Open Access Article
Open Access ArticleA metabolomic study for chronic heart failure patients based on a dried blood spot mass spectrometry approach
Gaowa Zhaoab,
Dong Chengab,
Yu Wangac,
Yalan Caoad,
Shuting Xiangad and
Qin Yu *a
*a
aDepartment of Cardiology, Affiliated Zhongshan Hospital of Dalian University, Jiefang Street 6, Zhongshan District, Dalian 116001, China. E-mail: yuqin@dlu.edu.cn; dryuqin060621@sina.com; Fax: +86-411-62893555; Tel: +86-411-62887018
bMedical College, Dalian University, Dalian 116622, China
cDalian Medical University, Dalian 116044, China
dZunyi Medical University, Zunyi, 563003, China
First published on 22nd May 2020
Abstract
Objective: a dried blood spot (DBS) method integrated with direct infusion mass spectrometry (MS) focused on a metabolomic analysis was applied to detect and compare the difference of metabolites between the heart failure (HF) patients and non-HF patients in order to facilitate the early detection of heart failures, provide targeted intervention and offer prognostic insights. Methods: the method we used was an untargeted metabolic approach. The dry blood spot mass spectrometry (DBS) was used to analyze 23 types of amino acids and 26 types of carnitine in blood samples. In the current study, 49 metabolites were selected to establish the PLS-DA model to compare the differences between the 117 HF patients and 118 non-HF patients, which inclined to detect the difference between the two groups. Multiple algorithms were run for selecting different metabolites as potential biomarkers. Ten-fold cross validation method was used to verify and evaluate the selected potential biomarkers. Results: through significant analysis of the microarrays (SAM) and analysis of 9 parameters, 8 metabolites showed significant discrepancies between the HF and non-HF groups. Among these metabolites, the levels of 5 metabolites were increased, and the other 3 metabolites were decreased in the HF group compared with the non-HF group. However, 7 metabolites including Asn, C0, C14, C4DC, C5-OH, C6 and Glu were selected to distinguish the HF group from the non-HF group with specificity and sensitivity of 0.8475 and 0.8974, respectively. Conclusion: metabolomic study for chronic heart failure (CHF) patients based on the dried blood spot mass spectrometry approach would be beneficial to understand the metabolic pathway of HF, and probably work as biomarkers to predict the prognosis of HF and provide the basis for an individualized treatment.
Introduction
Heart Failure (HF) is a global public health problem with high morbidity and mortality, involving 26 million worldwide population, particularly occupying approximately 1% to 2% of the adult population.1,2 From an epidemiological perspective, timely diagnosis and early interventions in HF probably have far-reaching impacts on the healthcare economics and public health. From a microscopic perspective, HF is a manifestation of metabolic derangements on the genetic, cellular, proteomic, and metabolic levels;3,4 however, the metabolomic mechanism of HF is not completely understood. According to statistics, more than 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 unique metabolites have been identified in the Human Metabolome Database.5 Therefore, exploring the specific metabolomics and related biomarkers in the HF patients would be beneficial to early diagnosis, prediction, prognosis and individualized treatments.
000 unique metabolites have been identified in the Human Metabolome Database.5 Therefore, exploring the specific metabolomics and related biomarkers in the HF patients would be beneficial to early diagnosis, prediction, prognosis and individualized treatments.
Metabolomics is used to detect low-molecular metabolites produced during the cellular metabolism processes.6 MS could detect and quantify low-abundance metabolite sensitively.7,8 As an important metabolic analytical technology, mass spectrometry (MS) has been widely used in the field of HF including the detection of metabolic spectra, screening of biomarkers and exploring the metabolic changes both in human blood and urine samples.9–11 Furthermore, the direct infusion MS technology could meet the requirements of high-throughput and rapid collection of fragmentation data and it has been applied in the metabolomics studies of various diseases.12,13 Compared to the traditional venous blood sampling technology, a dried blood spot (DBS) sample technology was usually used to collect blood in new born screening (NS), which makes samples easy to be transported and stored.14,15 In the current study, we combined direct infusion MS with the DBS samples to compare the metabolomic profiles of HF and non-HF patients, which has potential in prevention, detection, evaluation, and management of HF in patients.
Materials and methods
Sample information
HF and non-HF patients were obtained from Affiliated Zhongshan Hospital of Dalian University. All the clinical parameters were obtained from the targeted population including age, height, weight, and blood pressure (BP) levels. In addition, the medical histories were collected. DBS samples were composed of 117 HF patients as the case group. In addition, 118 non-HF patients were taken into the control group without cardiovascular diseases. The HF patients enrolled in the study should meet the following inclusion criteria for the diagnosis of chronic heart failure (CHF), who should meet the Framingham heart failure diagnostic criteria; ESC (European society of Cardiology) guidelines for the diagnosis and treatment of HF 2016; New York Heart Association (NYHA) class II to IV. Exclusion criteria: patients with malignant hypertension, severe arrhythmia, acute myocardial infarction (<72 h), acute myocarditis, aortic dissection, hemodynamic instability, and malignant tumor combined with severe hepatorenal dysfunction; other specific oxidative stress related diseases. In the control group, patients with non-HF were excluded from diabetes, digestive diseases, chronic infectious diseases, hypertension, kidney diseases, autoimmune diseases, endocrine disorders and cancer. The average age of the case and control groups is 54 (23–91) years and 73 (26–96) years, respectively. The information for HF patients and non-HF patients are shown in Table 1. There was no age difference between the case and control groups (p = 0.2975, t-test). All the participants were used to build a training set for the prediction model. Ten-fold cross validation was executed to build the test set for checking the applicability of the prediction model. The study was approved by Affiliated Zhongshan Hospital of Dalian University. Also, all the participants have understood and signed the informed consent.| Clinical indicator | Control | Case | p-Value |
|---|---|---|---|
| a The clinical indicators were compared between case and control groups. Continuous variables were analyzed by the unpaired t-test. Chi-squared test was used to count variables. | |||
| Age (year) | 54 ± 11.42 | 73 ± 12 | <0.0001 |
| Gender (M/F) | 56/61 | 65/53 | 0.2975 |
| Height (m) | 1.68 ± 8.71 | 1.68 ± 8.32 | 0.9015 |
| Weight (kg) | 64.51 ± 9.53 | 69.64 ± 14.80 | 0.0018 |
| BMI (kg m−2) | 22.89 ± 3.61 | 24.60 ± 4.59 | 0.0017 |
| SBP (mmHg) | 122.82 ± 11.93 | 135.32 ± 22.12 | <0.0001 |
| DBP (mmHg) | 76.43 ± 7.58 | 76.42 ± 12.27 | 0.9927 |
Chemicals
We acquired high-purity acetonitrile of high-performance liquid chromatography grade from Thermo Fisher (Waltham, MA), and obtained 1-butanol and acetyl chloride from Sigma-Aldrich (St Louis, MO). Also, we utilized isotope-labeled internal standards, which were obtained from Cambridge Isotope Laboratories (Tewksbury, MA) for 12 amino acids (NSK-A) and 8 acyl carnitines (NSK-B) to make absolute quantification. We prepared these standards by dissolving in 2 ml pure methanol and storing them at 4 °C. Besides, we diluted the working solution 100-fold for metabolite extraction. We purchased the quality control (QC) standards for amino acids and carnitine from Chromsystems (Grafelfing, Germany) and injected the QC standards into the real sample analysis queue to ensure analysis stability.Sample preparation
A DBS filter paper (equivalent to 3.2 μl whole blood) was placed into a 96-well plate. This disc was transferred to a Millipore Multi Screen HV 96-well plate (Millipore, Billerica, MA, USA) for metabolite extraction. Briefly, 100 μl of the working solution was dispensed into one well containing a DBS disc and gently shaken for 20 min at room temperature and centrifuged at 1500 × g for 2 min to collect the filtrate into a new flat bottom 96-well plate. The filtrate and QC solutions were subjected to drying by pure nitrogen gas at 50 °C. Each dried sample was derivatized in a 60 μl butanol and acetyl chloride mixture (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, v/v) at 65 °C for 20 min. The derivatized solutions were dried by pure nitrogen gas at 50 °C again. Every 100 μl mobile phase solution was added into each well prior to the metabolomics analysis.16
10, v/v) at 65 °C for 20 min. The derivatized solutions were dried by pure nitrogen gas at 50 °C again. Every 100 μl mobile phase solution was added into each well prior to the metabolomics analysis.16
Metabolomic analysis
The direct injection MS metabolomic analysis was conducted using an AB Sciex 4000 QTrap system (AB Sciex, Framingham, MA, USA). The equipped ion source was an electrospray ionization source. All the analytes were scanned under a positive model.17 The positive model, we used, was linked with abundant detection of the metabolites, while use of a negative model could not detect the metabolites, so only “positive model” was used in the current study.We injected 20 μl sample for each run, and used the 80% acetonitrile aqueous solution as the mobile phase at an initial flow rate of 0.2 ml min−1. The flow rate was subsequently decreased to 0.01 ml min−1 within 0.08 min and remained for 0.5 min. We set an ion spray voltage at 4.5 kV, a curtain gas pressure to 20 psi, the pressure for ion source gas 1 and gas 2 to 35 psi, and an auxiliary gas temperature at 350 °C.
Analyst v1.6.0 software (AB Sciex) was used to execute the raw data collection and system control. Data preprocessing was achieved by the ChemoView 2.0.2 software (AB Sciex). Partial least squared discriminant analysis (PLS-DA) and principal component analysis (PCA) were used to distinguish the HF patients from non-HF individuals. In addition, importance in projection (VIP) was calculated by SIMCA-P v12.0 (Umetrics, Umea, Sweden). This variable represents the contribution of variables towards disease classification. Therefore, VIP values were used to select the variables. In order to check the rationality of the building model, a 200-time permutation test was used. The normality for all the variables in the HF and non-HF groups was inspected using the Shapiro–Wilk test. T-test was used to check the discrepancy between the HF and non-HF groups. For the non-parametric variables, the Mann–Whitney test was applied. In addition, for adjusting the p-values towards multiple hypothesis testing, a Benjamini Hochberg-based false discovery rate (FDR) was applied.18 For selecting variables, the VIP values (VIP > 1) and adjusted p-values (adjusted p < 0.05) were applied with targeted values over 1 and lower than 0.05, respectively. In addition, the significant analysis of microarrays (SAM) method was used, which could inspect the variables with significant difference between the HF and non-HF groups. For further selection of the parameters, stepwise selection methods were applied to select the final variables, and the selected variables could be used to build the prediction model. At last, the binary logistic regression method was used to build the HF prediction model. SAS software was used to conduct the statistical analysis.
Results
Metabolomic differences between HF and non-HF groups
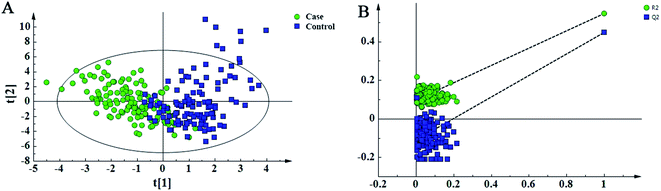
In this study, 49 metabolites including 23 amino acids and 26 carnitines were used to build the PLD-DA model.19 This model showed that the differences between the HF and non-HF groups were significant (Fig. 1A). A permutation test indicated that over-fitting was almost impossible to exist (Fig. 1B). R2 and Q2 values are given in the study: R2 = (0.0, 0.0954), Q2 = (0.0, −0.116). In addition, a PCA scatter plot for metabolomics data demonstrated that the heart failure group could be distinguished from the non-heart failure group (Fig. 2). However, from the analysis results, the difference between the two groups was not obvious.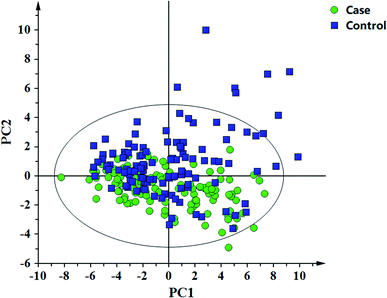 | ||
| Fig. 2 Principal component analysis (PCA) for the metabolomic data of the heart failure and non-heart failure groups. | ||
Selecting differential metabolites
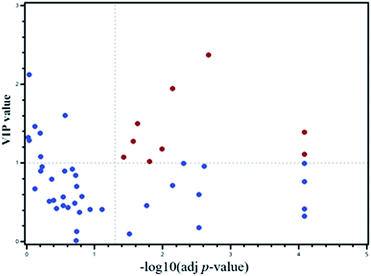
A PLS-DA model was applied to select the vital contribution variables to distinguish the HF from non-HF groups (Fig. 1A). A total of 16 parameters with VIP > 1.0 were selected for further analyses. Significant differences for all the variables were calculated using the Mann–Whitney test and t-test. In addition, false discovery rate was adjusted. There were altogether 20 variables retained. Together, a total of 9 metabolites were selected using the calculations of VIP and p-values (Fig. 3).For further defining the biomarkers with significant discrepancies between the HF and non-HF groups, SAM was analyzed (Fig. 4). The results were shown in Fig. 4. For the selected 9 parameters, the differences for 8 metabolites between the HF and non-HF groups were significant (Table 2). Among these, the levels of 5 metabolites were increased, and the levels of other 3 metabolites were decreased in the HF group compared with non-HF group.
| No | Parameters | Control (mean ± SD) | Case (mean ± SD) | Status | p-Value | Adjusted p-value |
|---|---|---|---|---|---|---|
| 1 | Asn | 73.8558 ± 23.0247 | 65.5182 ± 20.6377 | ↓ | 0.0048 | 0.0157 |
| 2 | C0 | 35.8565 ± 28.4928 | 68.1325 ± 70.5508 | ↑ | <0.0001 | <0.0001 |
| 3 | C14 | 0.0858 ± 0.0353 | 0.1117 ± 0.0683 | ↑ | 0.0011 | 0.0049 |
| 4 | C4DC | 0.6045 ± 0.3144 | 0.4548 ± 0.2128 | ↓ | 0.0003 | 0.0021 |
| 5 | C5-OH | 0.2585 ± 0.1031 | 0.2197 ± 0.0946 | ↓ | 0.0029 | 0.0101 |
| 6 | C6 | 0.0617 ± 0.0281 | 0.1059 ± 0.0538 | ↑ | <0.0001 | <0.0001 |
| 7 | Cit | 19.5681 ± 6.8929 | 24.1283 ± 11.8882 | ↑ | 0.0081 | 0.0233 |
| 8 | Glu | 102.5858 ± 36.0924 | 122.2345 ± 55.8465 | ↑ | 0.0099 | 0.0269 |
Prediction regression model
A stepwise regression method was used to further select the variables. Finally, 7 biomarkers were selected to establish the binary logistic regression model, and these biomarkers were Asn, C0, C14, C4DC, C5-OH, C6 and Glu (Fig. 5). This model is adjusted by gender, age, body mass index (BMI), systolic blood pressure (SBP), and diastolic blood pressure (DBP) with the logic probability being given by (Logit probability = 0.3582 − 0.828 × gender + 2.0622 × age + 0.7273 × BMI + 1.16 × SBP − 0.2799 × DBP − 0.3078 × Asn + 0.4063 × C0 + 0.9498 × C14 − 0.3275 × C4DC − 0.5561 × C5-OH + 1.4037 × C6 + 0.2495 × Glu). The receiver operating characteristic (ROC) curve analysis was executed to assess the diagnosis potential of the 7 biomarkers, and an ROC curve is shown in Fig. 6. The AUC value for this ROC curve is 0.9549. The ROC curve displayed sensitivity of 0.8475 and specificity of 0.9487 for distinguishing HF from the non-HF groups. Ten-fold cross validation was applied to test the diagnostic potential for the 7 selected biomarkers. The AUC value for the ROC curve of the ten-fold cross validation model was 0.9386. The sensitivity and specificity for this validation model were 0.8475 and 0.8974, respectively.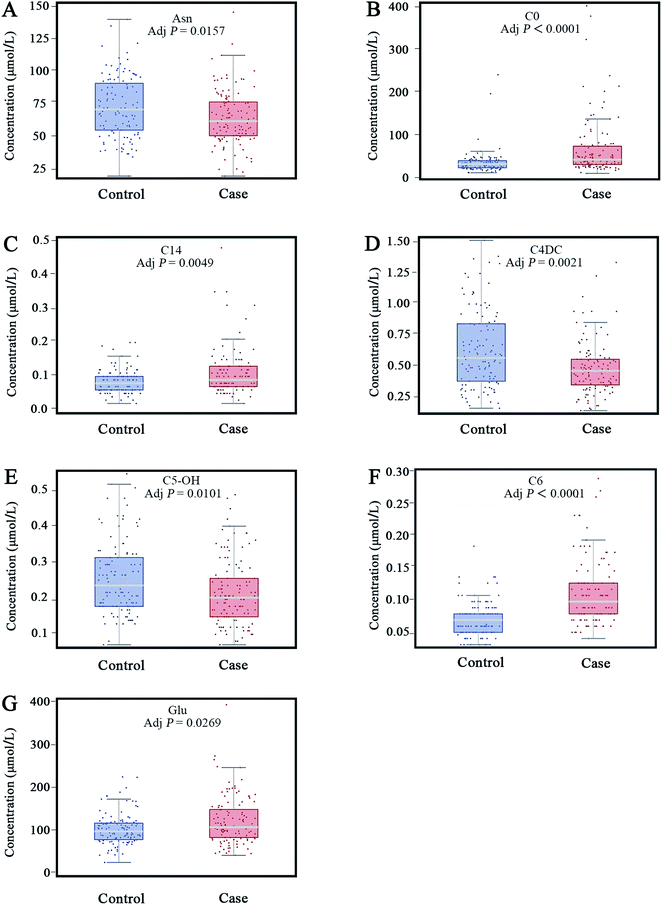 | ||
| Fig. 5 Levels of seven metabolites were used for building binary the logistic regression model (A–G). | ||
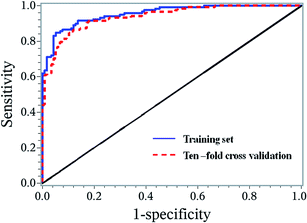 | ||
| Fig. 6 ROC curves for the training set and ten-fold cross validation set based on the logistic regression model. | ||
Discussion
HF is a global medical problem with poor prognosis. Although the HF treatment has been constantly updated with development in medicines and technology, the difference in metabolomics between HF and non-HF patients remains unclear. Metabolomic strategies could be used in probing the metabolic aspects of HF, which is beneficial to understand the altered metabolic pathways of HF. In the current study, the metabolites including amino acids, carnitine and acylcarnitines in the HF and non-HF groups were measured using the direct infusion MS detection method and DBS sample technology. After systematic selections, the differences for 8 metabolites between the HF and non-HF groups were significant. Ultimately, 7 metabolites as novel potential HF biomarkers taken from the stepwise regression process could be used in classification for non-HF patients.There are many reasons for heart failure, and the common causes of heart failure are high blood pressure, coronary atherosclerotic disease, myocardial ischemia, myocardial infarction, hyperglycemia, impaired glucose tolerance, diabetes, obesity and so on. However, during the development of heart failure, a complex network of compensatory mechanisms is activated on macro and micro structural, cellular, and molecular levels to maintain the heart function.20 The heart needs and consumes more energy than any other organ of the human body. To reach this enormous demand, the heart converts chemical energy stored in the fatty acids and glucose into mechanical energy. If these mechanisms do not reach the demand of the heart, cardiac malfunction, mechanical failure of the heart occurs. The “energy starvation” hypothesis subsumes the altered mechanisms of myocardial energetics, which leads to the energy depletion. From the perspective of metabolism, heart failure is the manifestation of a abnormal myocardial energy metabolism, including the metabolism of fatty acids, glucose, amino acids.
Amino acids are an important part of the human body and the basic components of structural and functional proteins.21 They are also used as substrates for generating energy.22 Previous metabolomics studies have shown HF to be associated with abnormal amino acid metabolism.23 In the study, Asn level was found to be decreased, whereas the Glu and Cit levels increased for HF patients, all of them being the non-essential amino acids. Asn can be used as a carrier of K+ and Mg2+ to deliver the electrolytes to the myocardium. Previous studies have verified the process involved in improving the myocardial systolic function, reducing oxygen consumption and effecting the myocardium protection when the coronary artery circulation is impaired by hypoxia. In addition, it is involved in the ornithine cycle, promoting the production of urea from oxygen and carbon dioxide and reducing the amount of nitrogen and carbon dioxide in the blood. It seems reasonable that the level of Asn is decreased in HF. Glu is the main component of protein and one of the basic amino acids for nitrogen metabolism in the body, which has been demonstrated to have the effect on vasodilatation, promoting blood circulation and improving tissue energy metabolism. The previous study showed a decreased expression of the mitochondrial protein 9030617O03Rik in the heart failure mouse model.24 The accumulation of glutamate in the heart of 9030617O03Rik-deficient mice suggests that glutamate is metabolized in the mammalian hearts. In our study, increased glutamate levels in the patients with HF may be associated with impaired myocardial cells and impaired glutamate metabolism.25
Cit is produced in the body when NOS catalyzes arginase reactions through the arginine NO pathway to produce nitric oxide (NO).21 NO acts on the vascular endothelial cells to expand the peripheral blood vessels, and has a variety of effects such as reducing blood pressure, inhibiting platelet aggregation, and antioxidant. The activity of arginase was increased in the previous studies, leading to the decrease in the NO formation and increasing the production of reactive oxygen species. Therefore, the increased Cit level in HF may be due to the increased arginase activities, which leads to the increased ornithine level and thus the increased citrulline level.26 So, the level of Cit could serve as a biomarker for the endothelial function.
Carnitine is a micronutrient that plays a key role in lipid metabolism.27 Carnitine exists as both free carnitine and acylcarnitine in the human body and present in all body fluids, which is mainly in tissues with high energy demand, skeletal muscle and myocardium, and only a small part in liver, brain, kidney and extracellular fluid (such as plasma and urine). Carnitine and its acyl derivatives play an important role in the fatty acid oxidation. In the current study, the levels of two acylcarnitines, including C4DC and C5-OH were obviously decreased in the HF patients; however, the levels of three carnitine and acylcarnitines, including C14, C6 and C0, were significantly increased in the HF patients. C4DC and C5DC are belonging to short-chain acylcarnitines, which might promote the expression of gene and protein related to oxidative stress. Hence, the decrease in the short-chain acylcarnitine in the HF patients may be related to the regulation of oxidative stress factors.28,29 The long-chain acylcarnitine are closely related to fatty acid β-oxidation, which is an important source of energy for the body. The heart is the most metabolically active organ in the body and primarily utilizes free fatty acids (FFAs) as energy substrates. Healthy myocardium primarily meets its requirements for energy through the oxidation of long-chain fatty acids (LCFA), where carnitine plays a key role as a carrier.30 In the HF group, changes occur in myocardial mitochondrial functions that result in a shift towards the preferential use of glucose for metabolism rather than FFAs.31 LCFAs are activated by the esterification of CoA at the outer mitochondrial membrane. The inner mitochondrial membrane is impermeable to the acyl-CoA esters. The “carnitine shuttle” regulates the flux of acyl-CoA esters into the mitochondria. Many clinical and experimental studies have demonstrated that the failing heart undergoes metabolic remodeling and develops a metabolic inflexibility, switching to glucose utilization at the expense of fatty acid oxidation.32 These findings are consistent with the notion that the syndrome of HF may be characterized by a general state of metabolic inflexibility and mitochondrial inefficiency that leads to the accumulation of the metabolic intermediates of fatty acid oxidation such as the long-chain acylcarnitines.33 C0 is an amino acid derivative, and is necessary for the entry of long-chain fatty acids into the mitochondria for β-oxidation and protects the myocardium against ischemic injury. When HF occurs, myocardial cells are damaged, accompanied with abnormal mitochondrial metabolism, long-chain fatty acids in the cytoplasm are difficult to enter the mitochondria for β-oxidation, myocardial energy is insufficient and lipid accumulation in the cytoplasm further damages the myocardial cells, forming a vicious cycle. The increased level of C0 in the plasma of HF patients might be associated with the leakage of C0 into plasma through damaged myocardial cells,34 which might explain the difference between the HF and non-HF patients.
A binary logistic regression model was constructed to test if the significantly changed metabolites could be used in differentiating the HF patients from non-HF individuals. Seven metabolites (Asn, C0, C14, C4DC, C5-OH, C6, Glu) were used to build the regression model. The ROC curve based on the seven parameters showed significant classification between the HF patients and healthy subjects with the AUC value of 0.9386. The sensitivity and specificity for this validation model were 0.8475 and 0.8974, respectively (Fig. 5). This suggested that excellent performance of classification between the HF and non-HF groups could be realized via the seven decisive parameters.
Conclusion
In the current study, the direct infusion MS analysis combined with the DBS sample approach supply us a convenient, accurate and effective metabolomic technology to detect metabolite changes in HF. A binary logistic regression model was built based on seven decisive metabolites. It is indicated that this prediction model could be used as an alternative method for the screening of HF, which would be beneficial to understand the metabolic pathway of HF, finding out the biomarkers to predict the prognosis of HF and providing the basis for individualized treatments.Abbreviations
| DBS | Dried blood spot |
| HF | Heart failure |
| MS | Mass spectrometry |
| CHF | Chronic heart failure |
| Cit | Citrulline |
| Glu | Glutamic acid |
| Asn | Asparagine |
| C4DC | Succinyl/methylmalonylcarnitine |
| C5-OH | 3-Hydroxyisovalerylcarnitine |
| C6 | Hexanoylcarnitine |
| C14 | Myristoyl-carnitine |
| C0 | Free carnitine |
| BP | Blood pressure |
| PLS-DA | Partial least squared discriminant analysis |
| PCA | Principal component analysis |
| SAM | Significant analysis of microarrays |
| VIP | Variable importance in projection |
| NS | Newborn screening |
| FDR | False discovery rate |
| ROC | Receiver operating characteristic |
| AUC | Area under the curve |
| BMI | Body mass index |
| SBP | Systolic blood pressure |
| DBP | Diastolic blood pressure |
Ethic statement
The study was approved by the ethics committee of Affiliated Zhongshan Hospital of Dalian University. Also, all the participants have understood and signed the informed consent. All the experiments were performed according to the national health and Family Planning Commission “biomedical research involving human ethics review approach” (the eleventh National Health Commission Order.), “Helsinki declaration” and “human CIOMS international ethical guidelines for biomedical research” ethical principle.Funding statement
This study was supported by National Natural Science Funds of China (No. 81770405).Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.References
- E. Tanai and S. Frantz, Pathophysiology of Heart Failure, Compr. Physiol., 2015, 6, 187–214 CrossRef PubMed.
- L. Papadimitriou, C. E. Hamo and J. Butler, Heart failure guidelines: what's new?, Trends Cardiovasc. Med., 2017, 27, 316–323 CrossRef PubMed.
- C. L. Albert and W. H. W. Tang, Metabolic Biomarkers in Heart Failure. Heart Fail, Clin, 2018, 14, 109–118 Search PubMed.
- A. T. Turer, Using metabolomics to assess myocardial metabolism and energetics in heart failure, J. Mol. Cell. Cardiol., 2013, 55, 12–18 CrossRef CAS PubMed.
- D. S. Wishart, T. Jewison, A. C. Guo, M. Wilson and C. Knox, et al., HMDB 3.0--The Human Metabolome Database in 2013, Nucleic Acids Res., 2013, 41, D801–D807 CrossRef CAS PubMed.
- K. Agnieszka, P. Magdalena, K. Zenon and M. Jan, Application of metabolomic tools for studying low molecular-weight fraction of animal venoms and poisons, Toxins, 2018, 10, E306 CrossRef PubMed.
- A. Zhang, H. Sun, G. Yan, P. Wang and X. Wang, Mass spectrometry-based metabolomics: applications to biomarker and metabolic pathway research, Biomed. Chromatogr., 2014, 30, 7–12 CrossRef PubMed.
- H. Guo, H. Peng and A. Emili, Mass spectrometry methods to study protein-metabolite interactions, Expert Opin. Drug Discovery, 2017, 12, 1271–1280 CrossRef CAS PubMed.
- M. Z. Israr, L. M. Heaney and T. Suzuki, Proteomic biomarkers of heart failure, Heart Fail. Clin., 2018, 14, 93–107 CrossRef PubMed.
- E. Holmes, R. L. Loo, J. Stamler, M. Bictash and I. K. Yap, et al., Human metabolic phenotype diversity and its association with diet and blood pressure, Nature, 2008, 453, 396–400 CrossRef CAS PubMed.
- Y. Liu, T. Chen, Y. Qiu, Y. Cheng and Y. Cao, et al., An ultrasonication-assisted extraction and derivatization protocol for GC/TOFMS-based metabolite profiling, Anal. Bioanal. Chem., 2011, 400, 1405–1417 CrossRef CAS PubMed.
- A. D. Southam, R. J. Weber, J. Engel, M. R. Jones and M. R. Viant, A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics, Nat. Protoc., 2016, 12, 310–328 CrossRef PubMed.
- S. Anand, S. Young, M. S. Esplin, B. Peaden and H. D. Tolley, et al., Detection and confirmation of serum lipid biomarkers for preeclampsia using direct infusion mass spectrometry, J. Lipid Res., 2016, 57, 687–696 CrossRef CAS PubMed.
- A. M. Butler, W. Charoensiriwatana, P. Krasao, R. Pankanjanato and P. Thong-Ngao, et al., Newborn thyroid screening: influence of pre-analytic variables on dried blood spot thyrotropin measurement, Thyroid, 2017, 27, 1128–1134 CrossRef CAS PubMed.
- J. Deglon, A. Thomas, P. Mangin and C. Staub, Direct analysis of dried blood spots coupled with mass spectrometry: concepts and biomedical applications, Anal. Bioanal. Chem., 2012, 402, 2485–2498 CrossRef CAS PubMed.
- Z. Hu, Z. Zhu, Y. Cao, L. Wang and X. Sun, et al., Rapid and Sensitive Differentiating Ischemic and Hemorrhagic Strokes by Dried Blood Spot Based Direct Injection Mass Spectrometry Metabolomics Analysis, J. Clin. Lab. Anal., 2016, 30, 823–830 CrossRef CAS PubMed.
- Q. Wang, T. Sun, Y. Cao, P. Gao and J. Dong, et al., A dried blood spot mass spectrometry metabolomic approach for rapid breast cancer detection, OncoTargets Ther., 2016, 9, 1389–1398 CAS.
- Y. Benjamini and Y. Hochberg, Controlling the False Discovery Rate-a Practical and Powerful Approach to Multiple Testing, J. Roy. Stat. Soc. B Stat. Methodol., 1995, 57, 289–300 Search PubMed.
- Q. Wang, T. Sun, Y. Cao, P. Gao and J. Dong, et al., A dried blood spot mass spectrometry metabolomic approach for rapid breast cancer detection, OncoTargets Ther., 2016, 9, 1389–1398 CAS.
- E. Tanai and S. Frantz, Pathophysiology of Heart Failure, in Comprehensive Physiology, ed. R. Terjung, 2015 Search PubMed.
- N. Morotomi, M. Saitoh, N. Ishii, K. Ohno and M. Nagayama, et al., Relation between change in exercise capacity and change in blood amino acids in patients with chronic heart failure, J. Phys. Ther. Sci., 2017, 29, 425–431 CrossRef PubMed.
- Q. Bai, B. Peng, X. Wu, Y. Cao and X. Sun, et al., Metabolomic study for essential hypertension patients based on dried blood spot mass spectrometry approach, IUBMB Life, 2018, 70, 777–785 CrossRef CAS PubMed.
- R. Aquilani, M. T. La Rovere and D. Corbellini, et al., Plasma Amino Acid Abnormalities in Chronic Heart Failure. Mechanisms, Potential Risks and Targets in Human Myocardium Metabolism, Nutrients, 2017, 9(11), 1251 CrossRef PubMed.
- M. Ariyoshi, M. Katane and K. Hamase, et al., D-Glutamate is metabolized in the heart mitochondria, Sci. Rep., 2017, 7, 43911 CrossRef PubMed.
- M. Ariyoshi, M. Katane and K. Hamase, et al., (D)-Glutamate is metabolized in the heart mitochondria, Sci. Rep., 2017, 7, 43911 CrossRef PubMed.
- D. E. Lanfear, J. J. Gibbs, J. Li, R. She and C. Petucci, et al., Targeted Metabolomic Profiling of Plasma and Survival in Heart Failure Patients, JACC Heart Fail., 2017, 5, 823–832 CrossRef PubMed.
- X. Song, H. Qu, Z. Yang, J. Rong and W. Cai, et al., Efficacy and Safety of L-Carnitine Treatment for Chronic Heart Failure: A Meta-Analysis of Randomized Controlled Trials, BioMed Res. Int., 2017, 2017, 6274854 Search PubMed.
- W. G. Hunter, J. P. Kelly, R. W. McGarrah 3rd, M. G. Khouri, D. Craig, C. Haynes, O. Ilkayeva, R. D. Stevens, J. R. Bain, M. J. Muehlbauer, C. B. Newgard, G. M. Felker, A. F. Hernandez, E. J. Velazquez, W. E. Kraus and S. H. Shah, Metabolomic Profiling Identifies Novel Circulating Biomarkers of Mitochondrial Dysfunction Differentially Elevated in Heart Failure With Preserved versus Reduced Ejection Fraction: Evidence for Shared Metabolic Impairments in Clinical Heart Failure, J. Am. Heart Assoc., 2016, 5(8), 003190 Search PubMed.
- B. Lefort, E. Gouache, C. Acquaviva, M. Tardieu and J. F. Benoist, et al., Pharmacological inhibition of carnitine palmitoyltransferase 1 restores mitochondrial oxidative phosphorylation in human trifunctional protein deficient fibroblasts, Biochim. Biophys. Acta, Mol. Basis Dis., 2017, 1863, 1292–1299 CrossRef CAS PubMed.
- P. Schoenfeld and L. Wojtczak, Short- and medium-chain fatty acids in the energy metabolism: the cellular perspective, J. Lipid Res., 2016, 57, 943–954 CrossRef CAS PubMed.
- M. Ruiz, F. Labarthe, A. Fortier, B. Bouchard and J. T. Legault, et al., Circulating acylcarnitine profile in human heart failure: a surrogate of fatty acid metabolic dysregulation in mitochondria and beyond, Am. J. Physiol.: Heart Circ. Physiol., 2017, 313, H768–H781 CrossRef CAS PubMed.
- S. Neubauer, The failing heart--an engine out of fuel, N. Engl. J. Med., 2007, 356(11), 1140–1151 CrossRef PubMed.
- T. Ahmad, J. P. Kelly, R. W. McGarrah, A. S. Hellkamp, M. Fiuzat, J. M. Testani, T. S. Wang, A. Verma, M. D. Samsky, M. P. Donahue, O. R. Ilkayeva, D. E. Bowles, C. B. Patel, C. A. Milano, J. G. Rogers, G. M. Felker, C. M. O'Connor, S. H. Shah and W. E. Kraus, Prognostic Implications of Long-Chain Acylcarnitines in Heart Failure and Reversibility With Mechanical Circulatory Support, J. Am. Coll. Cardiol., 2016, 67(3), 291–299 CrossRef CAS PubMed.
- T. Ueland, A. Svardal, E. Oie, E. T. Askevold and S. H. Nymoen, et al., Disturbed carnitine regulation in chronic heart failure — increased plasma levels of palmitoyl-carnitine are associated with poor prognosis, Int. J. Cardiol., 2013, 167, 1892–1899 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |