates were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min, then 100 μL of the supernatant was used to obtain absorbance reading at 540 nm. Water and PBS were used as positive and negative control respectively. The hemolysis percentage was calculated with the equation (absorbance of test compound − absorbance of the negative control)/(absorbance of water − absorbance of the negative control) × 100%, and a rate > 5% indicates hemolysis.39 Each assay was carried out in triplicates.
000 rpm for 5 min, then 100 μL of the supernatant was used to obtain absorbance reading at 540 nm. Water and PBS were used as positive and negative control respectively. The hemolysis percentage was calculated with the equation (absorbance of test compound − absorbance of the negative control)/(absorbance of water − absorbance of the negative control) × 100%, and a rate > 5% indicates hemolysis.39 Each assay was carried out in triplicates.
4.13. In vitro evaluation of potential drug resistance development
To further evaluate potential development of drug resistance induced these compounds and antibiotics (ciprofloxacin, vancomycin, rifampin) against bacterial strains (S. aureus ATCC 29213, B. subtilis 168, E. coli ATCC 25922), we refer to reported methods for drug resistance studies.34,40 The bacterial culture at a concentration of 0.5 × MIC in a 96-well microtiter plate of the initial MIC test was diluted to 5 × 105 CFU mL−1, and then various concentrations of compounds were added to the corresponding bacteria, the MICs were determined after incubated 24 h at 37 °C. This process was repeated for 35 times.4.14. Molecular docking study
Molecular modeling procedures were carried out using Discovery Studio (DS) version 2016 X-ray crystal structure of SaFtsZ downloaded from the PDB database (PDB code: 4DXD). The ligands were sketched using the tools of small molecules in DS. After removing water molecules and co-crystal ligands, the protein was subjected for docking using an automated procedure of DS-LibDock protocol.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the support from the National Natural Science Foundation of China (81703333, 81670480 and 81473082), Natural Science Foundation of Guangdong Province, China (2020A1515011326, 2017A030313078), Guangzhou Key Laboratory Fund (201905010004), and Major Scientific Research Projects at Provincial Level in Guangdong General University (2017KZDXM068). The authors are also grateful to the support from Department of Agriculture and Rural Affairs of Guangdong Province, China (2018LM2175), and Science and Technology Program of Guangzhou Municipal Health Commission (20192A011018).References
- K. K. Kumarasamy, M. A. Toleman, T. R. Walsh, J. Bagaria, F. Butt, R. Balakrishnan, U. Chaudhary, M. Doumith, C. G. Giske, S. Irfan, P. Krishnan, A. V. Kumar, S. Maharjan, S. Mushtaq, T. Noorie, D. L. Paterson, A. Pearson, C. Perry, R. Pike, B. Rao, U. Ray, J. B. Sarma, M. Sharma, E. Sheridan, M. A. Thirunarayan, J. Turton, S. Upadhyay, M. Warner, W. Welfare, D. M. Livermore and N. Woodford, Lancet Infect. Dis., 2010, 10, 597–602 CrossRef CAS PubMed.
- T. R. Walsh, J. Weeks, D. M. Livermore and M. A. Toleman, Lancet Infect. Dis., 2011, 11, 355–362 CrossRef PubMed.
- D. J. Payne, Science, 2008, 321, 1644–1645 CrossRef CAS PubMed.
- N. Sun, Y. J. Lu, F. Y. Chan, R. L. Du, Y. Y. Zheng, K. Zhang, L. Y. So, R. Abagyan, C. Zhuo, Y. C. Leung and K. Y. Wong, Front. Microbiol., 2017, 8, 855 CrossRef PubMed.
- G. D. Wright, Chem. Biol., 2012, 19, 3–10 CrossRef CAS PubMed.
- D. N. Margalit, L. Romberg, R. B. Mets, A. M. Hebert, T. J. Mitchison, M. W. Kirschner and D. RayChaudhuri, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 11821–11826 CrossRef CAS PubMed.
- Q. Huang, F. Kirikae, T. Kirikae, A. Pepe, A. Amin, L. Respicio, R. A. Slayden, P. J. Tonge and I. Ojima, J. Med. Chem., 2006, 49, 463–466 CrossRef CAS PubMed.
- P. Sass and H. Brotz-Oesterhelt, Curr. Opin. Microbiol., 2013, 16, 522–530 CrossRef CAS PubMed.
- P. Domadia, S. Swarup, A. Bhunia, J. Sivaraman and D. Dasgupta, Biochem. Pharmacol., 2007, 74, 831–840 CrossRef CAS PubMed.
- P. N. Domadia, A. Bhunia, J. Sivaraman, S. Swarup and D. Dasgupta, Biochemistry, 2008, 47, 3225–3234 CrossRef CAS PubMed.
- D. J. Haydon, N. R. Stokes, R. Ure, G. Galbraith, J. M. Bennett, D. R. Brown, P. J. Baker, V. V. Barynin, D. W. Rice, S. E. Sedelnikova, J. R. Heal, J. M. Sheridan, S. T. Aiwale, P. K. Chauhan, A. Srivastava, A. Taneja, I. Collins, J. Errington and L. G. Czaplewski, Science, 2008, 321, 1673–1675 CrossRef CAS PubMed.
- M. Kaul, L. Mark, Y. Zhang, A. K. Parhi, Y. L. Lyu, J. Pawlak, S. Saravolatz, L. D. Saravolatz, M. P. Weinstein, E. J. LaVoie and D. S. Pilch, J. Antimicrob. Chemother., 2015, 59, 4845–4855 CrossRef CAS PubMed.
- E. L. White, W. J. Suling, L. J. Ross, L. E. Seitz and R. C. Reynolds, J. Antimicrob. Chemother., 2002, 50, 111–114 CrossRef CAS PubMed.
- N. Sun, F. Y. Chan, Y. J. Lu, M. A. Neves, H. K. Lui, Y. Wang, K. Y. Chow, K. F. Chan, S. C. Yan, Y. C. Leung, R. Abagyan, T. H. Chan and K. Y. Wong, PLoS One, 2014, 9, e97514 CrossRef PubMed.
- N. Sun, Y. Y. Zheng, R. L. Du, S. Y. Cai, K. Zhang, L. Y. So, K. C. Cheung, C. Zhuo, Y. J. Lu and K. Y. Wong, Medchemcomm, 2017, 8, 1909–1913 RSC.
- A. Janas and P. Przybylski, Eur. J. Med. Chem., 2019, 182, 111662 CrossRef CAS PubMed.
- S. Singh, G. Kaur, V. Mangla and M. K. Gupta, J. Enzyme Inhib. Med. Chem., 2015, 30, 492–504 CrossRef CAS PubMed.
- N. Sun, L. Ban, M. Li, Z. Y. Fang, X. M. Li, W. X. Yao, J. H. Pan, Y. J. Lu, Z. H. Liu and W. L. Wong, J. Pharmacol. Sci., 2018, 138, 83–85 CrossRef CAS PubMed.
- N. Sun, R. L. Du, Y. Y. Zheng, Q. Guo, S. Y. Cai, Z. H. Liu, Z. Y. Fang, W. C. Yuan, T. Liu, X. M. Li, Y. J. Lu and K. Y. Wong, J. Enzyme Inhib. Med. Chem., 2018, 33, 879–889 CrossRef CAS PubMed.
- Y. J. Lu, Q. Deng, J. Q. Hou, D. P. Hu, Z. Y. Wang, K. Zhang, L. G. Luyt, W. L. Wong and C. F. Chow, ACS Chem. Biol., 2016, 11, 1019–1029 CrossRef CAS PubMed.
- W. Long, Y. J. Lu, K. Zhang, X. H. Huang, J. Q. Hou, S. Y. Cai, Y. Li, X. Du, L. G. Luyt, W. L. Wong and C. F. Chow, Dyes Pigm., 2018, 159, 449–456 CrossRef CAS.
- J. Meletiadis, C. Antachopoulos, T. Stergiopoulou, S. Pournaras, E. Roilides and T. J. Walsh, J. Antimicrob. Chemother., 2007, 51, 3329–3337 CrossRef CAS PubMed.
- F. C. Bi, D. Song, N. Zhang, Z. Y. Liu, X. J. Gu, C. Y. Hu, X. K. Cai, H. Venter and S. T. Ma, Eur. J. Med. Chem., 2018, 159, 90–103 CrossRef CAS PubMed.
- J. M. Andreu, C. Schaffner-Barbero, S. Huecas, D. Alonso, M. L. Lopez-Rodriguez, L. B. Ruiz-Avila, R. Nunez-Ramirez, O. Llorca and A. J. Martin-Galiano, J. Biol. Chem., 2010, 285, 14239–14246 CrossRef CAS PubMed.
- M. Kaul, L. Mark, Y. Z. Zhang, A. K. Parhi, E. J. LaVoie and D. S. Pilch, Biochem. Pharmacol., 2013, 86, 1699–1707 CrossRef CAS PubMed.
- Y. Li, J. Hsin, L. Zhao, Y. Cheng, W. Shang, K. C. Huang, H.-W. Wang and S. Ye, Science, 2013, 341, 392–394 CrossRef CAS PubMed.
- F. Y. Chan, N. Sun, M. A. Neves, P. C. Lam, W. H. Chung, L. K. Wong, H. Y. Chow, D. L. Ma, P. H. Chan, Y. C. Leung, T. H. Chan, R. Abagyan and K. Y. Wong, J. Chem. Inf. Model., 2013, 53, 2131–2140 CrossRef CAS PubMed.
- F. H. Stootman, D. M. Fisher, A. Rodger and J. R. Aldrich-Wright, Analyst, 2006, 131, 1145–1151 RSC.
- X. Xie, B. Choi, E. Largy, R. Guillot, A. Granzhan and M. P. Teulade-Fichou, Chem.–Eur. J., 2013, 19, 1214–1226 CrossRef CAS PubMed.
- T. K. Beuria, P. Singh, A. Surolia and D. Panda, Biochem. J., 2009, 423, 61–69 CrossRef CAS PubMed.
- T. K. Beuria, M. K. Santra and D. Panda, Biochemistry, 2005, 44, 16584–16593 CrossRef CAS PubMed.
- Z. J. Zheng, N. Tharmalingam, Q. Z. Liu, E. Jayamani, W. Kim, B. B. Fuchs, R. J. Zhang, A. Vilcinskas and E. Mylonakis, J. Antimicrob. Chemother., 2017, 61, e00686-17 CrossRef PubMed.
- D. J. Haydon, N. R. Stokes, R. Ure, G. Galbraith, J. M. Bennett, D. R. Brown, P. J. Baker, V. V. Barynin, D. W. Rice, S. E. Sedelnikova, J. R. Heal, J. M. Sheridan, S. T. Aiwale, P. K. Chauhan, A. Srivastava, A. Taneja, I. Collins, J. Errington and L. G. Czaplewski, Science, 2008, 321, 1673–1675 CrossRef CAS PubMed.
- H. K. Lui, W. Gao, K. C. Cheung, W. B. Jin, N. Sun, J. W. Y. Kan, I. L. K. Wong, J. Chiou, D. Lin, E. W. C. Chan, Y.-C. Leung, T. H. Chan, S. Chen, K.-F. Chan and K.-Y. Wong, Eur. J. Med. Chem., 2019, 163, 95–115 CrossRef CAS PubMed.
- M. Kaul, Y. Z. Zhang, A. K. Parhi, E. J. LaVoie and D. S. Pilch, Biochem. Pharmacol., 2014, 89, 321–328 CrossRef PubMed.
- Z. Q. Qin, J. Zhang, B. Xu, L. L. Chen, Y. Wu, X. M. Yang, X. Shen, S. Molin, A. Danchin, H. L. Jiang and D. Qu, BMC Microbiol., 2006, 6, 96 CrossRef PubMed.
- A. Mukherjee and J. Lutkenhaus, J. Bacteriol., 1999, 181, 823–832 CrossRef CAS PubMed.
- Z. Y. Fang, L. Ban, Y. A. Li, W. C. Yuan, Z. H. Liu, T. Liu, X. M. Li, K. Y. Wong, Y. J. Lu, N. Sun and X. G. Yao, J. Pharmacol. Sci., 2018, 137, 283–289 CrossRef CAS PubMed.
- S. Lim, S. Lee, S. H. Yi, Y. S. Son, S. M. Choi and Y. K. Kim, Evid.-Based Complementary Altern. Med., 2010, 7, 259–264 CrossRef PubMed.
- S. M. Lin, J. J. Koh, T. T. Aung, W. L. W. Sin, F. H. Lim, L. Wang, R. Lakshminarayanan, L. Zhou, D. T. H. Tan, D. R. Cao, R. W. Beuerman, L. Ren and S. P. Liu, J. Med. Chem., 2017, 60, 6152–6165 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00691b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |