Open Access Article
Open Access ArticleAn electrochemical aptasensor for lead ion detection based on catalytic hairpin assembly and porous carbon supported platinum as signal amplification†
Huali Jina,
Di Zhanga,
Yong Liu b and
Min Wei
b and
Min Wei *a
*a
aCollege of Food Science and Technology, Henan Key Laboratory of Cereal and Oil Food Safety Inspection and Control, Henan University of Technology, Zhengzhou 450001, PR China
bCollege of Chemistry and Chemical Engineering, Henan University, Kaifeng, 475004, PR China. E-mail: wei_min80@163.com
First published on 12th February 2020
Abstract
An electrochemical aptasensor is fabricated for lead ion (Pb2+) detection based on catalytic hairpin assembly as signal amplification. Biomass porous carbon derived from soybean straw (PCs) is used to load platinum nanoparticles (PtNPs), which are introduced as mimetic enzymes to catalyze the hydroquinone–H2O2 system. In the presence of Pb2+, the complementary DNA (cDNA) releases from aptamer-cDNA and hybridizes with hairpin DNA1 (HP1) on the electrode surface. After bio-hairpin DNA2 (bio-HP2) is added, hybridization between HP1 and bio-HP2 further releases cDNA, which participates in the next cycle and triggers amplification, eventually forming a large number of bio-HP2/HP1 on the electrode surface. Then PtNPs@PCs immobilize on the electrode surface by specific binding of streptavidin with biotin, and catalyze the oxidation of hydroquinone in the presence of H2O2. The produced electrochemical signal depends on the concentration of Pb2+. The developed biosensor exhibits a wide linear range from 50 pM to 1000 nM with a detection limit of 18 pM, and high selectivity for Pb2+ over other environmentally relevant metal ions at concentration ratios of 100. The recoveries of Pb2+ in real samples were 93.5–108%. Thus, the proposed biosensor can provide an efficient method for sensitive and selective detection of Pb2+ in real samples.
1. Introduction
As one of the most hazardous heavy metal contaminants, lead ions (Pb2+) can exist in the environment for a long time and accumulate in the body through skin absorption, the digestive tract or food chain, leading to nerve disorders, memory decay, or anemia even at very low level exposure, particularly in children.1,2 The World Health Organization (WHO) and the US Environmental Protection Agency (EPA) regulate that the maximum acceptable concentration of Pb2+ in drinking water is 48 nM and 72 nM, respectively.3,4 Recent studies reported that the adverse effects of Pb2+ are widely recognized as toxic at concentrations as low as 1 pM.5,6 Thus, it is of great value to construct simple, ultrasensitive and rapid methods for Pb2+ detection. Many methods such as atomic absorption spectrometry (AAS), inductively coupled plasma atomic emission spectrometry (ICP-AES), and inductively coupled plasma mass spectrometry (ICP-MS) have been reported for the detection of Pb2+.7–9 Although these conventional analytical methods are sensitive and accurate, they are labor-intensive, time-consuming, and expensive, which restrict their wider applications.10 At present, electrochemical sensors with different signal amplification strategies have been widely utilized in the detection of Pb2+ owing to its simple instrumentation, rapid response, low cost and high sensitivity.11–13As a kind of novel recognition element, aptamers exhibit many advantages, such as easy synthesis, excellent stability and strong specificity.14,15 Based on these unique properties, many electrochemical sensors based on aptamers have been fabricated for ultrasensitive detection of Pb2+.16,17
In order to further amplify response signal and enhance the sensitivity, different signal amplification techniques including palindromic molecular beacon (PMB),18 rolling circle amplification (RCA),19 DNA walker,20 hybridization chain reaction (HCR)21 and catalytic hairpin assembly (CHA)22 have attracted increasing attention. Among them, as isothermal nucleic acid amplification strategy, with the simple reaction conditions, high sensitivity and the low costs, CHA is powerful for amplifying and transducing signals at the terminus of nucleic acid amplification reactions,23,24 and has been broadly applied in electrochemical sensing platforms.25,26 The presence of initiator DNA can trigger CHA process to achieve signal amplification.27 DNAzyme with the existence of Pb2+ can act as initiator for triggering CHA process, and has been successfully used for Pb2+ detection.28,29 However, Pb2+ dependent DNAzyme cleaves RNA irreversibly, and leads to the destroyed structure, high cost and hard reuse, which limit its wider applications.30,31 Instead, aptamer only changes the structure and can be reusable. To our best knowledge, aptamer-based CHA for Pb2+ detection have not been reported. Herein, CHA based on Pb2+ aptamer is designed to realize the amplification strategy.
In recent years, compared with natural enzymes, due to the advantages of broader reaction conditions, higher stability, lower production cost, and better catalytic activity, mimetic enzymes based on different nanomaterials have been developed to construct biosensors.32–34 Among them, with the advantages of good electrical conductivity, larger surface activity, favorable catalytic ability, and wider range of pH and temperature conditions, Pt-based nanomaterials such as Pt nanotubes and Pt nanoparticles (PtNPs) have been widely used in electrochemical biosensors to obtain the better performance.35–37 In order to avoid the aggregation of Pt-based nanomaterials and improve the catalytic activity, different nanomaterials such as metal–organic frameworks38 and reduced graphene oxide39 were used as carriers to enhance the dispersity. Due to the unique properties of pore structure, high surface area, good conductivity, low production cost and environment-friendly preparation, biomass porous carbons (PCs) derived from soybean straw can be a good choice to support PtNPs for electrochemical sensors.
Herein, a novel label-free electrochemical aptasensor is developed to detect Pb2+. CHA based on Pb2+ aptamer is designed to realize the amplification strategy. PCs produced by soybean straw is introduced as the carrier of PtNPs to prepare PtNPs@PCs, which acts as peroxide mimetic enzyme to catalyze the oxidation of hydroquinone in the presence of hydrogen peroxide and realize the second signal amplification.
2. Experimental
The “Apparatus and electrochemical measurement” was provided in the ESI.†2.1. Reagents and materials
Streptavidin (SA) was purchased from Solarbio Science and Technology Co., Ltd. (Beijing, China). 6-Mercapto-1-hexanol (MCH) was purchased from Aladdin Co., Ltd. (Shandong province, China). Tris(hydroxymethyl)aminomethane (Tris) was obtained from Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). Tris(2-carboxyethyl)phosphine (TCEP) was purchased from shanghai yuanye Bio-Technology Co., Ltd. (Shanghai, China). Hydroquinone (HQ) and lead nitrate (Pb(NO3)2) were ordered from Kermel Co., Ltd. (Tianjin, China). H2O2 (30%, w/w) was obtained from Haohua Co., Ltd. (Luoyang, Henan Province, China). Soybean straw comes from farmland (Qi County, Henan Province, P. R. China). All used reagents were of analytical grade. The ultrapure water was used throughout the experiments. All DNA sequences were synthesized by Sangon Biotechnology (Shanghai, China) and listed below:Aptamer: 5′-GGG TGG GTG GGT GGG TAT-3′
Complementary DNA (cDNA): 5′-TCA TAC CCA CCC ACC-3′;
Hairpin DNA 1 (HP1): 5′-TTT TGG GTG GGT ATG ACC ACC GCC CAC CCA-3′
Bio-Hairpin DNA 2 (bio-HP2): 5′-bio-TAT GAC CAC CTG GGT GGG CGG TGG TCA TAC CCA C-3′
Each HP was heated at 95 °C for 5 min and then slowly cooled to room temperature before using. All oligonucleotide solutions for DNA hybridization reaction were prepared by using 20 mM Tris–HCl (pH 7.4) containing 140 mM NaCl and 5 mM MgCl2. 0.1 M phosphate buffered solution (PBS, pH 7.4) was obtains by mixing the 0.1 M Na2HPO4 and 0.1 M KH2PO4 as working buffer.
2.2. Preparation of PtNPs@PCs-SA
The honeycomb-like structure PCs was prepared by one-step pyrolysis using soybean straw as the precursor and magnesium oxide as the template. 1.0 g soybean straw was placed in the vibration mill together with 2.0 g magnesium oxide. After even grinding, the mixtures were carbonized at high temperature of 800 °C for 2 h with heating rate of 3 °C min−1 under the N2 atmosphere in a tube furnace. The obtained black solid was treated with 1 M HCl in order to remove pore-forming agent magnesium oxide. Then, the examples were washed with distilled water until the solution pH reached 7, and were dried at 60 °C for 12 h in the oven. So, PCs was obtained. PtNPs@PCs were prepared using the ethylene glycol (EG) reduction method according to the reported literatures.40 Finally, 2 mg of PtNPs@PCs was dispersed in PBS buffer, and then 2 mg of streptavidin was added with gently stirring for 12 h at 4 °C to sufficiently immobilize onto the surface of PtNPs@PCs via the strong Pt–N affinity.412.3. Fabrication of the electrochemical biosensor
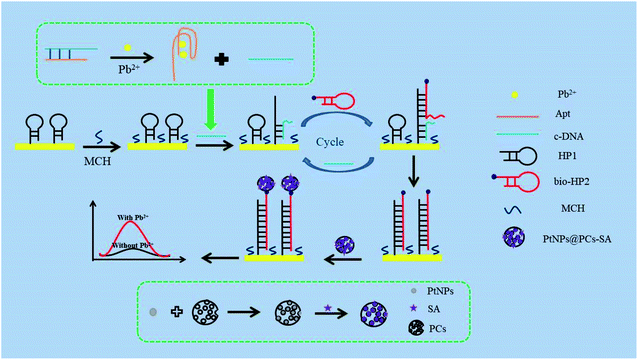
Firstly, the mixture of 3 μL of 4 μM Apt and 3 μL of 4 μM cDNA were heated to 95 °C for 5 min then slowly cooled to room temperature to form double-stranded structure. Then Pb2+ with different concentrations was added into above solution for 40 min at 37 °C to obtain the mixture of Pb2+/Apt and cDNA. 5 μL of 3.5 μM HP1 was incubated with 10 mM TCEP for 2 h to block the formed disulfide bonds, and then was added onto the surface of AuE at room temperature for 12 h to obtain the HP1/AuE. After washing with Tris–HCl buffer (pH 7.4), the HP1/AuE was blocked with MCH for 30 min at 37 °C. Then, the mixture of Pb2+/Apt and cDNA were added to electrode for 1.5 h at 37 °C. Next, 5 μL of 4 μM bio-HP2 was dropped onto the electrode and incubated for 1.5 h at 37 °C. Finally, 5 μL of PtNPs@PCs-SA was added to the electrode and incubated at 37 °C for 2 h.As shown in Scheme 1, the signal amplification strategy is conducted as follows: firstly, in the presence of Pb2+, the combination of Pb2+ and aptamer forms stable G-quadruplex structure, leading to the release of cDNA. Then, the released cDNA hybridizes with HP1 on the AuE surface. After bio-HP2 is added onto the AuE surface, its hairpin structure opens and hybridizes with HP1. Thus, the cDNA is replaced by bio-HP2 and released from HP1 for the next cycle. When the cyclic processes complete, there are a large number of capture probe (bio-HP2/HP1) left on the electrode. Finally, PtNPs@PCs-SA are immobilized on the electrode by the specific recognition between biotin and streptavidin. Thus, in the presence of H2O2, PtNPs@PCs acts as peroxide mimetic enzyme to catalyze the oxidation of HQ and realize the second signal amplification.
3. Results and discussion
3.1. Characterization of PtNPs@PCs
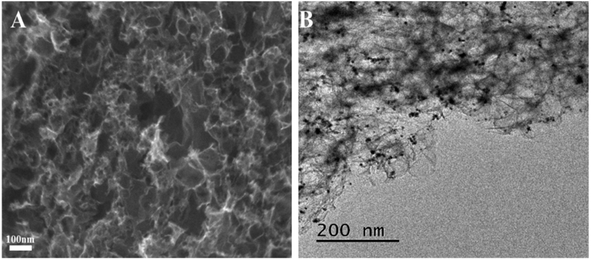
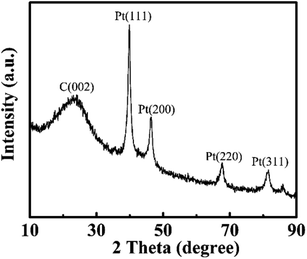
The prepared PCs and PtNPs@PCs were characterized by SEM and TEM, and the results were shown in Fig. 1. The SEM image of PCs (Fig. 1A) indicated that the obtained carbon materials were porous structure monolith after the removal of MgO template. As shown in Fig. 1B, it can be seen that PtNPs were uniformly spread on the surface of the porous of PCs.Fig. 2 showed the XRD patterns of PtNPs@PCs. The diffraction peak at 2θ of 39.9°, 45.5°, 67.7°, and 81.4° correspond to (110), (200), (220), and (311) crystalline planes of face-centered cubic structure of Pt, respectively. The diffraction peak at about 23.5° observed can be attributed to the C (002) facet of graphitic carbon.
In addition, DPV was used to characterize the mimic enzyme performance of PtNPs@PCs (Fig. S1†). When the bare electrode was placed in the PBS buffer containing HQ, the oxidation peak was measured at 0.136 V and the oxidation peak current was 22.09 μA (curve a). However, when the bare electrode was put into the PBS buffer with HQ and H2O2, the oxidation peak potential shifted negatively, and the oxidation peak current increased to 32.63 μA (curve b), indicating that H2O2 can promote the oxidation of HQ. When the PtNPs@PCs-modified electrode was placed in PBS buffer with HQ, the peak current increased significantly to 91.54 μA and the peak potential shifted negatively to 0.064 V (curve c), indicating that PtNPs@PCs has the effect of accelerating electron transfer. When the PtNPs@PCs-modified electrode was immersed in the PBS buffer with HQ and H2O2 (curve d), the peak current further increased to 113.4 μA and the peak potential shifted negatively to 0.044 V, which was ascribed that larger surface activity and good electrical conductivity of PtNPs@PCs make it act as peroxidase-like catalyst to catalyze H2O2 and accelerate the oxidation of HQ. Moreover, due to its good stability, PtNPs@PCs can be reusable and retains its catalytic activity in a wider range of pH and temperature, making it a great potential application.
3.2. Characterization of the proposed biosensor
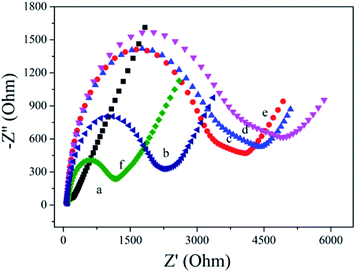
EIS is used to characterize the fabrication of biosensor. As shown in Fig. 3, the bare AuE showed a low Ret value (curve a). After HP1 was assembled on the AuE surface, the Ret value increased obviously (curve b), which is ascribed to the mutual repulsion between negatively charged phosphate group and redox probe. When the MCH was immobilized onto the HP1/AuE, the Ret value further increased (curve c), indicating that MCH blocks nonspecific sites on the electrode surface. When cDNA was introduced, the Ret value increased due to the negative charge of DNA (curve d). Similarly, the incubation of bio-HP2 also led to further increase of Ret value (curve e). With the introduction of PtNPs@PCs-SA (curve f), the Ret value apparently decreased due to that the formation of PtNPs@PCs-SA on the electrode promote the electron transfer.3.3. Electrochemical behavior of the biosensor
The prepared sensor is applied to the detection of Pb2+, and the result is shown in Fig. 4. In the absence of Pb2+, cDNA and Apt hybridizes to form the double-stranded DNA structure. Therefore, cDNA cannot reach the AuE surface to hybridize with HP1. The cycle amplification of CHA cannot be performed. So, the initial peak current was 0.65 μA (curve a). In the presence of 1 μM Pb2+, Apt combines with Pb2+ to form a G-quadruplex structure, resulting in that cDNA detached from the double-stranded DNA structure and hybridized with HP1 on the AuE surface, thereby opening the hairpin structure of HP1. With the introduction of bio-HP2, cDNA released from HP1 due to the greater number of complementary bases between bio-HP2 and HP1. The released cDNA participated the next cycle and further hybridized with HP1 to realize the catalytic hairpin assembly and achieve the amplification strategy. When the cyclic processes completed, there were a large number of bio-HP2/HP1 formed on the electrode. By combining with bio-HP2 by the specific recognition between biotin and streptavidin, PtNPs@PCs-SA can catalyze the oxidation of HQ in the presence of H2O2, thereby enhancing the peak current signal and realizing the detection of Pb2+. So, the peak current of 5.84 μA was found (curve b). The produced electrochemical signal depends on the concentration of Pb2+, which can be used for the detection of Pb2+.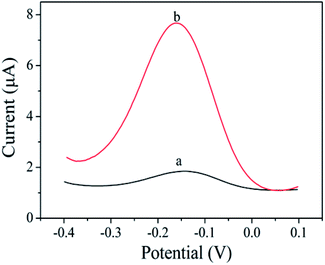 | ||
| Fig. 4 DPV curves of without Pb2+ (a) and with Pb2+ (1 μM) (b) in PBS (pH 7.4) buffer containing 10 mM HQ and 10 mM H2O2. | ||
3.4. Optimization of the experimental conditions
The experimental conditions including bio-HP2 concentration, HQ concentration and PtNPs@PCs incubation time are optimized. As depicted in Fig. 5A, by increasing the bio-HP2 concentration, more PtNPs@PCs were modified on the electrode surface by the combination of biotin and streptavidin, and the current response gradually increased. When the concentration of bio-HP2 was 4 μM, the current response reached the maximum. As the concentration continued to increase, the current value became stable. Thus, 4 μM was chosen as the optimal concentration of bio-HP2 for the following experiments. As shown in Fig. 5B, with the concentration of HQ increased, the current response gradually increased and tended to be steady after 10 mM. Thus, 10 mM was used as the optimal concentration of HQ. The incubation time of PtNPs@PCs was optimized, and the results were shown in Fig. 5C. As the incubation time increased, the current value gradually increased, and when the incubation time was 120 min, the current value reached the maximum. As the incubation time continued to increase, the current value slightly reduced. Therefore, 120 min was chosen as the optimal incubation time.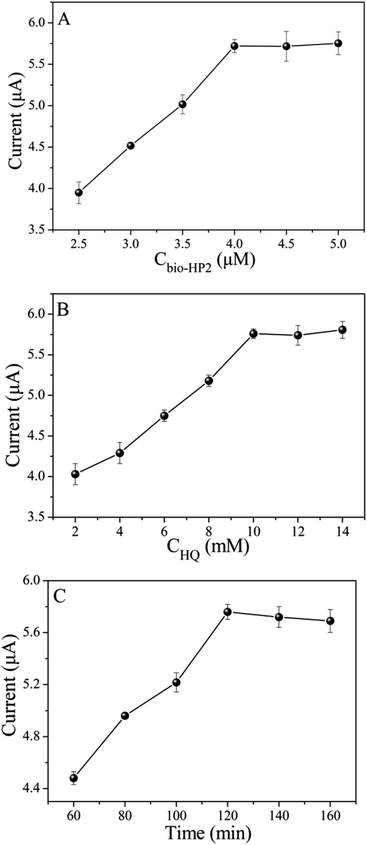 | ||
| Fig. 5 The influence of (A) bio-HP2 concentration, (B) HQ concentration, and (C) the incubation time of PtNPs@PCs on the biosensor. | ||
3.5. Analytical performance of biosensor
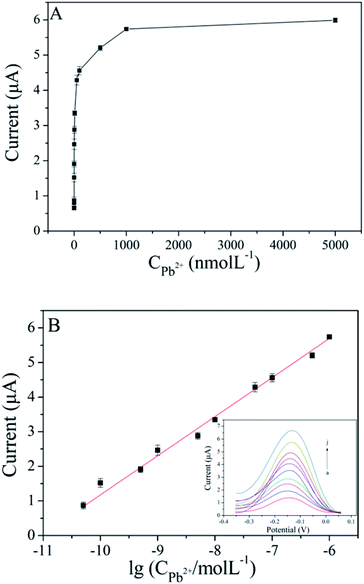
Different concentrations of Pb2+ are detected by the prepared biosensor. As shown in Fig. 6A, the peak current increased gradually with increasing Pb2+ concentration in the range of 0.05–5000 nM. From the results showed in Fig. 6B, a linear relationship between peak current and the logarithm of Pb2+ concentration from 0.05 nM to 1000 nM, and the linear regression equation was I = 1.13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CPb2+ + 12.47 (R2 = 0.995). The detection limit of 0.018 nM was calculated on the basis of 3 times the standard deviation above the blank. As shown in Table S1,† the performance of the proposed aptasensor is better or comparable with that of previously reported Pb2+ sensors.
CPb2+ + 12.47 (R2 = 0.995). The detection limit of 0.018 nM was calculated on the basis of 3 times the standard deviation above the blank. As shown in Table S1,† the performance of the proposed aptasensor is better or comparable with that of previously reported Pb2+ sensors.
3.6. Selectivity, reproducibility and stability of the biosensor
Under the same experimental conditions, some metal ions such as Cu2+, Co2+, Zn2+, Cd2+, Ni2+, Hg2+, Ba2+ and Fe3+ are used to evaluate the selectivity of the biosensor, and the results are shown in Fig. 7. The concentration of other metal ions is 100 μM, and that of Pb2+ is 1 μM. In the absence of Pb2+, the current response of other metal ions had no significant change compared with that of the measured blank solution. The current response of the mixture with Pb2+ was similar to that with Pb2+ only. These results indicated that the biosensor had an excellent selectivity for Pb2+ detection.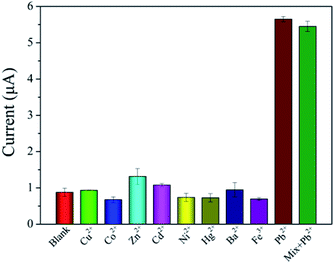 | ||
| Fig. 7 Selectivity of the prepared biosensor for Pb2+compared with other metal ions (n = 3). The concentration of Pb2+ is 1 μM and that of other metal ions are 100 μM. | ||
The inter-assay precision was evaluated by five times parallel measurements of 1 μM Pb2+, and relative standard deviation (RSD) value of 1.48% was obtained. The intra-assay precision was determined by five times repeated measurements on the same aptasensor, and RSD value of 4.69% was obtained. After a 15 day storage period at 4 °C, the aptasensor retained 88.1% of its initial current response, indicating the acceptable stability.
3.7. Analysis of Pb2+ in real samples
Different concentrations of Pb2+ at 500 nM, 10 nM and 0.1 nM were respectively added into tap water samples and lake samples, which were detected using the proposed biosensor. As shown in Table 1, the recoveries were in the range of 93.5–108%, suggesting that proposed biosensor can be used for Pb2+ detection in real samples.| Samples | Added (nM) | Found (nM) | Recovery (%) | RSD (%) (n = 3) |
|---|---|---|---|---|
| Tap water | 500.0 | 496.9 | 99.4 | 5.02 |
| 10.0 | 9.35 | 93.5 | 5.10 | |
| 0.1 | 0.105 | 104.9 | 4.71 | |
| Lake water | 500.0 | 477.3 | 95.5 | 4.38 |
| 10.0 | 10.8 | 108 | 3.11 | |
| 0.1 | 0.939 | 93.9 | 12.21 |
4. Conclusion
In summary, an electrochemical biosensor was fabricated for Pb2+ detection by catalytic hairpin assembly and PtNPs@PCs as signal amplification strategy. The developed biosensor exhibited higher sensitivity, good selectivity, and favorable recovery in water samples. Therefore, the proposed electrochemical biosensor has a potential application for Pb2+ detection in the environment, and can be expanded for the detection of other metal ions.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This study was funded by the National Key Research and Development Program of China (2018YFC1602800), Key Scientific and Technological Project of Henan Province (192102310255), and the Fundamental Research Funds for the Henan Provincial Colleges and Universities in Henan University of Technology (2016RCJH04).References
- J. N. Ding, Y. Liu, D. W. Zhang, M. L. Yu, X. J. Zhan, D. Zhang and P. Zhou, Microchim. Acta, 2018, 185, 545 CrossRef PubMed.
- L. Y. Wang, X. L. Peng, H. J. Fu, C. Huang, Y. P. Li and Z. M. Liu, Biosens. Bioelectron., 2020, 147, 111777 CrossRef CAS PubMed.
- F. L. Wang, Z. Wu, Y. X. Lu, J. Wang, J. H. Jiang and R. Q. Yu, Anal. Biochem., 2010, 405, 168–173 CrossRef CAS PubMed.
- L. Zhang, H. M. Deng, R. Yuan and Y. L. Yuan, Microchim. Acta, 2019, 186, 709 CrossRef PubMed.
- F. Long, A. N. Zhu and H. C. Wang, Anal. Chim. Acta, 2014, 849, 43–49 CrossRef CAS PubMed.
- E. Rossi, Clin. Biochem. Rev., 2008, 29, 63–70 Search PubMed.
- Z. H. Hao, J. Z. Yao, R. L. Tang, X. M. Zhang, W. G. Li and Q. Zhang, Spectrosc. Spectral Anal., 2015, 35(2), 527–533 CAS.
- Y. K. Wang, S. T. Gao, X. H. Zang, J. C. Li and J. J. Ma, Anal. Chim. Acta, 2012, 716, 112–118 CrossRef CAS PubMed.
- S. Wang, X. Dong, B. Y. Dai, M. F. Pan, S. Y. He and J. P. Wang, J. AOAC Int., 2015, 98(1), 218–224 CrossRef CAS PubMed.
- Y. G. Wang, G. H. Zhao, Q. Zhang, H. Wang, Y. Zhang, W. Cao and Q. Wei, Sens. Actuators, B, 2019, 288, 325–331 CrossRef CAS.
- J. J. Shi, J. C. Zhu, M. Zhao, Y. Wang, P. Yang and J. He, Talanta, 2018, 183, 237–244 CrossRef CAS PubMed.
- B. Zhang, J. F. Chen, B. Q. Liu and D. P. Tang, Biosens. Bioelectron., 2015, 69, 230–234 CrossRef CAS PubMed.
- Q. Zhou, Y. X. Lin, Y. P. Lin, Q. H. Wei, G. N. Chen and D. P. Tang, Biosens. Bioelectron., 2016, 78, 236–243 CrossRef CAS PubMed.
- C. Y. Yang, Q. Wang, Y. Xiang, R. Yuan and Y. Chai, Sens. Actuators, B, 2014, 197, 149–154 CrossRef CAS.
- F. L. Li, Y. M. Guo, X. Y. Wang and X. Sun, Biosens. Bioelectron., 2018, 115, 7–13 CrossRef CAS PubMed.
- S. M. Taghdisi, N. M. Danesh, P. Lavaee, M. Ramezani and K. Abnous, Sens. Actuators, B, 2016, 234, 462–469 CrossRef CAS.
- F. Gao, C. Gao, S. Y. He, Q. X. Wang and A. Q. Wu, Biosens. Bioelectron., 2016, 81, 15–22 CrossRef CAS PubMed.
- R. J. Zeng, Z. B. Luo, L. S. Su, L. J. Zhang, D. P. Tang, R. Niessner and D. Knop, Anal. Chem., 2019, 91, 2447–2454 CrossRef CAS PubMed.
- H. X. Ji, F. Yan, J. P. Lei and H. X. Ju, Anal. Chem., 2012, 84, 7166–7171 CrossRef CAS PubMed.
- S. Z. Lv, K. Y. Zhang, Y. Y. Zeng and D. P. Tang, Anal. Chem., 2018, 90, 7086–7093 CrossRef CAS PubMed.
- X. Y. Xie, Y. Q. Chai, Y. L. Yuan and R. Yuan, Anal. Chim. Acta, 2018, 1034, 56–62 CrossRef CAS PubMed.
- J. Y. Dai, Z. J. Duan, M. Z. Cao, M. R. Hao, H. F. He and D. Xiao, Talanta, 2018, 181, 142–146 CrossRef CAS PubMed.
- S. S. Tang, Y. Gu, H. T. Lu, H. F. Dong, K. Zhang, W. H. Dai and X. J. Zhang, Anal. Chim. Acta, 2018, 1004, 1–9 CrossRef CAS PubMed.
- Y. Jiang, B. L. Li, J. N. Milligan, S. Bhadra and A. D. Ellington, J. Am. Chem. Soc., 2013, 135(20), 7430–7433 CrossRef CAS PubMed.
- X. Y. Wu, Y. Q. Chai, P. Zhang and R. Yuan, ACS Appl. Mater. Interfaces, 2014, 7(1), 713–720 CrossRef PubMed.
- H. L. Shuai, K. J. Huang, L. L. Xing and Y. X. Chen, Biosens. Bioelectron., 2016, 86, 337–345 CrossRef CAS PubMed.
- Y. Wang, N. Gan, Y. Zhou, T. H. Li, F. T. Hu, Y. T. Cao and Y. J. Chen, Biosens. Bioelectron., 2017, 97, 100–106 CrossRef CAS PubMed.
- X. Y. Huang, J. L. Li, Q. Y. Zhang, S. Chen, W. Xu, J. Y. Wu, W. C. Niu, J. J. Xue and C. R. Li, J. Electroanal. Chem., 2018, 816, 75–82 CrossRef CAS.
- Y. D. Ma, C. Yu, Y. J. Yu, J. Chen, R. F. Gao and J. L. He, Microchim. Acta, 2019, 186(10), 677 CrossRef PubMed.
- H. Wang, Y. Liu and G. Liu, Microchim. Acta, 2018, 185, 142 CrossRef PubMed.
- D. Wang, C. C. Ge, K. P. Lv, Q. S. Zou, Q. Liu, L. P. Liu, Q. H. Yang and S. Y. Bao, Chem. Commun., 2018, 54, 13718–13721 RSC.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42(14), 6060–6093 RSC.
- D. P. Cormode, L. Gao and H. Koo, Trends Biotechnol., 2018, 36(1), 15–29 CrossRef CAS PubMed.
- S. Chen, P. Liu, K. W. Su, X. Li, Z. Qin, W. Xu, J. Chen, C. R. Li and J. F. Qiu, Biosens. Bioelectron., 2018, 99, 338–345 CrossRef CAS PubMed.
- R. J. Zeng, Z. B. Luo, L. J. Zhang and D. P. Tang, Anal. Chem., 2018, 90, 12299–12306 CrossRef CAS PubMed.
- Z. Z. Yu, Y. Tang, G. N. Cai, R. R. Ren and D. P. Tang, Anal. Chem., 2018, 91, 1222–1226 CrossRef PubMed.
- Q. Yu, Y. M. Wu, Z. Liu, S. Lei, G. P. Li and B. X. Ye, Biosens. Bioelectron., 2018, 107, 178–183 CrossRef CAS PubMed.
- C. L. Zhang, J. L. He, Y. C. Zhang, J. Chen, Y. L. Zhao, Y. Z. Niu and C. Yu, Biosens. Bioelectron., 2018, 102, 94–100 CrossRef CAS PubMed.
- Y. M. Wu, L. N. Zou, S. Lei, Q. Yu and B. X. Ye, Biosens. Bioelectron., 2017, 97, 317–324 CrossRef CAS PubMed.
- Y. M. Zhang, Y. Liu, W. H. Liu, X. Y. Li and L. Q. Mao, Appl. Surf. Sci., 2017, 407, 64–71 CrossRef CAS.
- W. Cai, S. B. Xie, Y. Tang, Y. Q. Chai, R. Yuan and J. Zhang, Talanta, 2017, 163, 65–71 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00022a |
| This journal is © The Royal Society of Chemistry 2020 |