Open Access Article
Open Access ArticleRapid and selective recognition of Vibrio parahaemolyticus assisted by perfluorinated alkoxysilane modified molecularly imprinted polymer film†
Kaiyue Fu‡
ab,
Huiwen Zhang‡a,
Yuanyuan Guo a,
Juan Li
a,
Juan Li *a,
Heran Niec,
Xiuling Songa,
Kun Xu
*a,
Heran Niec,
Xiuling Songa,
Kun Xu a,
Juan Wang
a,
Juan Wang a and
Chao Zhao
a and
Chao Zhao *a
*a
aSchool of Public Health, Jilin University, Changchun, Jilin 130021, PR China. E-mail: li_juan@jlu.edu.cn; czhao0529@jlu.edu.cn
bHebi Center for Disease Control and Prevention, Hebi, Henan 458030, PR China
cDalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, Liaoning 116023, PR China
First published on 7th April 2020
Abstract
Molecular imprinting technology offers a means of tailor-made materials with high affinity and selectivity for certain analysts. However, the recognition and separation of specific bacteria in complex matrices are still challenging. Herein, a bacteria-imprinted polydimethylsiloxane (PDMS) film was facilely prepared and modified with 1H,1H,2H,2H-perfluorooctyltriethoxysilane (POTS). Employing Vibrio parahaemolyticus as a model bacterium, the imprinted surface exhibited three-dimensionality cavities with mean size of 1000 × 800 nm in square and 100 nm in depth. After incubation for 2 h with 6 × 107 CFU mL−1 of V. parahaemolyticus, the imprinted polymer film can reach a 62.9% capture efficiency. Furthermore, the imprinted POTS-modified PDMS film based solid phase extraction combined with polymerase chain reaction and agarose gel electrophoresis allows for detecting 104 CFU mL−1 with excellent selectivity in fresh oyster samples. As a result, the developed selective sample pretreatment method using molecular imprinting technology provides a promising platform for separation, identification, and analysis of pathogens.
Introduction
Tissue infection, which is often caused by bacteria, is a major public health concern and its detection still faces limitations in clinical settings such as specificity, costs, and speed. For example, food poisoning and infectious diseases caused by foodborne pathogenic bacteria are serious public health concerns.1 Vibrio parahaemolyticus (V. parahaemolyticus) is a Gram-negative halophilic bacterium widely distributed in coastal and marine waters and frequently isolated from a variety of seafood, including codfish, sardine, mackerel, flounder, clam, octopus, shrimp, crab, lobster, crawfish, scallop and oyster.2 V. parahaemolyticus contamination can cause typical gastroenteritis reactions such as abdominal cramping, diarrhea, nausea, vomiting, fever and other typical gastroenteritis.3–6 The first reported outbreak of seafood borne disease due to V. parahaemolyticus was in Japan in 1950 in which 20 people were reported dead while over 270 people were likewise hospitalized.7 There also be outbreak in China, Spain, Chile, Peru and other coastal state. A survey about the incidence rate of V. parahaemolyticus from 1998–2013 in China, the positive rate of intestinal pathogenic bacteria of V. parahaemolyticus is up to 83%.8 Associated with health risk, this pathogen is a key indicator in the national monitoring plan for seafood in China.9 Traditionally, plate-culture and colony-counting9–11 is available to provide accurate validation and often recommended in national standards, but the method is tedious, laborious, and time consuming. In order to address these problems, polymerase chain reaction (PCR) and enzyme-linked immune-sorbent assay (ELISA) have been developed and applied to detect foodborne pathogens.12 PCR-based methods have become popular due to their reliability, specificity, and speed. Nevertheless, a lengthy culture-enrichment step for DNA extraction and purification is often still required prior to PCR.13 Based on the strong antigen–antibody interaction, the ELISA is a sensitive and selective method for pathogen screening. However, screening of antibodies is laborious, costly, time consuming. In addition, the natural antibodies are inherently fragile in a harsh environment.14 Therefore, it is critical to design and fabrication of artificial antibodies with easy availability and high stability, as alternatives to the natural antibodies.15,16Molecular imprinting technology (MIT) has been employed as a promising approach to synthesize artificial antibodies,17 which can create molecularly imprinted polymers (MIPs) with tailor-made binding sites complementary to the size, shape, and surface functional groups of template molecules.18 The synthesized artificial antibodies exhibit a natural antibody-like binding affinity and selectivity. To date, MIPs against small template molecules (MW < 1000) are widely applied for the development of biosensors and other fields,19 benefit from their easy availability and operability, high stability to harsh chemical and physical conditions, and potential reusability.20 However, imprinting supramolecular structures, like viruses and entire cells, is still a challenging field of research due to the fact that these entities cannot diffuse through a bulk imprinted material.21
To overcome these limitations, molecularly imprinted polymer film (IPF) has been used and further extended to bacteria-imprinted polymers (BIPs), which possess a bacterial durable and robust recognition property. Several different polymer types have been investigated as bacteria-imprinted materials,22–24 such as the silica.25–27 Among these, polydimethylsiloxane (PDMS) has attracted the most attention as a monomer for pathogen imprinting,28–30 because PDMS is an inexpensive and commercially available polymer with good biocompatibility. But the main drawback of PDMS is that many proteins may tend to adsorb onto the surface due to its the high intrinsic hydrophobicity.31 As a result, these non-specific interactions can lead to “false positives”. Therefore, the choice of the right substrate to modify the PDMS surface is one of the most critical factors for a successful bacterial imprinting.
With these insights, in this study, we fabricated bacteria-imprinted PDMS film with POTS-modified for V. parahaemolyticus enrichment and detection. Through the excellent adsorption property of glass slide, V. parahaemolyticus were coated onto its surface using generally Pickering emulsion polymerization without any additional pressure applied. This way can not only maintain the morphology of the bacteria, but also the operation was simple, convenient and easy to realize. As shown in Fig. 1, V. parahaemolyticus imprinted PDMS film were prepared on a microslide glass surface by microcontact imprinting method with the bacteria stamp. POTS as a fluorinated reagent was preferred to functionalize the film surface using an evaporation-deposition method, leading to the non-specific adhesion of bacteria significantly reduced. The capability of the imprinted film as an absorbent for efficient capture of V. parahaemolyticus was investigated, which was further compared with the non-imprinted PDMS film and the unmodified PDMS imprinted film. Additionally, based on the dual role of POTS-modified imprinted PDMS film for the capture of pathogens and DNA extraction, a method for rapid and selective detection of V. parahaemolyticus was performed with fresh oyster samples.
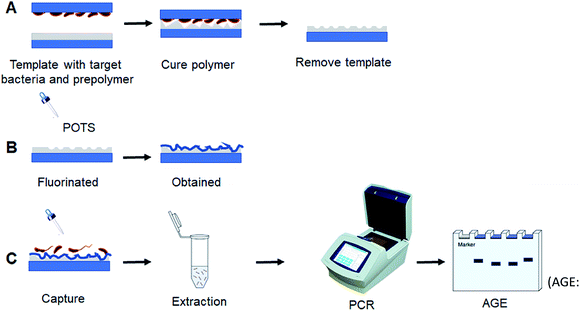 | ||
| Fig. 1 Schematic diagram for bacteria-imprinted film fabrication and PCR detection of V. parahaemolyticus (AGE: agarose gel electrophoresis). | ||
Materials and methods
Materials and reagents
PDMS was purchased from Momentive Performance Materials Inc. (NY, USA). Cyclohexane was obtained from Beijing Chemical Reagent Co., Ltd (Beijing, China). Glass microscope slides were supplied by Citotest Labware Manufacturing Co., Ltd (Haimen, China). POTS was purchased from Sigma-Aldrich (Shanghai, China). The commercial PCR kit, DNA molecular weight marker ranging from 25 to 500 bp and the primers in PCR experiments (Table 1) were obtained from Sangon BioTech Co., Ltd (Shanghai, China). Double-distilled water and phosphate-buffered saline (PBS, 0.01 mol L−1, pH 7.4) were prepared by us. All other the chemicals and reagents employed were of analytical grade and were used without any further purification.| Primer | Sequence (5′ to 3′) | Target gene | Amplicon size (bp) |
|---|---|---|---|
| V. parahaemolyticus-1 | GTAAAGGTCTCTGACTTTTGGAC | tdh | 269 |
| V. parahaemolyticus-2 | TGGAATAGAACCTTCATCTTCACC | ||
| E. coli O157:H7-1 | TTGACCCACACTTTGCCGTAA | uidA | 227 |
| E. coli O157:H7-2 | GCGAAAACTGTGGAATTGGG |
Bacteria culture
All the bacterial strains listed in Table 2 were stored at −80 °C with 15% glycerol and revived by streaking on Luria–Bertani (LB) agar plates. They were cultivated in LB medium with/without 3% NaCl at 37 °C for 18 h to 24 h with shaking at 180 rpm, respectively. Subsequently, the cultures were harvest by centrifugation at 4000 rpm for 20 min at room temperature. 100 μL of the diluted suspensions was plated onto appropriate agar plates by spread plating, and bacterial counts were assessed and reported as CFU per mL.| Bacteria | Abbreviation | ATCC no. |
|---|---|---|
| Vibrio parahaemolyticus | V. parahaemolyticus | 17802 |
| Salmonella typhimurium | S. typhimurium | 13311 |
| Listeria monocytogenes | L. monocytogenes | 19111 |
| Escherichia coli O157:H7 | E. coli O157:H7 | 25922 |
| Staphylococcus aureus | S. aureus | 25923 |
Preparation of bacterial template
The bacterial template was prepared according to the previous literatures with fewer modifications.28,30 Firstly, glass microscope slide (75 × 25 mm) was washed with absolute ethyl alcohol and deionized water for 15 min, respectively, and then was treated with acidic Piranha solution (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, H2SO4/H2O2, v/v) for 1 h. A 120 μL suspension of the formalin-inactivated V. parahaemolyticus (109 CFU mL−1) was spread on a precleaned glass microscope slide and kept at 4 °C for 30 min to settlement of bacteria. And then the glass microscope slide was placed on a spin coater and further centrifuged at 1500 rpm for 1 min to remove excess solvent. The ready-made bacterial template was used as a stamp in the next step.
1, H2SO4/H2O2, v/v) for 1 h. A 120 μL suspension of the formalin-inactivated V. parahaemolyticus (109 CFU mL−1) was spread on a precleaned glass microscope slide and kept at 4 °C for 30 min to settlement of bacteria. And then the glass microscope slide was placed on a spin coater and further centrifuged at 1500 rpm for 1 min to remove excess solvent. The ready-made bacterial template was used as a stamp in the next step.
Fabrication of bacteria-imprinted film with POTS modification
The bacteria-imprinted PDMS film was prepared according to the method previously reported (Schirhagl, et al. 2012).30 Briefly, 10 g of pre-polymer PDMS monomer and 1 g of PDMS cross-linker was diluted with 5.5 g of cyclohexane. The mixture was then degassed by using a vacuum pump. A certain volume of pre-polymerization solution was spin-coated onto a precleaned glass microscope slide (75 × 25 mm) and subsequently pre-cured on a hot plate at 80 °C for 2 min to enhance the viscosity of the pre-polymer, preventing the bacteria from sinking too deeply into the PDMS film. Afterwards, the obtained bacterial template stamp with adhered V. parahaemolyticus was pressed into the pre-polymer. The polymer was cured at room temperature overnight, followed by further curing at 80 °C for 1 h. The imprinted film was cleaned with distilled water by sonicating for 15 min and dried in air. As a control, a non-imprinted PDMS film (NIP) was simultaneously prepared by the above method without the use of bacterial template. The fluorinated PDMS film was produced by evaporation-deposition method using POTS. Firstly, the imprinted film was placed in an airtight jar, which containing 800 μL of POTS solution at the bottom. Secondly, the jar was put into an oven and heated at 180 °C for 6 h. Finally, the slide was taken off and kept at 4 °C in a closed Petri dish until use.Detection of V. parahaemolyticus by PCR amplification
Bacteria samples with varying concentrations (0, 102, 103, 104, 105, 106, 107, and 108 CFU mL−1) were prepared by diluting the freshly cultured bacteria with sterile water. 1 mL of each V. parahaemolyticus standard solution and the imprinted POTS-modified PDMS film on a piece of slide (75 × 25 mm) were added into slide box, which containing 9 mL of water. In order to effectively capture the bacteria in a large volume of sample solutions, the suspension was gently vortexed with a rocking shaker at room temperature for 2 h. Next, the slide was taken off and rinsed three times with water to remove unbound bacteria.For PCR amplification, the template DNA was extracted by boiling method. Briefly, the imprinted film was peeled off from the glass microscope slide and transferred to a 5 mL sterile Eppendorf tube containing 2 mL of sterile water. The tube was placed in a boiling water bath for 10 min to lyse the bacterial cells. The mixture was thoroughly mixed by vortexing and then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C, and the supernatant was used as template. The PCR was carried out in 25 μL reaction mixtures containing 5 μL of template DNA, 1 μL of each 10 μM primer, 12.5 μL 2× SYBR green master mix, 4.5 μL of 25 mM MgCl2, and 1 μL PCR water, and the following cycling parameters were used in a DNA thermal cycler (PerkinElmer, Model 480): 35 cycles (denaturation at 94 °C for 30 s, primer annealing at 55 °C for 30 s, primer extension at 72 °C for 30 s), were performed after an initial denaturation at 94 °C for 4 min. Following the end of the amplification cycles, sample was kept at 72 °C for 10 min to allow the final extension of the incompletely synthesized DNA. The PCR products were analysed by 2% agarose gel electrophoresis.
000 rpm for 10 min at 4 °C, and the supernatant was used as template. The PCR was carried out in 25 μL reaction mixtures containing 5 μL of template DNA, 1 μL of each 10 μM primer, 12.5 μL 2× SYBR green master mix, 4.5 μL of 25 mM MgCl2, and 1 μL PCR water, and the following cycling parameters were used in a DNA thermal cycler (PerkinElmer, Model 480): 35 cycles (denaturation at 94 °C for 30 s, primer annealing at 55 °C for 30 s, primer extension at 72 °C for 30 s), were performed after an initial denaturation at 94 °C for 4 min. Following the end of the amplification cycles, sample was kept at 72 °C for 10 min to allow the final extension of the incompletely synthesized DNA. The PCR products were analysed by 2% agarose gel electrophoresis.
Detection of V. parahaemolyticus in real samples
The oyster samples were purchased from a local supermarket and contaminated with different concentrations of V. parahaemolyticus (104 to 108 CFU mL−1) as described in detail in our previous work (Liu, Zhao, Fu, et al. 2017;4 Liu, Zhao, Song et al. 2017).5 The detection protocol was described in section Detection of V. parahaemolyticus by PCR amplification, except that pure PBS buffer was replaced with oyster samples.Results and discussion
Characterization of bacteria-imprinted film with POTS modification
V. parahaemolyticus imprinted PDMS film with a thickness of 2 mm were prepared by microcontact imprinting method with the bacterial stamp and further surface modified by vapor deposition of POTS (Fig. 1). AFM images were shown in Fig. 2 for surface morphology of 3D bacteria-imprinted film at various fabrication stages. Besides, lower magnification AFM images of imprinted PDMS film and POTS-modified imprinted PDMS film were shown in Fig. S1 and S2† for measurement of the cavity size. After the removal of the template bacteria, Fig. 2A showed crater-like micron-sized imprint on the PDMS film. It's a common sense that V. parahaemolyticus are polymorphic, such as arcs, short rods, spheres, etc. Since V. parahaemolyticus used here were inactivated, they mainly exist in sphere or short rod in the non-physiological state,32,33 which was consistent with the cavity observed by AFM after the bacteria washed away. The cavity was measured to be about 1000 × 800 nm in square and 100 nm in depth, which is similar with the common size of target bacterium (Fig. 2B). Upon a layer of POTS vapor-deposited on PDMS film, the shape of the cavity remained (Fig. 2C), and the size of the cavity was further narrowed to 900 × 800 nm in square (Fig. 2D). After captured V. parahaemolyticus, the cavities in the film were filled (Fig. S3†). Furthermore, ATR-IR and SEM-EDS were performed to confirm the composition of POTS-modified imprinted PDMS film. ATR-IR spectrum was shown in Fig. S4.† As reported in the literature,34 the main absorption peaks at 920–905 cm−1, 1080–1050 cm−1, 1220–1200 cm−1 can be assigned to the Si–OH, Si–O–Si, and Si–CH2, respectively. Additionally, with respect to the new peak at around 1720 cm−1, it can be attributed to the characteristic absorption band of triethoxy, which indicated that POTS was successfully deposited onto the surface of the imprinted PDMS film. SEM-EDS was used to map chemical elements on the surface of POTS-modified imprinted PDMS film. As shown in Fig. S5,† the initial imprinted PDMS film exhibit the characteristic peaks of C, O, and Si (black line). After POTS modification, a new characteristic peak of F appeared (red line). All these results implied that POTS-modified imprinted PDMS film were expectedly prepared.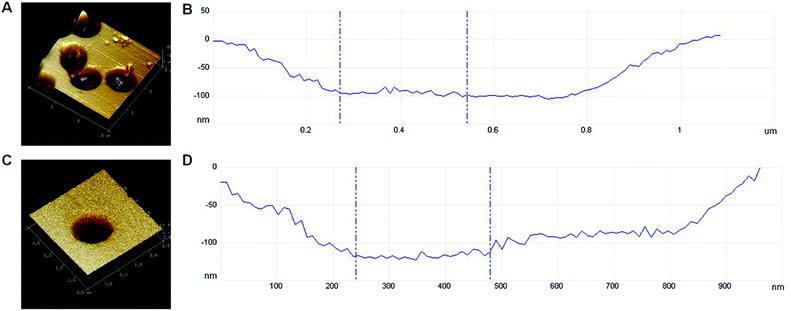 | ||
| Fig. 2 (A) AFM image of imprinted PDMS film; (B) cavity size of imprinted PDMS film; (C) AFM image of POTS-modified imprinted PDMS film; (D) cavity size of POTS-modified imprinted PDMS film. | ||
The adsorption capacity of the imprinted PDMS film with bacteria was based on the superhydrophobic effect.35 The film modified with POTS can increase surface roughness due to the Si–CH3 groups are replaced by CF3 by fluorination, which can reduce the hydrophobic interaction between the imprinted PDMS film and non-target bacteria leading to the inhibition of the non-specific adsorption effect. To verify the effect of POTS modification, static contact angle data were measured to characterize the hydrophobic properties (Fig. 3). Static contact angle values of the non-imprinted PDMS film (NIP) and the imprinted PDMS film were 118.1–118.0 (Fig. 3A) and 114.5–114.5 (Fig. 3C), respectively, that indicated both of them were hydrophobic. After POTS was fabricated to the surface of two kinds film, the static contact angle value was significant reduced to 107.7–107.6 (Fig. 3B) and 107.8 ± 0.4 (Fig. 3D), suggesting the hydrophobic effect decreased.
| Lag angle = advanced angle − back angle |
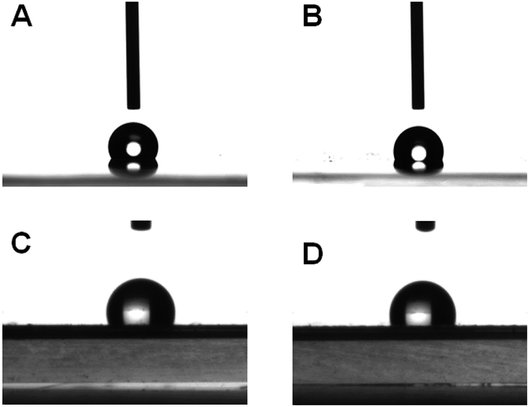 | ||
| Fig. 3 Static contact angles of (A) non-imprinted PDMS film (NIP), (B) POTS-modified non-imprinted PDMS film (POTS-modified NIP), (C) imprinted PDMS film, and (D) POTS-modified imprinted PDMS film. | ||
In addition, the dynamic contact angles of the films with/without modification were measured (Table 3). After modified with POTS, the lag angle values of POTS-modified imprinted PDMS film were reduced compared to the POTS-modified NIP. Because the lag angle represents the viscosity of the surface, the decreased viscosity demonstrated the affinity of POTS-modified imprinted PDMS film to bacteria were weakened. The above results suggested the non-target bacteria even in complex matrix could be easily washed off, whereas the target can be captured and kept in the cavity with special structure. The specific dynamic process of dynamic contact angles were shown in ESI.†
| Classification | Advanced angle | Back angle | Lag angle |
|---|---|---|---|
| NIP | 125.0857 | 95.59065 | 29.49505 |
| POTS-modified NIP | 110.8186 | 105.2685 | 5.5501 |
| Imprinted PDMS film | 119.7184 | 85.59501 | 34.12339 |
| POTS-modified imprinted PDMS film | 108.703 | 103.4097 | 5.2933 |
| POTS-modified imprinted PDMS film + V. parahaemolyticus | 116.4673 | 96.11665 | 20.35065 |
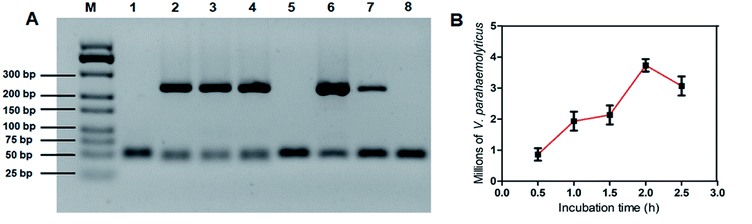
To confirm that our imprinted POTS-modified PDMS film can selectively recognition of target bacteria without non-specific affinity, both of V. parahaemolyticus and E. coli O157:H7 at 108 CFU mL−1 were monitored by PCR method. As shown in Fig. 4A, the PCR products of the two species of bacteria appeared in the electrophoretogram using unmodified imprinted PDMS film as an absorbent. In the contrast, the amplicon band pattern of E. coli O157:H7 was totally eliminated after fluorination, indicating their high selectivity.
Next, to estimate the adsorption capacity of the imprinted film, an excess of V. parahaemolyticus at 6 × 107 CFU mL−1 was adopted as a model. In our procedure, 10 mL of V. parahaemolyticus suspension was incubated with a piece of imprinted film at various time intervals. Negative control was tested without using the imprinted film under the same conditions. The supernatant was then plated with proper dilution on the LB agar for culture and counting colonies. According to previous work,4,5 corresponding adsorption efficiency percentages were calculated based on the following formula:
| Adsorption efficiency = (N0 − Ns)/N0 × 100% | (1) |
Detection of V. parahaemolyticus
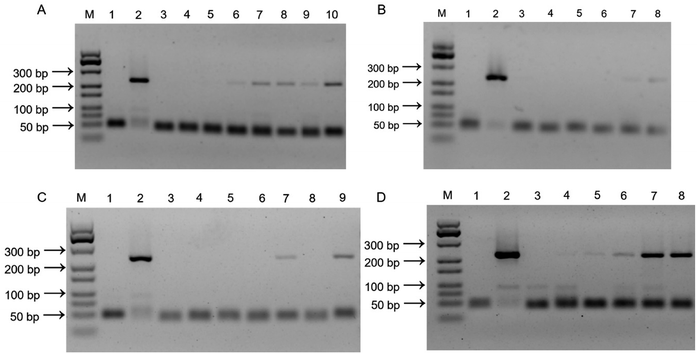
The performance of the PCR-based method using the imprinted film as an adsorbent for the enrichment of V. parahaemolyticus was experimented. Fig. 5A shows the electropherograms of a GeneRuler 500 bp DNA ladder, the PCR products from negative control, positive control, and different concentration of V. parahaemolyticus from 102 to 108 CFU mL−1, respectively. The limit of detection (LOD) for V. parahaemolyticus was determined to be 104 CFU mL−1, which showed a distinct difference in 269 bp band from that of the negative control sample. With increasing of the concentration of V. parahaemolyticus, the 269 bp band of target amplicon dramatically changed from light to deep. The non-imprinted film was also chosen as a control. As presented in the images in Fig. 5B, none of the target gene tdh was amplified with V. parahaemolyticus concentration range of 104 to 107 CFU mL−1. Only a weak band appeared at the bacteria concentration of 108 CFU mL−1. Because the PDMS arrangement on the NIP was not imprinted by bacteria template, there was a weak binding force for bacterial adhesion. Based on the above results, it can be concluded that the bacteria-templated cavity is decisive factor in bacterial recognition.Herein, the specific adsorption capacity of the imprinted film was examined against 4 other common pathogenic bacteria, including two Gram-positive bacteria (L. monocytogenes and S. aureus) and two Gram-negative bacteria (E. coli O157:H7 and S. typhimurium). Fig. 5C shows the samples contained interfering bacteria and negative control sample displayed no characteristic DNA band in the agarose gel electrophoresis due to inadequate interaction between the imprinted film and interfering bacteria. On the contrary, a significant 269 bp band of target amplicon was observed in the presence of V. parahaemolyticus alone, V. parahaemolyticus mixed with other bacteria, and positive control sample. These results proved that the imprinted film was highly specific for V. parahaemolyticus and can distinguish V. parahaemolyticus from other bacteria because of the POTS modification, contributed to the bacterial outer surface templated PDMS arrangement.
Detection of V. parahaemolyticus in real samples
It was difficult to obtain fresh oysters from the market that already carried the target bacteria. In order to verify the feasibility of the imprinted film used as an absorbent for enrichment of V. parahaemolyticus in real samples, a series of V. parahaemolyticus concentration (104 to 108 CFU mL−1) was spiked to the sterile oyster samples and detected by PCR-based method. As shown in Fig. 5D, the 269 bp band of target amplicon appeared in the positive samples, which is the amplification product of the specific nuclease gene tdh of V. parahaemolyticus. The LOD and working range were not interfered, indicating that using our proposed imprinted film modified with POTS has strong anti-interference ability for capture V. parahaemolyticus in complex food matrices. The lengthy culture-enrichment step can be averted prior to PCR. Simultaneously, the elution step of bacteria from the imprinted film was eliminated. The entire detection process only took an average of 5 h, including the capture of bacteria (2 h), lysis of bacteria and DNA extraction (10 min), PCR (2 h), agarose gel electrophoresis (50 min). The developed method was compared with traditional PCR-based methods aiming at detection of bacteria,36–39 as shown in Table 4. The method has considerable advantages over most of traditional PCR-based methods in terms of analytical time with a similar detection limit.| Methods | Pre-enrichment time | Detection limit (CFU mL−1) | References |
|---|---|---|---|
| Multiplex PCR | 6 h | 104 | Federici et al. 2018 (ref. 3) |
| Reverse transcriptase-PCR-denatured gradient gel electrophoresis (ReVT-PCR-DGGE) | 24 h | 102 | Chahorm and Prakitchaiwattana 2018 (ref. 36) |
| Colorimetric integrated PCR | 12 h | 2.9 × 104 | Cheng et al. 2016 (ref. 37) |
| Multiplex touchdown PCR | 16 h | 103 | Wei et al. 2014 (ref. 38) |
| POTS-modified imprinted PDMS film-PCR-agarose gel electrophoresis | 2 h | 104 | This work |
Conclusions
In summary, a bacteria-imprinted PDMS film with surface modification by POTS for highly specific bacteria recognition was synthesized which had a thickness of 2 mm. The increase of roughness and the decrease of viscosity caused by POTS modification reduced the possibility of non-specific adsorption. Overall, the merit of high specificity for the POTS-modified imprinted film was added on the base of simplicity, rapidity and low cost. Employing V. parahaemolyticus as a model bacterium, the imprinted surface exhibited three-dimensionality cavities with mean size of 1000 × 800 nm in square and 100 nm in depth. As a result, after incubation for 2 h with 6 × 107 CFU mL−1 of V. parahaemolyticus, the imprinted polymer film can reach a 62.9% capture efficiency. Furthermore, a PCR method for detection of V. parahaemolyticus in fresh oyster samples was established based on the solid phase extraction of the imprinted polymer film. This method processes excellent selectivity and allows for detecting 104 CFU mL−1 of Vibrio parahaemolyticus with a wide linear arrange from 104 to 108 CFU mL−1. Therefore, we envision that this fabrication strategy may open up a new avenue for designing novel recognition platform for biomacromolecules.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the financial support from the National Natural Science Foundation of China (Grant No. 81602895, 81602894, and 81872668), China Postdoctoral Science Foundation (Grant No. 2017T100214 and 2016M591492), the Science and Technology Development of Jilin Province (Grant No. 20170204003SF and 20180520132JH), and the Education Department of Jilin Province (Grant No. JJKH20170873KJ and JJKH20180240KJ).Notes and references
- A. A. Mostafa, A. A. Al-Askar, K. S. Almaary, T. M. Dawoud, E. N. Sholkamy and M. M. Bakri, Saudi J. Biol. Sci., 2018, 25(2), 361–366 CrossRef PubMed.
- T. H. T. Tran, H. Yanagawa, K. T. Nguyen, Y. Hara-Kudo, T. Taniguchi and H. Hayashidani, J. Vet. Med. Sci., 2018, 80(11), 1737–1742 CrossRef CAS PubMed.
- S. Federici, D. I. Serrazanetti, M. E. Guerzoni, R. Campana, E. Ciandrini, W. Baffone and A. Gianotti, J. Food Sci. Technol., 2018, 55(2), 749–759 CrossRef CAS PubMed.
- Y. Liu, C. Zhao, K. Fu, X. Song, K. Xu, J. Wang and J. Li, Food Control, 2017, 80, 380–387 CrossRef CAS.
- Y. Liu, C. Zhao, X. Song, K. Xu, J. Wang and J. Li, Microchim. Acta, 2017, 184(12), 4785–4792 CrossRef CAS.
- J. Tang, J. Jia, Y. Chen, X. Huang, X. Zhang, L. Zhao and Z. Wu, Curr. Microbiol., 2018, 75(1), 20–26 CrossRef CAS PubMed.
- O. A. Odeyemi, SpringerPlus, 2016, 5(1), 464 CrossRef PubMed.
- X. L. Qi, H. X. Wang, S. R. Bu, X. G. Xu, X. Y. Wu and D. F. Lin, J. Infect. Dev. Countries, 2016, 10(2), 127–133 CrossRef PubMed.
- Y. Wang, X. S. Shen, R. R. Gu, Y. F. Shi and L. L. Tian, J. Food Saf., 2015, 35(1), 26–31 CrossRef CAS.
- L. Wang, J. Zhang, H. Bai, X. Li, P. Lv and A. Guo, Appl. Biochem. Biotechnol., 2014, 173(5), 1073–1082 CrossRef CAS PubMed.
- J. Zeng, H. Wei, L. Zhang, X. Liu, H. Zhang, J. Cheng and L. Liu, Int. J. Food Microbiol., 2014, 174, 123–128 CrossRef CAS PubMed.
- B. Priyanka, R. K. Patil and S. Dwarakanath, Indian J. Med. Res., 2016, 144(3), 327–338 CrossRef CAS PubMed.
- Y. Liu, Y. Cao, T. Wang, Q. Dong, J. Li and C. Niu, Front. Microbiol., 2019, 10, 222 CrossRef PubMed.
- K. A. Janes, Sci. Signal., 2020, 13(616), eaaz8130 CrossRef CAS PubMed.
- J. J. Xu, H. Miao, J. Wang and G. P. Pan, Small, 2020, e1906644 CrossRef PubMed.
- Z. Zhang, X. Yu, J. Zhao, X. Shi, A. Sun, H. Jiao, T. Xiao, D. Li and J. Chen, Chemosphere, 2020, 246, 125622 CrossRef CAS PubMed.
- T. Takeuchi and H. Sunayama, Chem. Commun., 2018, 54(49), 6243–6251 RSC.
- R. Li, Y. Feng, G. Pan and L. Liu, Sensors, 2019, 19(1), 177 CrossRef PubMed.
- Y. Gu, J. Wang, H. Shi, M. Pan, B. Liu, G. Fang and S. Wang, Biosens. Bioelectron., 2019, 128, 129–136 CrossRef CAS PubMed.
- Z. Liu, X. Zhang, A. Liang, L. Sun and A. Luo, Sepu, 2019, 37(3), 287–292 CAS.
- R. Schirhagl, Anal. Chem., 2014, 86(1), 250–261 CrossRef CAS PubMed.
- C. Alexander and E. N. Vulfson, Adv. Mater., 1997, 9(9), 751–755 CrossRef CAS.
- O. Hayden, D. Podlipna, X. Chen, S. Krassnig, A. Leidl and F. L. Dickert, Mater. Sci. Eng., C, 2006, 26(5–7), 924–928 CrossRef CAS.
- Y. Saylan, F. Yilmaz, E. Ozgur, A. Derazshamshir, H. Yavuz and A. Denizli, Sensors, 2017, 17(4), 898 CrossRef PubMed.
- W. Cai, H.-H. Li, Z.-X. Lu and M. M. Collinson, Analyst, 2018, 143(2), 555–563 RSC.
- D. Sen, J. Bahadur, A. Das, S. Mazumder, J. S. Melo, H. Frielinghaus and R. Loidl, Colloids Surf., B, 2015, 127, 164–171 CrossRef CAS PubMed.
- M. A. Hamed Jafari, E. Abdi, S. Latifi Navid, J. Bouckaert, R. Jijie, R. Boukherroub and S. Szunerits, Biosens. Bioelectron., 2019, 124–125, 161–166 CrossRef PubMed.
- A. Karthik, K. Margulis, K. Ren, R. N. Zare and L. W. Leung, Nanoscale, 2015, 7(45), 18998–19003 RSC.
- K. Ren, N. Banaei and R. N. Zare, ACS Nano, 2013, 7(7), 6031–6036 CrossRef CAS PubMed.
- R. Schirhagl, E. W. Hall, I. Fuereder and R. N. Zare, Analyst, 2012, 137(6), 1495–1499 RSC.
- D. Wang, M. Douma, B. Swift, R. D. Oleschuk and J. H. Horton, J. Colloid Interface Sci., 2009, 331(1), 90–97 CrossRef CAS PubMed.
- S.-Y. Chen, W.-N. Jane, Y.-S. Chen and H. Wong, Int. J. Food Microbiol., 2009, 129(2), 157–165 CrossRef CAS PubMed.
- C.-P. Su, W.-N. Jane and H. Wong, Int. J. Food Microbiol., 2013, 160(3), 360–366 CrossRef PubMed.
- V. Sunkara and Y.-K. Cho, ACS Appl. Mater. Interfaces, 2012, 4(12), 6537–6544 CrossRef CAS PubMed.
- J. K. Sakkos, B. R. Mutlu, L. P. Wackett and A. Aksan, ACS Appl. Mater. Interfaces, 2017, 9(32), 26848–26858 CrossRef CAS PubMed.
- K. Chahorm and C. Prakitchaiwattana, Int. J. Food Microbiol., 2018, 264, 46–52 CrossRef CAS PubMed.
- K. Cheng, D. Pan, J. Teng, L. Yao, Y. Ye, F. Xue and W. Chen, Sensors, 2016, 16(10), 1600 CrossRef PubMed.
- S. Wei, H. Zhao, Y. Xian, M. A. Hussain and X. Wu, Diagn. Microbiol. Infect. Dis., 2014, 79(2), 115–118 CrossRef CAS PubMed.
- J. F. Yin, M. Y. Wang, Y. J. Chen, H. Q. Yin, Y. Wang, M. Q. Lin and C. J. Hu, Surg. Infect., 2018, 19(1), 48–53 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00306a |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |