Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Diagnostically analyzing 1H NMR spectra of sub-types in chaetoglobosins for dereplication†
Chen Lin‡
ab,
Jian-chun Qin‡c,
Yong-gang Zhang*d and
Gang Ding*a
aKey Laboratory of Bioactive Substances and Resources Utilization of Chinese Herbal Medicine, Ministry of Education, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100193, P. R. China. E-mail: gding@implad.ac.cn
bZhengzhou Key Laboratory of Synthetic Biology of Natural Products, Henan Joint International Research Laboratory of Drug Discovery of Small Molecules, Huanghe Science and Technology College, Zhengzhou, Henan 450063, P. R. China
cCollege of Plant Sciences, Jilin University, Changchun, Jilin 130062, P. R. China
dBiology Institute, Qilu University of Technology (Shandong Academy of Sciences), Jinan 250103, Shandong Province, P. R. China. E-mail: zhangygcq@163.com
First published on 9th January 2020
Abstract
1H-NMR spectra provide abundant diagnostic information including chemical shift values, splitting patterns, coupling constants, and integrals. Thus some key functional groups, and even planar structures could be elucidated on the basis of carefully analyzing the corresponding 1H NMR spectrum. In this paper, the different sub-types of chaetoglobosins are classified according to the structural features, of which the 1H NMR spectra are systematically summed up. Thus diagnostically analyzing the 1H-NMR spectra could identify possible sub-types of chaetoglobosins, which could be used for dereplication. According to the analysis of this report, it implies that different new sub-types or new sub-type combinations in the key skeleton of chaetoglobosins might exist in nature. More importantly, dereplication based on 1H NMR spectral analysis will not only provide a useful approach to determine the chaetoglobosins structures quickly, but also could set a good example for structural dereplication of other NPs.
1. Introduction
Natural products (NPs) play a pivotal role in drug discovery and development, and many drugs in prescriptions directly or indirectly originate from NPs.1,2 Nowadays purification of NPs is not a laborious task due to the combination of different chromatographic technologies, whereas structural elucidation of NPs is still a manual job, which needs much time (even several months) to characterize the complex structures. To date, several techniques including HR-MS (High Resolution Mass), MSn, NMR (Nuclear Magnetic Resonance) spectroscopy, X-ray diffraction, and even bioinformatics approaches are used to elucidate the structures of NPs. HR-MS can give exact molecular weight leading to the molecular formula, and provides unsaturation degrees of target NPs; MSn can produce precursor and product ions, which help to deduce possible structural fragments. Combination of HR-MS and MSn techniques is akin to piecing together a complex jigsaw puzzle by assembling the molecular weight, molecular formula and structural fragments to elucidate structures. Most of the time this approach could not establish the planar structure especially for complex structures; X-ray diffraction can obtain the direct structural information including planar, relative and absolute configurations, whereas not all NPs could be grown to be a suitable crystal for X-ray diffraction experiment. Recently, bioinformatics approaches were also used to predict the possible substrates of secondary metabolites such as polyketide synthases (PKSs) or nonribosomal peptide synthetases (NRPSs) biosynthesized compounds.3 Considering the different post-modification reactions in the biosynthetic pathways, using bioinformatic way to elucidate the NPs structures is still not a reliable method now. NMR techniques such as 1D NMR (1H, and 13C NMR spectra), and 2D NMR such as HMQC, 1H 1H-COSY, HMBC, and NOESY spectra are the main tools to elucidate the structures of different NPs. Actually, 1H NMR spectral analysis is the first and key step in the structural elucidation of NPs. Compared with other NMR spectra, 1H NMR spectrum has a wealth of diagnostic information including chemical shift values, splitting patterns, coupling constants, and integrals, which reflect the structural features.4Chemical shift values are the direct reflection of different functionalities in the structure. For example, usually the chemical shift values of aromatic or olefinic proton are 6.0–9.5 or 4.5–7.5 ppm, respectively.5 If there are electron withdrawing groups (such as keto-group) near the aromatic proton or olefinic proton, the chemical shift values of these protons nearby will be shifted to be more down-fielded, vice versa. Thus according to different chemical shift values, the possible functional groups in the structure could be predicted.
Splitting patterns can tell us how many protons connecting the corresponding groups. If a proton is a triplet (t) or a doublet doublet (dd), implying that a –CH2– unit or two different protons might be connected near by the proton.
Analysis of coupling constants can help determine the configuration of corresponding protons with others. Usually big coupling constant indicates trans-configuration, vice versa. For example, if the coupling constant of olefinic protons is over 15.0 Hz, indicating the Z-configuration of the double bond, otherwise E-configuration.
Integrals could give the proton numbers of a natural product, but the intergrals are different in different deuterated solvents. Usually, the integrals (in DMSO-d6) could reflect the total proton numbers including exchangeable protons.
Then detailed analysis of chemical shift values, splitting patterns, coupling constants, and integrals in the 1H NMR spectrum can characterize structural fragments, and even establish the possible planar structures. If the deduced structures are known compounds by searching different data bases such as SCIfinder, Dictionary of Natural Products (DNP) or others, there is no need to process the next steps (13C, HMQC, 1H 1H-COSY, HMBC, and NOESY spectra) further, which can save a lot of time and resources. In this paper, the different sub-types of chaetoglobosins are classified according to the structural features, of which the 1H NMR spectral are systematically summed up. Finally, two examples are given in details for dereplication by diagnostically analyzing the 1H NMR spectra on this member of mycotoxins (other eight examples are included in ESI†).
2. Results and discussion
2.1 Structural features of chaetoglobosins
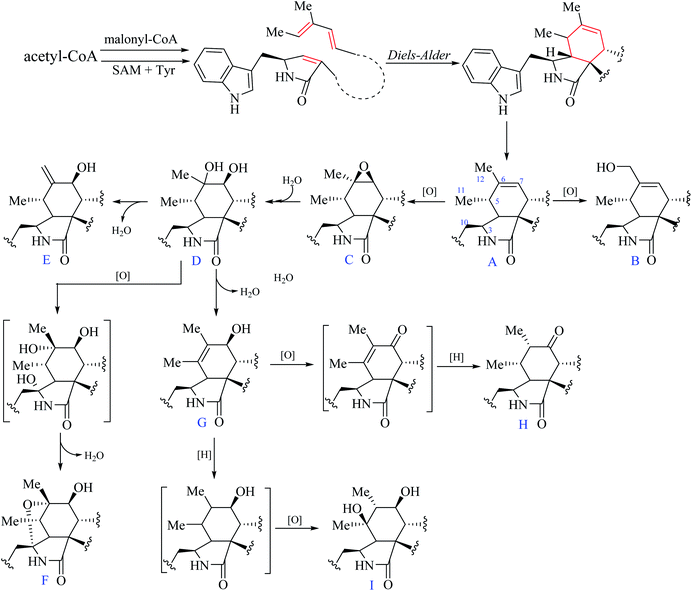
Chaetoglobosins are a large member of mycotoxins biosynthesized by a PKS−NRPS hybrid megasynthetase.6,7 The structural features of these compounds are a highly substituted perhydro-isoindolone moiety bearing an (indol-3-yl) methyl group to which typically a macrocyclic ring – is fused (Fig. 1). The different substitutions, connectivity patterns and oxygenation sites on the perhydro-isoindolone moiety and macrocycle ring increase the chemical diversity. Up to date, 56 analogs (See ESI†) containing the key nucleus of chaetoglobosin (perhydro-isoindolone moiety and macrocycle ring Fig. 1) were isolated different fungi mainly from Chaetomium spp.7,8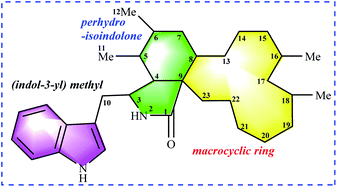 | ||
| Fig. 1 Chaetoglobosin nucleus: (indol-3-yl) methyl unit, perhydro-isoindolone moiety, and macrocyclic ring. | ||
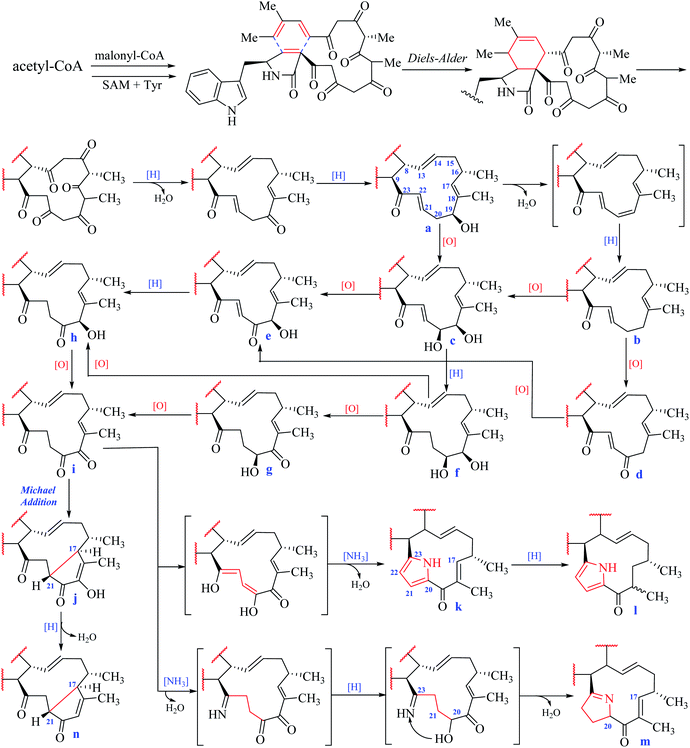
According to the structural features of chaetoglobosins, 9 sub-types in the perhydro-isoindolone moiety are summed up on account of the different variations at C-3, C-5, C-6, C-7 and C-12 (Fig. 2), whereas 14 sub-types in the macrocycle ring are classified according to the diverse variations at C-17, C-18, C-19, C-20, C-21, C-22 and C-23 (Fig. 3).
2.2 1H NMR spectral characteristics of perhydro-isoindolone moiety
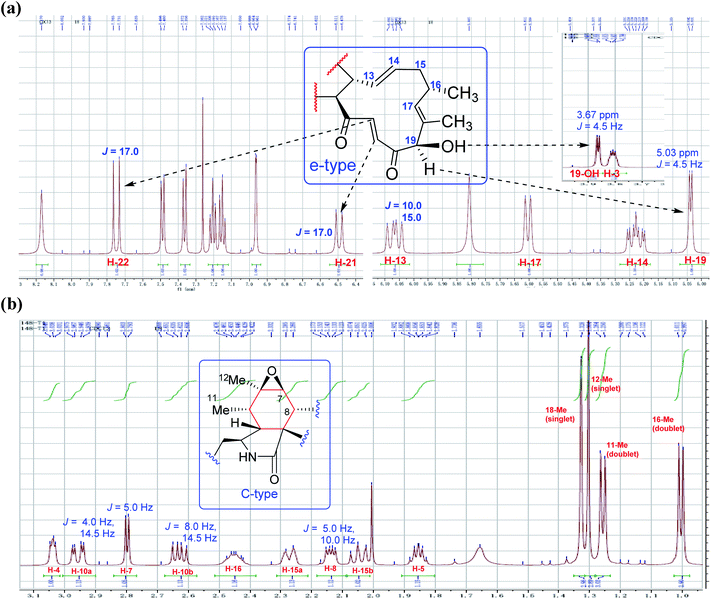
From the postulated biogenetic pathway of chaetoglobosins, the Diels–Alder reaction forms the key cyclohexene ring (C-4-C-5-C-6-C-7-C-8-C-9), and then a series of post-modification reactions mainly oxygenations at C-3, C-5, C-6, C-7 and C-12 construct nine sub-types in the perhydro-isoindolone part. This leads to chemical environment variations of different protons in the perhydro-isoindolone ring, which produces different chemical shift values, splitting patterns, coupling constants (of H-7, –CH2-10, CH3-11, and CH3-12), and different integrals. Thus according to different chemical shift values, splitting patterns, coupling constants, and integrals of those protons, it could, in return, deduce the structural fragments on the perhydro-isoindolone moiety.2.3 1H NMR characteristics of macrocyclic ring
In the macrocyclic ring of chaetoglobosins, C-8, C-9, C-13, C-14, C-16, 16-Me, and 18-Me are usually not changed. Considering different oxygenation degrees at C-19 and C-20, the double bond formation at C-21/C-22, the carbon–carbon connectivity at C-17/C-21, or pyrrole ring formed by C-20, 21, 22 and 23 in the macrocylic ring, 14 sub-types (a–n) are summed up (Fig. 3), which are distinct from each other according to their diagnostic 1H NMR features.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C-22–C
C-22–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O-23 with chemical shift values at 6.80 ppm (m, H-21) and 6.89 ppm (d, J = 16.0 Hz, H-22).
O-23 with chemical shift values at 6.80 ppm (m, H-21) and 6.89 ppm (d, J = 16.0 Hz, H-22).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O-19), the chemical shift value of H-17 in the 1H NMR spectrum is in down-fielded zone between 6.00–6.50 ppm compared with the a–f types (<5.70 ppm).
O-19), the chemical shift value of H-17 in the 1H NMR spectrum is in down-fielded zone between 6.00–6.50 ppm compared with the a–f types (<5.70 ppm).According to the classification of different sub-types in chaetoglobosins, theoretically, there exist 126 analogs (9 × 14), which include all the 56 known chaetoglobosins possessing the nucleus: indol-3-yl methyl unit, perhydro-isoindolone moiety, and macrocyclic ring (Fig. 1). All these known chaetoglobosins are the different assembly of A–I types in perhydro-isoindolone moiety combined with a–n types in macrocyclic ring. Thus according to the 1H-NMR rules of sub-types, analyzing the 1H-NMR spectra of chaetoglobosins can deduce the corresponding structures fast. The purpose of the following 10 examples (eight examples are included in ESI†) is to explain how to analyze 1H NMR spectral to elucidate the structures of chaetoglobosins, and then to dereplicate known analogs.
2.4 Analysis of 1H NMR spectra to dereplicate known chaetoglobosins
| Chaetoglobsin A (1) = (C-type + e-type) |
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–21CH
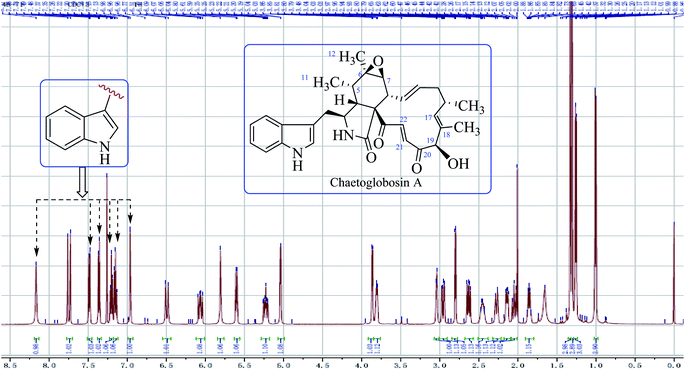
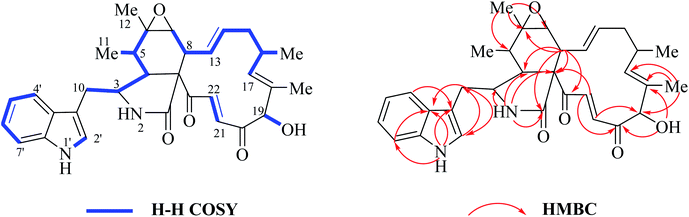
O–21CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) 22CH). The signal at 5.04 ppm (J = 4.5 Hz, H-19) coupled the one at 3.86 ppm (J = 4.5 Hz, 19-OH) revealed the structural features of 19-OH and 20-keto-group in the macro-ring. In addition, the olefinic protons H-13/H-14/H-17 together with the 16-Me (doublet) and 18-Me (singlet) in high field were observed in the 1H NMR spectrum, which confirms that this compound must possess the e-type of macrocyclic ring (Fig. 5a and b). The proton signals of H-7 (δH = 2.80, d, J = 5.0 Hz) along with the 11-Me (δH = 1.26, d) and 12-Me (δH = 1.30, s) in the up-field indicated the existence of a peroxide at C-6 and C-7. In addition, the H-3, H-4, H-5, H-8, and –CH2-10 were also were observed in the 1H NMR spectrum, which completely matched the C-type in perhydro-isoindolone moiety. Thus detailed analysis of the 1H NMR spectrum revealed that this chaetoglobosin belongs to the combination of e-type plus C-type (Fig. 5a and b). Searching the structural types of all known analogs, it demonstrated that only does chaetoglobosin A possess the combination of e-type and C-types.16 Finally, this conclusion was confirmed by 2D NMR spectra (Fig. 6).
22CH). The signal at 5.04 ppm (J = 4.5 Hz, H-19) coupled the one at 3.86 ppm (J = 4.5 Hz, 19-OH) revealed the structural features of 19-OH and 20-keto-group in the macro-ring. In addition, the olefinic protons H-13/H-14/H-17 together with the 16-Me (doublet) and 18-Me (singlet) in high field were observed in the 1H NMR spectrum, which confirms that this compound must possess the e-type of macrocyclic ring (Fig. 5a and b). The proton signals of H-7 (δH = 2.80, d, J = 5.0 Hz) along with the 11-Me (δH = 1.26, d) and 12-Me (δH = 1.30, s) in the up-field indicated the existence of a peroxide at C-6 and C-7. In addition, the H-3, H-4, H-5, H-8, and –CH2-10 were also were observed in the 1H NMR spectrum, which completely matched the C-type in perhydro-isoindolone moiety. Thus detailed analysis of the 1H NMR spectrum revealed that this chaetoglobosin belongs to the combination of e-type plus C-type (Fig. 5a and b). Searching the structural types of all known analogs, it demonstrated that only does chaetoglobosin A possess the combination of e-type and C-types.16 Finally, this conclusion was confirmed by 2D NMR spectra (Fig. 6).
The 1H NMR spectra analysis of other known analogues (2–9) were provided in ESI.†
2.5 Analysis of 1H NMR spectra to target new chaetoglobosin analog
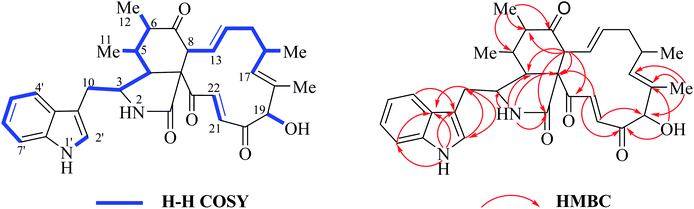
Analyzing the 1H NMR spectrum of chaetoglobosins can discriminate the differently combinatory types of the 56 known analogs. Thus new characteristics or combinations in the 1H NMR spectra different from those of 56 analogs will imply new chaetoglobosins. Recently, we isolated a series chaetoglobosins from OE::CgLaeA C. globosum (CBS148.51).26 Detailed analyzing the 1H NMR spectra of these compounds revealed that one of analogs contained different combination of sub-types, and it implied that this compound might be a new chaetoglobosin. This hypothesis was further supported by 2D NMR spectra. The following example will explain the characteristics of 1H NMR spectrum of this new compound with different sub-types found in this analog.| Chaetoglobosin Z (10) = (D-type + e-type) |
In the 1H NMR spectrum of chaetoglobosin Z (10) (Fig. 7a), the diagnostic proton signals of an indole-ring are observed in the down-fielded zone (Fig. 7b). The trans-olefinic protons with chemical shift values at 8.06 ppm (H-22, J = 16.5 Hz) and 6.59 ppm (H-21, J = 16.5 Hz) implied that C-20 must possess a keto-group leading the chemical shift value of H-22 to be significant down-field (Fig. 7c). Three signals of olefinic protons including H-13 (5.95 ppm, dd, J = 15.0, 10.0 Hz), H-14 (5.15 ppm, ddd, J = 15.0, 11.0, 3.5 Hz) and H-17 (d) are observed between 5.00–6.00 ppm. Considering the chemical shift value of H-17 (5.65 ppm), this revealed that C-19 do not possess the keto-group (if C-19 is oxidized to be corresponding keto-group, which will lead the chemical shift value of H-17 to be more down-fielded, < 6.00 ppm). The oxymethine proton (5.10 ppm, J = 3.5 Hz) coupled with the corresponding proton (3.88 ppm, J = 3.5 Hz) confirms that the C-19 might contain a free hydroxyl group same as those of chaetoglobosins A, B, J and others (Fig. 7d). On accounting of the fact that signals of 16-Me (doublet) and 18-Me (singlet) are not changed in the upfield zone, together considering the information above-mentioned, it implied that chaetoglobosin Z (10) must contain the e-type in macrocyclic ring.
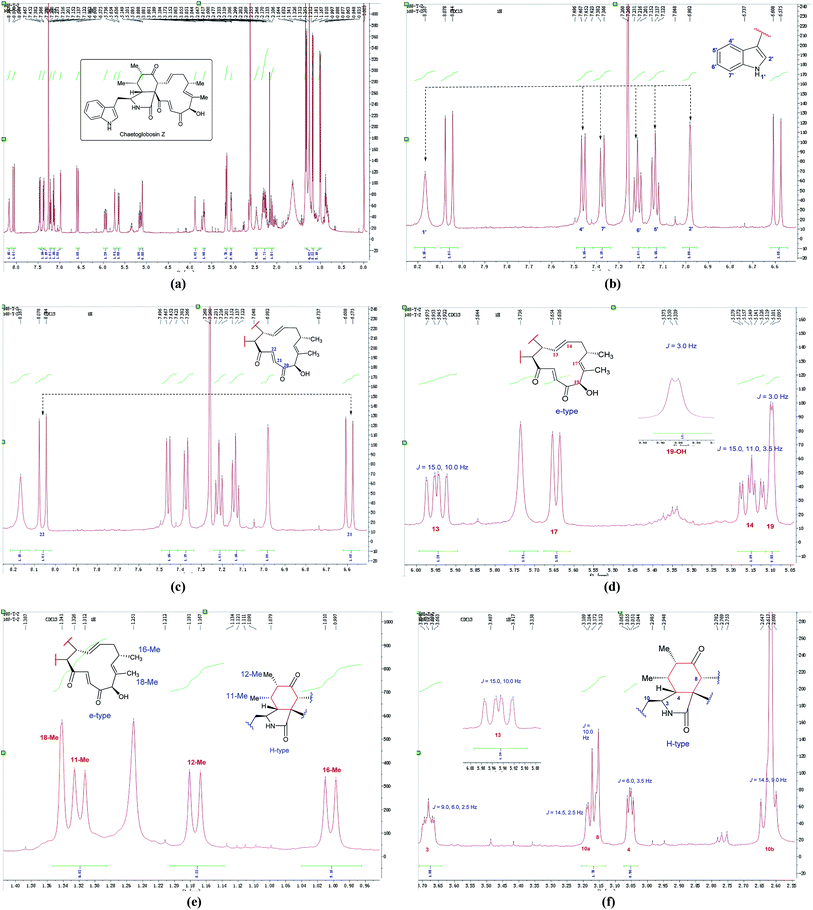 | ||
| Fig. 7 (a) The full spectrum of chaetoglobosin Z (10) in CDCl3. (b–f) Expanded 1H NMR spectrum of chaetoglobosin Z (10) in CDCl3. | ||
Two doublet peaks (11-Me and 12-Me) except 16-Me (doublet) and 18-Me (singlet) in the high field are observed (Fig. 7e), which implies that the C-5 and C-6 must be two methine groups. In addition, the chemical shift value of H-8 is down-fielded to 3.16 ppm as a doublet peak (J = 10.0 Hz) coupled with olefinic proton H-13 (5.95 ppm, dd, J = 15.0, 10.0 Hz) (Fig. 6e), which denotes that the C-7 must be oxidized to be the corresponding keto-group. The splitting patterns of H-3, H-4 and –CH2-10 are not changed, and the chemical shift values of these protons (H-3, H-4 and –CH2-10) show small differences (Fig. 7f), which reveals that perhydro-isoindolone moiety in chaetoglobosin Z (10) must be D-type. Thus, chaetoglobosin Z (10) contains the combination of D-type plus e-type, which is different from those found in other 56 known chaetoglobosins, implying that this compound might be a new chaetoglobosin analog. Finally, the planar, and relative configuration of this new compound were determined by 2D NMR spectra (Fig. 8).26
3. Conclusion
In conclusion, compared with other techniques, 1H NMR spectrum contains a wealth of diagnostic information, and analysis of 1H NMR spectrum is the first and key step, which can infer the structural fragments and even characterize the possibly planar structures. In this report, the different sub-types of perhydro-isoindolone moiety (A–I types, 9 sub-types) and of macrocyclic ring (a–n, 14 sub-types) in the chaetoglobosins are classified according to their possibly biogenetic pathway and structural features. Attempts to summarize the relationship between structural features and 1H NMR spectral are tried, which can dereplicate known analogs, and target new ones. According to the structural features of chaetoglobosins in this paper, new chaetoglobosins with different sub-type combinations or new sub-types are waiting to be found in future. More importantly, the results in this report will not only provide a useful approach to determine the different chaetoglobosins structures fast, but also could set a good example for other NPs structural elucidation.4. Experimental section
4.1 Samples
The compounds in this report have been isolated and characterized previously in the authors' laboratories.26–284.2 NMR Spectroscopy
1H, 13C and 2D NMR data were acquired with a Bruker 600 spectrometer using solvent signals (DMSO-d6: δH 2.50/δC 39.9; CDCl3: δH 7.26/δC 77.6) as references.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We gratefully acknowledge financial support from the National Key Research and Development Program of China “Research and Development of Comprehensive Technologies on Chemical Fertilizer and Pesticide Reduction and Synergism” (2017YFD0201402), CAMS Initiative for Innovative Medicine (2017-I2M-4-004), the National Major Increase or Decrease Project-Construction of the Sustainable Utilization Capacity of Famous Traditional Chinese Medicine Resources (2060302), National Natural Science Foundation of China (31570340, 31670011 and 31870332) and the Key Project at Central Government Level: The Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources (2060302).References
- J. N. David and M. C. Gordon, J. Nat. Prod., 2007, 70, 461–477 CrossRef PubMed.
- J. N. David and M. C. Gordon, J. Nat. Prod., 2012, 75, 311–335 CrossRef PubMed.
- P. N. Leão, H. Nakamura, M. Costa, A. R. Pereira, R. Martins, V. Vasconcelos, W. H. Gerwick and E. P. Balskus, Angew. Chem., Int. Ed., 2015, 54, 11063–11067 CrossRef PubMed.
- M. Badertscher, P. Bühlmann and E. Pretsch, 1H NMR Spectroscopy, Structure Determination of Organic Compounds, 2000, pp. 1–86 Search PubMed.
- G. F. Pauli, S. N. Chen, D. C. Lankin, J. Bisson, R. J. Case, L. R. Chadwick, T. Gödecke, T. Inui, A. Krunic, B. U. Jaki, J. B. McAlpine, S. Mo, J. G. Napolitano, J. Orjala, J. Lehtivarjo, S. P. Korhonen and M. Niemitz, J. Nat. Prod., 2014, 77, 1473–1487 CrossRef CAS PubMed.
- J. Schuemann and C. Hertweck, J. Am. Chem. Soc., 2007, 129, 9564–9565 CrossRef CAS PubMed.
- K. Scherlach, D. Boettger, N. Remme and C. Hertweck, Nat. Prod. Rep., 2010, 27, 869–886 RSC.
- Q. Zhang, H. Q. Li, S. C. Zong, J. M. Gao and A. L. Zhang, Mini-Rev. Med. Chem., 2012, 12, 127–148 CrossRef PubMed.
- S. Sekita, K. Yoshihira, S. Natori and H. Kuwano, Tetrahedron Lett., 1977, 32, 2771–2774 CrossRef.
- W. Jiao, Y. Feng, J. W. Blunt, A. L. Cole and M. H. Munro, J. Nat. Prod., 2004, 67, 1722–1725 CrossRef CAS PubMed.
- I. Chika, Y. Takeshi, I. Yoshinori, I. Katsuhiko and M. N. Atsushi, Tetrahedron, 2001, 57, 2997–3004 CrossRef.
- C. M. Cui, X. M. Li, C. S. Li, P. Proksch and B. G. Wang, J. Nat. Prod., 2010, 73, 729–733 CrossRef CAS PubMed.
- H. Oikawa, Y. Murakami and A. Lchihara, J. Chem. Soc., Perkin Trans. 1, 1992, 21, 2949–2953 RSC.
- C. M. Chen, H. C. Zhu, J. P. Wang, J. Yang, X. Y. Li, J. Wang, K. L. Chen, Y. Y. Wang, Z. W. Luo, G. M. Yao, Y. B. Xue and Y. H. Zhang, Eur. J. Org. Chem., 2015, 14, 3086–3094 CrossRef.
- C. Chen, Q. Tong, H. Zhu, D. Tan, J. Zhang, Y. Xue, G. Yao, Z. Luo, J. Wang, Y. Wang and Y. Zhang, Sci. Rep., 2016, 6, 18711 CrossRef CAS PubMed.
- S. Sekita, K. Yoshihira, S. Natori and H. Kuwano, Chem. Pharm. Bull., 1982, 30, 1618–1628 CrossRef CAS.
- S. Sekita, K. Yoshihira, S. Natori and H. Kuwano, Chem. Pharm. Bull., 1982, 30, 1629–1638 CrossRef CAS.
- G. Ding, Y. C. Song, J. R. Chen, C. Xu, H. M. Ge, X. T. Wang and R. X. Tan, J. Nat. Prod., 2006, 69, 302–304 CrossRef CAS PubMed.
- O. Hideaki, M. Yasunobu and I. Akitami, Biosci., Biotechnol., Biochem., 1993, 57, 628–631 CrossRef.
- A. Numata, C. Takaheshic, Y. Ito, K. Minoura, T. Yamada, C. Matsuda, T. Yamada, C. Matsuda and K. Nomoto, J. Chem. Soc., Perkin Trans. 1, 1995, 3, 239–245 Search PubMed.
- J. Zhang, H. M. Ge, R. H. Jiao, J. Li, Y. R. Wang, J. H. Wu, Y. C. Song and R. X. Tan, Planta Med., 2010, 76, 1910–1914 CrossRef CAS PubMed.
- A. Ichihara, K. Katayama, H. Teshima, H. Oikawa and S. Sakamura, Biosci., Biotechnol., Biochem., 1996, 60, 360–361 CrossRef CAS PubMed.
- M. Xue, Q. Zhang, J. M. Gao, H. Li, J. M. Tian and G. Pescitelli, Chirality, 2012, 24, 668–674 CrossRef CAS PubMed.
- S. Thohinung, S. Kanokmedhakul, K. Kanokmedhakul, V. Kukongviriyapan, O. Tusskorn and K. Soytong, Arch. Pharmacal Res., 2010, 33, 1135–1141 CrossRef CAS PubMed.
- Q. C. Zheng, M. Z. Kong, Q. Zhao, G. D. Chen, H. Y. Tian, X. X. Li, L. D. Guo, J. Li, Y. Z. Zheng and H. Gao, Fitoterapia, 2014, 93, 126–131 CrossRef CAS PubMed.
- T. Jiang, M. H. Wang, L. Li, J. G. Si, B. Song, C. Zhou, M. Yu, X. Wang, Y. Zhang, G. Ding and Z. M. Zou, J. Nat. Prod., 2016, 79, 2487–2494 CrossRef CAS PubMed.
- Q. F. Guo, Z. H. Yin, J. J. Zhang, W. Y. Kang, X. W. Wang and G. Ding, Molecules, 2019, 24, 3240 CrossRef CAS PubMed.
- Q. F. Guo, L. Chen, Z. H. Yin, J. J. Zhang, W. Y. Kang, X. W. Wang and G. Ding, Mycosystema, 2019, 38, 134–144 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra10434h |
| ‡ These authors made equal contributions to this work. |
| This journal is © The Royal Society of Chemistry 2020 |