Open Access Article
Open Access ArticleEfficient biodegradation of petroleum n-alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa san ai with multidegradative capacity†
Ana Medića,
Marija Lješevićb,
Hideyuku Inui c,
Vladimir Beškoskid,
Ivan Kojiće,
Ksenija Stojanovićd and
Ivanka Karadžić
c,
Vladimir Beškoskid,
Ivan Kojiće,
Ksenija Stojanovićd and
Ivanka Karadžić *a
*a
aDepartment of Chemistry, Faculty of Medicine, University of Belgrade, Višegradska 26, 11000 Belgrade, Serbia. E-mail: ivanka.karadzic@med.bg.ac.rs; Tel: +381113607067
bDepartment of Chemistry, Institute of Chemistry, Technology and Metallurgy, University of Belgrade, Njegoševa 12, 11000 Belgrade, Serbia
cBiosignal Research Center, Kobe University, 1-1 Rokkodaicho, Nada-ku, Kobe, Hyogo 657-8501, Japan
dFaculty of Chemistry, University of Belgrade, Studentski trg 12-16, 11000 Belgrade, Serbia
eInnovation Center of the Faculty of Chemistry, University of Belgrade, Studentski trg 12-16, 11000 Belgrade, Serbia
First published on 7th April 2020
Abstract
Pseudomonas aeruginosa san ai, an alkaliphilic, metallotolerant bacterium, degraded individual selected petroleum compounds, i.e., n-alkanes (n-hexadecane, n-nonadecane) and polycyclic aromatic hydrocarbons (fluorene, phenanthrene, pyrene) with efficiency of 80%, 98%, 96%, 50% and 41%, respectively, at initial concentrations of 20 mg L−1 and in seven days. P. aeruginosa san ai showed a high biodegradative capacity on complex hydrocarbon mixtures, the aliphatic and aromatic fractions from crude oil. The efficiency of P. aeruginosa san ai degradation of crude oil fractions in seven days reached stage 3–4 of the oil biodegradation scale, which ranges from 0 (no biodegradation) to 10 (maximum biodegradation). Identified metabolites concomitant with genomic and enzymatic data indicated the terminal oxidation pathway for the n-alkane degradation, and the salicylate and phthalate pathways for fluorene biodegradation. Polyextremophilic P. aeruginosa san ai, as a biosurfactant producer with multidegradative capacity for hydrocarbons, can be used in an improved strategy for environmental bioremediation of hydrocarbon-contaminated sites, including extreme habitats characterized by low or elevated temperatures, acidic or alkaline pH or high concentrations of heavy metals.
1. Introduction
Widespread use of crude oil/petroleum causes serious environmental problems in the forms of effluents from oil refinery plants and accidental oil spills. While some crude oil compounds are readily degraded, long-chain alkanes, and particularly polycyclic aromatic hydrocarbons (PAHs), are relatively resistant to biodegradation. Because of their carcinogenic and mutagenic properties, some PAHs are classified by regulatory agencies as high-priority pollutants that pose risks to humans and wildlife.1 Bioremediation techniques based on the use of microorganisms capable of heterogeneous contaminant degradation have been developed as an alternative to chemical and physical techniques for cleaning up polluted environments, as they have a comparative advantage because they are economically viable and less harmful to the environment.1 During bioremediation, organic pollutants are transformed into compounds with reduced toxicity or are completely degraded/mineralized. These techniques do not generate any waste, and the cultivated land can recover its natural biological activity.2,3Several pseudomonads have been reported to degrade hydrocarbons with various chain lengths and structures. Pseudomonas putida was found to degrade n-alkanes,4 P. aeruginosa W10 preferentially utilized n-C16, but also degraded naphthalene, phenanthrene, fluoranthene and pyrene,5 Pseudomonas F274 was found to utilize fluorene,6 Pseudomonas NCIB 9816-4 had the ability to degrade fluorene, dibenzofuran and dibenzothiophene,7 and P. aeruginosa DQ8, isolated from petroleum-contaminated soil, could degrade PAHs such as fluorene, phenanthrene, fluoranthene and pyrene.8 Being able to utilize a broad spectrum of hydrocarbons, Pseudomonas genus has a key role in the removal of these compounds from petroleum-polluted sites.9 This is particularly important because hydrocarbon-degrading extremophiles can tolerate a wide range of conditions and could be used for bioremediation of polluted extreme habitats.10,11
There are four oxidation pathways of n-alkane degradation: (i) the monoterminal/terminal pathway where terminal methyl produces the primary alcohols further oxidized to fatty acid;12 (ii) the biterminal pathway in which the termini of the n-alkane undergo oxidation to the corresponding fatty acids;12 (iii) the subterminal pathway where the subterminal carbon is oxidized to an ester,13 and; (iv) a unique pathway postulated for Acinetobacter sp. where n-alkanes are oxidized to fatty acids through n-alkyl hydroperoxides and aldehydes.14 As pathways for n-alkane catabolism vary in different microorganisms, the genetic bases, enzymes and metabolites need to be elucidated in order to characterize the specific mechanism of n-alkane degradation in each specific microorganism.
Although there are plenty of PAH degradation pathways depending on the number of fused rings, catabolism of PAHs usually starts with hydroxylation to activate the ring, followed by ring-cleaving and several transformations that lead to formation of two key intermediates, phthalate or salicylate, which are further degraded into metabolites of the tricarboxylic acid cycle (TCA).6,8,15–17 However, in spite of extensive investigation of PAH catabolism, there is a lack of knowledge about the mechanisms involved in degradation of different PAHs, as well as about the whole metabolic network that includes metabolites and enzymes that mediate the reactions. Among the PAHs, bacterial degradation of fluorene (FLU) has been particularly well studied because of its environmental significance. Three major degradation pathways of FLU have been described via angular and lateral dioxygenation or monooxygenation as the upper catabolic pathways.18,19 Products of the upper pathway can then be transformed by intradiol or extradiol ring-cleaving dioxygenases through either an ortho- or a meta-cleavage pathway, resulting in intermediates such as protocatechuates and catechols that are further funneled into the β-ketoadipate pathway.20–22
P. aeruginosa san ai was isolated from industrial alkaline mineral metal-cutting oil, as its natural habitat.22 This bacterium is a polyextremophile (alkaliphilic, metallotolerant)22–24 that produces several biotechnologically important compounds, such as extracellular enzymes23 and the biosurfactant classes of rhamnolipids25 and exopolysaccharides,26 which can facilitate the emulsification of hydrocarbons.
The goal of this study was to analyze the capacity of P. aeruginosa san ai to degrade and biotransform the individual, selected petroleum compounds, n-hexadecane (n-C16), n-nona-decane (n-C19), fluorene (FLU), phenanthrene (PHE) and pyrene (PYR), and complex mixtures of petroleum compounds, i.e. the aliphatic and aromatic hydrocarbon fractions isolated from crude oil. To confirm the multidegradative capacity of P. aeruginosa san ai, production of metabolites, the presence of genes and the activities of enzymes involved in degradation pathways of individual n-alkanes and PAHs were investigated. In addition, respiration of P. aeruginosa san ai grown on n-C16, n-C19, FLU, PHE and PYR was automatically measured to evaluate CO2 production and efficiency of degradation.
2. Materials and methods
Chemicals
n-C16, n-C19, FLU, PHE, PYR, and organic solvents were purchased from Sigma Aldrich (St. Louis, MO, USA) with purity above 99% and HPLC grade. All other chemicals used in this study were of pro analysis grade purchased from Merck (Darmstadt, Germany). Non-biodegraded, paraffinic crude oil used in this study was taken from the Turija-Sever oil field (South-East Pannonian Basin, Serbia).Media and growth conditions
The microorganism used for the degradation study of n-C16, n-C19, FLU, PHE, PYR and crude oil fractions was P. aeruginosa san ai, originally isolated from alkaline cutting oil rich in heavy metals.23 P. aeruginosa san ai grows well in laboratory conditions, in media with alkaline pH up to 9.8,23 it tolerates high concentrations of cadmium up to 7.3 mM,22 and chromium up to 5.0 mM.27 Consequently, P. aeruginosa san ai is an alkaliphilic and metallotolerant bacterium.24 The isolate is deposited in NCAIM (National Collection of Agricultural and Industrial Microorganisms), Faculty of Food Sciences, Corvinus University of Budapest, Hungary, labeled as NCAIM B.001380 and in the public collection ISS WDCM 375 (Institute of Soil Science, Belgrade, Serbia) with accession number ISS 619.Aliphatic and aromatic hydrocarbon fractions used in this study were isolated from non-biodegraded, paraffinic crude oil (Turija-Sever oil field, SE Pannonian Basin, Serbia) according to the procedure described by Bastow et al.28
P. aeruginosa san ai was activated on nutrient agar (Torlak, Belgrade, Serbia) at 30 °C for 24 h and transferred to 500 mL Erlenmeyer flasks, containing 100 mL of minimal salt medium (MSM) as previously reported,29,30 supplemented with different sources of carbon and energy, to achieve initial counts (in colony-forming units; CFU mL−1) of approximately 4 × 107 mL−1. n-C16, n-C19, FLU, PHE, PYR, and aliphatic and aromatic hydrocarbon fractions isolated from Turija-Sever crude oil were used as sole carbon sources, while sunflower oil was used as a control substrate for bacterial growth. An appropriate volume of a stock solution of each sole carbon source in n-hexane was injected individually into sterile 500 mL Erlenmeyer flasks to obtain 20 mg L−1 of the sole carbon source, after which n-hexane was allowed to evaporate under the airflow. After forming a thin film of n-C16, n-C19, FLU, PHE, PYR or crude oil fraction in the bottom of the flask, 100 mL of sterilized MSM was added into each flask31 to achieve the final concentration of the sole carbon source of 20 mg L−1, and each flask was shaken at 150 cycles min−1 using a horizontal shaker (Kuhner, Basel, Switzerland) at 30 °C for 7 days.
Sterile MSM with n-C16, n-C19, FLU, PHE, PYR, or aliphatic and aromatic hydrocarbon fraction isolated from crude oil were utilized as controls of chemical decomposition, and inoculated sterile MSM without any of the hydrocarbon substrates were prepared as controls of microbial growth. All experiments were performed in three independent replicates and average values are given in the study. Differences in results between replicates in all experiments did not exceed 1.2%.
Production of biosurfactants, i.e. rhamnolipid and exopolysaccharide, was determined on MSM supplemented with crude oil as a sole C-source.25
Microbial growth was measured at 580 nm using a UV-2600 spectrophotometer (Shimadzu, Japan). As the intensities of microbial growth on n-C16, n-C19, FLU, PHE, PYR, and the hydrocarbon fractions from crude oil were similar to the measured growth intensity on sunflower oil,25 any increase of absorbance of >0.05 after 4 days was considered as culture growth.31
Gas chromatography-mass spectrometry analysis of degradation of individual n-alkanes, PAHs, and crude oil fractions
Each whole flask culture was extracted with n-hexane three times. The extracts were dehydrated with anhydrous Na2SO4, and the solvents were evaporated under reduced pressure by a rotary evaporator (Senco GG17, Shanghai, China). The extraction residues were dissolved in n-hexane and analyzed by gas chromatography-mass spectrometry (GC-MS). A gas chromatograph Agilent 7890A GC (HP5-MS capillary column, 30 m × 0.25 mm, 0.25 μm film thickness, He carrier gas 1.5 cm3 min−1) coupled to an Agilent 5975C mass selective detector (70 eV) was used. The column was heated from 80 to 300 °C at a rate of 2 °C min−1, held at 300 °C for 20 minutes and then heated from 300 to 310 °C at a rate of 10 °C min−1, and held at 310 °C for 1 minute. The injector temperature was 250 °C. The spectrometer was operated in the EI (electron ionization) mode over a scan range from m/z 45 to 550. The individual peaks for n-C16, n-C19, FLU, PHE, PYR and their degradation products, as well as the compounds in the aliphatic and aromatic fractions of crude oil, were identified on the basis of their mass spectra (library NIST11). Quantification of the compounds in standard series, controls (sterile MSM with n-C16, n-C19, FLU, PHE, and PYR) and inoculated samples (P. aeruginosa san ai inoculated into sterile MSM supplemented with n-C16, n-C19, FLU, PHE, and PYR) were performed by integration of peak areas (software GCMS Data Analysis). For the calculations, a standard series of commercial n-C16, n-C19, FLU, PHE, and PYR solutions in n-hexane concentrations from 2 to 20 mg L−1 (2, 5, 7, 10, 15 and 20 mg L−1) were prepared and analyzed under the same GC-MS conditions as samples. The ratios of the peak areas to corresponding concentrations were linear, with very high correlations (r2 ≥ 0.991). The degradation efficiency of individual hydrocarbons was determined based on the concentrations of target compounds in control and inoculated samples, and is given in percent. The degradation efficiency of crude oil fractions was determined based on the mass loss of the fraction before and after microbial treatment and notable changes in distributions and abundances of individual compounds in total ion chromatograms (TICs) of control and inoculated aliphatic and aromatic fractions.Comprehensive two dimensional gas chromatography-quadrupole mass spectrometry analysis of fluorene metabolites
For analysis of FLU metabolites, ethyl acetate extracts were prepared in the same way as n-hexane extracts, except that the solvent was ethyl acetate. These extracts were derivatised with N,O-bis(trimethylsilyl)trifluoroacetamide with trimethylchlorosilane (BSTFA + TMCS) (Sigma-Aldrich) for 45 min at 60 °C and analyzed using a GC-MS-QP2010 Ultra (Shimadzu, Kyoto, Japan) comprehensive two dimensional gas chromatograph-quadrupole mass spectrometer (GC × GC-MS) with ZX2 thermal modulation system (Zoex Corp.). Two columns, a RtxR-1 (RESTEK, CrossbondR 100% dimethyl-polysiloxane, 30 m × 0.25 mm, 0.25 μm film thickness) and a BPX50 (SGE Analytical Science, 1 m × 0.1 mm, 0.1 μm film thickness), were connected through the GC × GC modulator as the first and second capillary columns, respectively. The oven was programmed at an initial temperature of 40 °C for 5 min and then ramped at 5 °C min−1 to 300 °C, which was held for 5 min. The modulation period was 6 s. The scan range was m/z 70–500. The data were analyzed using GCImage R2.8 (GCImage LLC), and metabolites were identified based on the comparison to the NIST library. Semiquantitative analysis of metabolites was estimated using the area over which analytes ranged in the GC × GC-MS chromatograms.Determinations of rhamnolipid and exopolysaccharide
Concentrations of rhamnolipid (RL) and exopolysaccharide (EPS) were determined spectrophotometrically using the UV-2600 (Shimadzu, Japan). The concentration of RL in the culture broth was determined by the orcinol assay.25 The reaction mixture of 0.15 mL of sample and 1.35 mL of orcinol reagent (0.19% orcinol in 53% sulfuric acid) was heated for 30 min at 80 °C, then cooled to room temperature and the absorbance was measured at 421 nm. The RL concentration was calculated from standard curves prepared with L-rhamnose and expressed as a rhamnose equivalent, where 1 μg rhamnose is equivalent to 2.5 μg RL.The concentration of EPS in the fermentation broth was determined by the phenol-sulfuric acid method.32 The reaction mixture consisted of 0.1 mL of supernatant, 0.9 mL of distilled water, 0.05 mL of 80% phenol solution and 2.5 mL of concentrated sulfuric acid. Reaction mixtures were vortexed and absorbances measured at 490 nm after 10 minutes. The EPS concentration was calculated from standard curves prepared with starch solution at concentrations from 0 to 100 mg L−1.
Respiration measurement
Microbial respiration activity of P. aeruginosa san ai was measured using a twelve-channel Micro Oxymax® respirometer (Columbus, USA) connected to a PC. The experiments were performed in Micro Oxymax light-proof 500 mL bottles containing 100 mL of MSM supplemented with 20 mg L−1 of n-C16, n-C19, FLU, PHE or PYR and stirred constantly (150 rpm) with a magnetic stirrer at 27 °C for four days. Produced carbon dioxide (μL) was determined. Cell respiration was measured every 300 min for four days. Data were evaluated by Micro Oxymax® software.Genome analysis
The databases Integrated Microbial Genomes (IMG)33 https://img.jgi.doe.gov/cgi-bin/m/main.cgi and Kyoto Encyclopedia of Genes and Genomes (KEGG)34 https://www.genome.jp/kegg/ were used to analyze the P. aeruginosa san ai genome. The genome was deposited in Gen Bank: BioProject PRJNA19571 with the accession number JMKR00000000.24Enzyme assays
Total proteins were isolated from biomass grown in MSM supplemented with different C-sources (n-C16, n-C19, FLU, PHE, or PYR) to early stationary phase, by the method described by Medić et al.29 Crude protein extract was used for enzyme assays after determination of protein content by Bradford's method.35The specific enzyme activity was calculated as enzyme activity per mg of protein.
1,2-Catechol dioxygenase (C12O) and 2,3-catechol dioxygenase (C23O) activities were measured spectrophotometrically at 260 nm and 375 nm,36,37 respectively, using the UV-2600, by a slightly modified procedure,29 and with FLU, PHE, or PYR as substrates. One unit of activity is defined as the amount of C12O and C23O enzyme, respectively, producing 1 μmol of cis,cis-muconate (ε260 = 1.6 × 104 mol−1 cm−1),37 or 2-hydroxymuconic semialdehyde (ε375 = 4.4 × 104 mol−1 cm−1),38 per minute at 25 °C.
Hydroxylase activity was measured spectrophotometrically as a decrease of absorbance of NADH at 340 nm by the slightly modified procedure of Jauhari et al.;39 therefore, we used 20 mM sodium phosphate instead of Tris–HCl buffer, pH 7.4.
Alcohol dehydrogenase was measured against ethanol, propanol and iso-propanol according to Jauhari et al.39
Hydroxylase and alcohol dehydrogenase activities were calculated using the NADH extinction coefficient of ε340 of 6220 mol−1 cm−1. One enzyme unit is defined as the amount of enzyme which in the presence of substrate causes the oxidation of 1 μmol NADH per min.
3. Results and discussion
Degradation of individual n-alkanes and PAHs by P. aeruginosa san ai
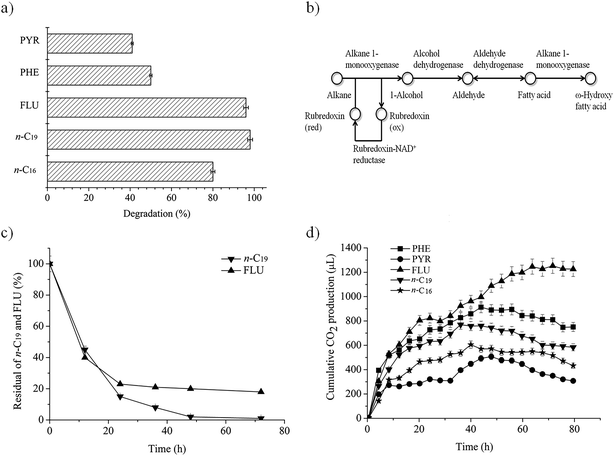
As Fig. 1a shows, P. aeruginosa san ai has a high capacity for degradation of n-alkanes (n-C16, n-C19) and PAHs (FLU, PHE, PYR). The degradation efficiency of individual hydrocarbons (initial concentrations of 20 mg L−1) reached 80%, 98%, 96%, 50% and 41%, respectively, over a period of seven days. Therefore, P. aeruginosa san ai can be considered as a multidegradative hydrocarbonoclastic bacterium with the ability to degrade n-alkanes and PAHs. PAHs with three rings (FLU, PHE) were significantly better degraded than PYR as a four-ring PAH (Fig. 1a), similar to the observation made by Zhang et al.8 FLU, as a naphthenoaromatic hydrocarbon (containing aromatic and methylenic moieties) was almost completely degraded. An increase in the number of fused rings results in increased hydrophobicity and decreased solubility that leads to reduced availability of such compounds for microbial degradation. FLU, the most soluble compound (1.992 mg L−1), was completely exhausted, while almost 60% of PYR remained non-degraded as a result of its lower solubility (0.135 mg L−1). Several other P. aeruginosa isolates were reported to degrade PAHs similarly to P. aeruginosa san ai. P. aeruginosa DQ8 completely degraded 40 mg L−1 of FLU and PHE in 7 days, and 34% of PYR (40 mg L−1) in 12 days.8 P. aeruginosa BZ-3 degraded 75% of PHE at an initial concentration of 50 mg L−1 in 7 days.31 P. aeruginosa completely removed PHE in concentrations up to 200 mg L−1 in 30 days.40 P. aeruginosa ATAI19 degraded 31% of PYR at an initial concentration of 50 mg L−1 in 9 days.41Complete degradation of low soluble n-alkanes (hexadecane – 0.023 mg L−1, nonadecane – 0.008 mg L−1) seems to be a surfactant-mediated process in which alkane-degrading bacteria produce diverse surfactants.42 In addition, biosurfactant producing P. aeruginosa S5 highly promoted the removal of PAHs, implying that hydrocarbons removal is a surfactant-mediated process.43 Indeed, P. aeruginosa san ai produces a mixture of the biosurfactants, rhamnolipids and exopolysaccharide, which could facilitate the emulsification of the hydrocarbons. Specifically, during P. aeruginosa san ai growth on medium supplemented with crude oil, both biosurfactants were detected (37.4 mg L−1 for RL and 39.7 mg L−1 for EPS).25,26 A similar affinity for medium-chain alkanes was found for P. aeruginosa B1,44 while P. aeruginosa SJTD-1,45 RM1, SK146 and D88 efficiently degraded medium- and long-chain n-alkanes. P. aeruginosa D8 completely removed n-C22, n-C30 and n-C40, concentration 100 mg L−1, in 7 days.
In spite of the limited bioavailability of n-alkanes and PAHs due to their low solubility, Pseudomonas has been regarded as critical for recycling organic carbon on the planet.47 Compared to other hydrocarbonoclastic bacteria, P. aeruginosa san ai has an additional advantage, as it has a particular ability to survive and to grow in extreme environments such as alkaline and heavy metal polluted sites.22,23 It seems that there are similarities in the efficacy and capacity to degrade hydrocarbons between extremophilic P. aeruginosa that have originated from different extreme conditions. So, for example, the thermophilic bacterium P. aeruginosa AP02-1 has a high multidegradative capacity, ranging from n-alkanes to PAH,48 while the halotolerant P. aeruginosa Asph2 successfully degrades different fractions of petroleum hydrocarbons49 similar to polyextremophilic P. aeruginosa san ai. Heavy metals (cadmium, chromium, lead, zinc and, particularly, nickel and vanadium) are present in crude oil, oil spills, and in alkaline oil refinery effluents.50 In conditions of crude oil pollution caused by spills or oil industry activity, the alkaliphilic, metallotolerant, hydrocarbonoclastic P. aeruginosa san ai could be an ideal microorganism that can be used for remediation of polluted environments.
Dynamics of n-nonadecane and fluorene degradation
To investigate the dynamics of degradation, n-C19 and FLU were selected as representative of alkanes and PAHs, as these two classes of substances were efficiently degraded by P. aeruginosa san ai (98% and 96%, respectively; Fig. 1a). As depicted in Fig. 1c, the degradation of both hydrocarbons fits first order kinetics, being in agreement with degradation kinetics of other n-alkanes and PAHs.6,31,39 Biodegradation rate constants were calculated to be 1.59 and 0.51 (per day) for n-C19 and FLU, respectively, using the following equation: ln[A] = ln[Ao] − kt, where [A] is the residual concentration of hydrocarbon (n-C19 and FLU) at a specific time, [Ao] is the initial hydrocarbon concentration, k is the biodegradation rate constant, and t is time (days). The biodegradation rate constant for n-C19 by P. aeruginosa san ai of 1.59 per day is higher than that published for Pseudomonas BP10 (0.11 for n-C26),39 indicating easier degradation of the shorter, odd number n-alkane. This result is in concordance with biodegradation in natural conditions, where both aerobic and anaerobic bacteria preferably utilize short-chain n-alkanes.51 Additionally, P. aeruginosa san ai has better capacity for PAH degradation (0.51 per day for FLU), than Pseudomonas BZ-3 (0.11 per day for PHE).31The half-life (t½) of n-C19 and FLU was calculated using the equation t½ = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/k, and it was estimated as 0.43 and 1.36 days, respectively. The higher constant rate and lower t½ for n-C19 indicate the easier biodegradability of n-alkanes than PAHs. Apparently, P. aeruginosa san ai quickly removed both hydrocarbons from the growth medium that contained 20 mg L−1 of n-C19 or FLU, clearly demonstrating the excellent potential of this bacterium for aerobic biodegradation of n-alkanes and PAHs. Grifoll et al.6 revealed intensive FLU utilization within the first three days of growth and almost complete degradation of FLU by Pseudomonas F274, which is in good agreement with our data.
2/k, and it was estimated as 0.43 and 1.36 days, respectively. The higher constant rate and lower t½ for n-C19 indicate the easier biodegradability of n-alkanes than PAHs. Apparently, P. aeruginosa san ai quickly removed both hydrocarbons from the growth medium that contained 20 mg L−1 of n-C19 or FLU, clearly demonstrating the excellent potential of this bacterium for aerobic biodegradation of n-alkanes and PAHs. Grifoll et al.6 revealed intensive FLU utilization within the first three days of growth and almost complete degradation of FLU by Pseudomonas F274, which is in good agreement with our data.
Respiration activity
P. aeruginosa san ai can use n-alkanes and PAHs in MSM as sole carbon and energy sources. The disappearance of hydrocarbons accompanied by CO2 production implies metabolic and respiratory activity of the microorganism during its growth on these carbon sources. Cumulative CO2 production, monitored continuously during the biodegradation of n-alkanes and PAHs, is given in Fig. 1d. All substrates caused intensive respiration, implying the active metabolism of the bacterium when growing on n-alkanes and PAHs. The most intensive respiration in all cases occurred in the first day, with cumulative CO2 production (μL) of: 280 (PYR), 470 (n-C16), 630 (n-C19), 750 (PHE) and 820 (FLU). The respiration levels did not entirely correlate with the degradation percentages from Fig. 1a, which is a result of differences in metabolism during the biodegradation of n-alkanes and PAHs. Salicylic and phthalic acids, as key intermediates of PAHs degradative pathways, both funnel into the β-ketoadipate pathway, the main products of which are succinate (C4) and adipate (C6) that go into central TCA metabolism.52 On the other hand, degradation of n-alkanes produces acetate (C2) as a result of β-oxidation, which induces the glyoxylate cycle to bypass the steps in the TCA cycle and to prevent the loss of CO2. Therefore, there is a difference between the TCA and the glyoxylate pathways in carbon balance, which is C2 → 2C1 for the TCA, while for the glyoxylate cycle, it is C2 + C2 → C4.53 Besides, key enzymes of the glyoxylate cycle, isocitrate lyase and malate synthase, are induced by n-alkane.54,55 Taking all these metabolic facts together, it becomes clear why the respiration levels of hydrocarbons do not entirely correlate with the degradation percentages.Biodegradation of crude oil fractions by P. aeruginosa san ai
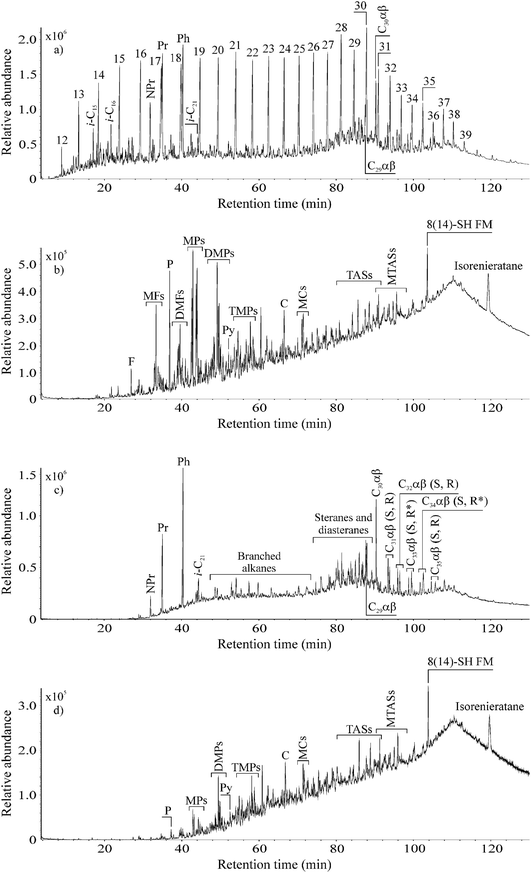
In order to study the degradation potential of P. aeruginosa san ai in complex hydrocarbon mixtures, experiments were carried out on aliphatic and aromatic fractions isolated from non-biodegraded, paraffinic crude oil. Oil was separated into the fractions since it contains a huge number of hydrocarbons, and without separation it is difficult to perform precise identification (particularly in TIC) due to the co-elution of more than one compound in the single GC-MS chromatographic peak. Moreover, some compounds that are present in lower amounts (particularly aromatic hydrocarbons) could be masked by compounds such as n-alkanes that are present in higher concentrations in crude oil. Nevertheless, both our aliphatic and aromatic fractions contained numerous hydrocarbons having different structures (Fig. 2a and b).The mass of the aliphatic hydrocarbon fraction was reduced by 45%, whereas the mass of the aromatic hydrocarbon fraction was reduced by 25%. In controls, no mass change of oil fraction was detected. This result clearly shows the lower resistance of aliphatic than aromatic hydrocarbons against biodegradation by P. aeruginosa san ai, as we observed in experiments with individual hydrocarbons (Fig. 1a and c), and as was previously proven in natural conditions i.e. oil reservoirs that are located at low depths usually up to 800 m and where temperatures do not exceed 70–80 °C.51
Comparison of TICs in control and inoculated aliphatic fraction revealed significant differences. The full series of n-alkanes from C12 to C39 which notably dominated the TIC of the aliphatic fraction in the control, was completely removed by P. aeruginosa san ai in 7 days (Fig. 2a and c). As a result, the inoculated aliphatic fraction was then dominated by isoprenoids, hopanes and steranes (Fig. 2c). Detailed examination of the inoculated aliphatic fraction using m/z 71 (the characteristic ion fragmentogram for n-alkanes) confirmed the complete absence of these hydrocarbons (Fig. S1, ESI†). n-Alkanols and fatty acids, which are possible products of n-alkane degradation, were not detected at all in our inoculated aliphatic fraction (non-derivatised n-hexane extract), even when using their specific ion fragmentograms, indicating complete mineralization of n-alkanes during microbial growth on complex hydrocarbon mixtures.
Comparison of TICs of the control and inoculated aromatic fractions showed that P. aeruginosa san ai completely degraded FLU and almost all the PHE in 7 days (Fig. 2b and d). This clearly confirms the high capability of P. aeruginosa for FLU degradation. The contents of methyl- and dimethyl-derivatives of FLU and PHE which prevailed in the control aromatic fraction were notably decreased in the inoculated crude oil aromatic fraction, whereas chrysene, methylchrysenes, triaromatic steroids, 8(14)-secohopanoid with fluorene moiety, and isorenieratane contents were unaltered (Fig. 2b and d). This is not surprising, since it is known that, for example, triaromatic steroids are the compounds that are most resistant to microbial attack in oil wells.51 More detailed inspection of ion chromatograms of naphthalene (m/z 142 + 156 + 170), FLU (m/z 166 + 180 + 194) and PHE (m/z 178 + 192 + 206) derivatives of control and inoculated aromatic fractions (Fig. S2, ESI†) shows that resistance of aromatic hydrocarbons against biodegradation increases in the following order: naphthalene derivatives < fluorene derivatives < phenanthrene derivatives. Furthermore, it is obvious that resistance of aromatic hydrocarbons to biodegradation increases with the degree of methylation. Both observations are in concordance with results from natural oil wells.56,57
The changes detected in distributions of aliphatic and aromatic hydrocarbons in control and inoculated fractions indicate the hydrocarbon degradation efficiency of P. aeruginosa san ai during 7 days corresponds to stage 3–4 of the oil biodegradation scale (in oil reservoirs), which ranges from 0 to 10 (0 indicates no biodegradation, whereas stage 10 equates to removal of almost all hydrocarbons from oil with the exception of oleanane, gammacerane, diasteranes, diahopanes and aromatic steroids, which are, however, also altered).51,58,59 For comparison, the same biodegradation degree (stage 3–4) in oil wells is attained after hundreds of thousands or millions of years.60 Therefore, the proven degradation capacity of P. aeruginosa san ai against complex hydrocarbon mixtures follows the biodegradation sequence in natural geological and environmental conditions.
Degradation of n-hexadecane and n-nonadecane
A collection of genes encoding for proteins involved in alkane degradation, i.e., alkane monooxygenase, rubredoxin, rubredoxin reductase and alcohol and aldehyde dehydrogenases, have been found in the P. aeruginosa san ai genome, as shown in Fig. 1b.12 In addition to extensively studied alkanes with an even number of carbon atoms, P. aeruginosa san ai has the ability to transform the odd number alkane n-C19 that, to the best of our knowledge, has not been reported yet for P. aeruginosa. P. aeruginosa san ai efficiently degraded both of the analyzed n-alkanes (Fig. 1a). Based on GC-MS analysis, the following n-C16 metabolites were identified: 1-hexadecanol (Mw = 242, characteristic m/z fragment ions: 55, 69, 83, 97, 111, 125) and n-hexadecanoic acid (Mw = 256, characteristic m/z fragment ions: 73, 60, 129, 213, 83, 97, 157, 171), unambiguously demonstrating a terminal oxidation metabolic pathway for n-alkane degradation in P. aeruginosa san ai. However, only 1-nonadecanol with Mw = 284 and fragment ions (m/z) 55, 69, 83, 97, 111, 125, 139 were found as a result of n-C19 degradation transformation. In addition to the detected 1-nonadecanol-specific activity of the first, activating enzyme in the metabolic pathway, alkane monooxygenase/hydroxylase was determined: 0.234 U mg−1 and 0.436 U mg−1 for n-C16 and n-C19, respectively, signifying hydroxylation of both alkanes and even more intensive hydroxylation of n-C19. Alcohol dehydrogenase activities towards ethanol, propanol and iso-propanol from biomass of P. aeruginosa san ai grown on n-C16 (0.281, 0.225 and 0.348 U mg−1) and n-C19 (0.421, 0.251 and 0.204 U mg−1) explicitly show high enzyme activity against alcohols. In accordance, several genes coding for alcohol dehydrogenases were found in the P. aeruginosa san ai genome, including a propanol-preferring enzyme, for which very broad substrate specificity toward primary and secondary alcohols has been referred (https://www.brenda-enzymes).Degradation of fluorene
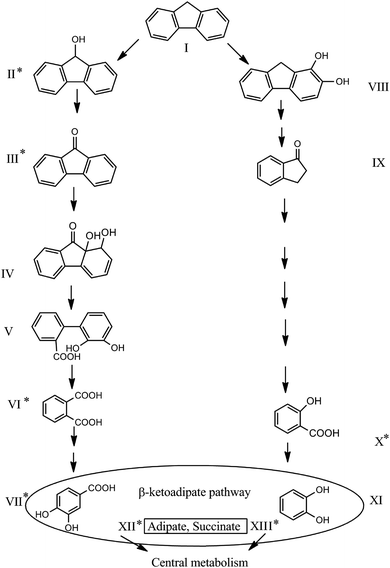
In this research, the catabolic pathway of FLU degradation was studied, as FLU is the simplest naphthenoaromatic, and bears structural relationships to carbazoles, dibenzothiophenes, dibenzofurans and dibenzodioxins, so thus, it can be a useful model for biodegradation studies. Moreover, FLU is an important component of fossil fuels and is one of the compounds on the EPA Priority Pollutants List. In the present study using GC × GC-MS, six metabolites were identified during FLU degradation by P. aeruginosa san ai (Table 1 and Fig. S3, ESI†). The genes encoding for proteins involved in the catabolism of aromatic compounds were identified in P. aeruginosa san ai, implying the potential of this bacterium for PAH degradation.24,29| Compound number referred to in Fig. 3 | Retention time (min) | m/z (% of relative intensity) | Metabolite identification according to NIST library | Duration of biodegradation by P. aeruginosa san ai | ||
|---|---|---|---|---|---|---|
| 18 h | 24 h | 48 h | ||||
| a + – present; *nd – not detected. | ||||||
| I | 34.80 | 166 (100), 165 (81), 167 (24), 164 (17), 82 (16), 163 (13) | Fluorene | + | + | + |
| II | 35.70 | 182 (100), 181 (78), 166 (29), 165 (28), 183 (24), 152 (18) | 9H-Fluoren-9-ol | + | + | + |
| III | 38.20 | 180 (100), 152 (39), 181 (22), 151 (19), 76 (17), 150 (11), 153 (8) | 9H-Fluoren-9-one | + | + | + |
| VI | 37.30 | 147 (100), 73 (63), 148 (19), 295 (14), 149 (9) | 1,2-Benzene-dicarboxylic acid, bis(trimethylsilyl)ester (phthalic acid, di-TMS) | + | + | + |
| VII | 40.40 | 73 (100), 193 (85), 370 (23), 194 (18), 74 (13), 355 (13) | Benzoic acid, 3,4-bis[(trimethylsilyl)oxy]-trimethylsilylester (protocatechuic acid, di-TMS) | nd* | nd | + |
| X | 36.00 | 73 (100), 267 (95), 268 (24), 74 (15), 91 (9) | Salicylic acid (2TMS) | + | + | + |
| XII | 32.90 | 73 (100%), 75 (38%), 111 (33%), 147 (25%) | Adipic acid (2TMS) | — | — | + |
| XIII | 27.70 | 147 (100%), 73 (65%), 148 (23%), 75 (18%) | Succinic acid (2TMS) | — | — | + |
The presence of 9H-fluoren-9-ol (II) and 9H-fluoren-9-one (III) implied that ring activation starts with monooxygenation at C-9, which is then dehydrogenated to 9H-fluoren-9-one (Fig. 3). In addition, high specific activities of hydroxylases (0.420, 1.286, and 0.846 U mg−1 for FLU, PHE and PYR, respectively), clearly demonstrated extensive enzymatic ring activation. Compounds IV (1,1a-dihydroxy-1-hydro-9-fluorenone) and V (2′-carboxy-2,3-dihydroxybiphenyl)6,16 were not found in the present study, but the final product of upper pathway transformation – phthalate (VI) – was detected (Fig. 3). As Fig. 3 shows, most of the metabolites identified in this study support well the proposed FLU degradation by the phthalate pathway.6,16 Salicylic acid (X) was also detected (Fig. 3), suggesting the existence of an additional pathway which starts with dioxygenation of FLU (compound VIII) and proceeds through several derivatives of 1-indanone (compound IX17). Furthermore, the GC × GC-MS peak ratio of phthalic to salicylic acids was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, implying an increased accumulation of salicylic acid over time, although metabolites of the salicylate pathway were not identified after 18 h of microbial growth. Substantial secretion and accumulation of intermediates in Pseudomonas growing on aromatic compounds were determined by the limited activity of some of the enzymes of the β-ketoadipate pathway51 – a central pathway to which many different peri-pheral pathways converge.20 Salicylate (X) funnels into catechol (XI), while phthalate (VI) funnels into protocatechuate (VII), both parts of the β-ketoadipate pathway (Fig. 3). Interestingly, catechol formation was identified as the bottleneck in the β-ketoadipate pathway,51 which is in good agreement with the amount of salicylic acid found in our study. The catechol and protocatechuate branches converge at the intermediate, β-ketoadipate enol-lactone, which gives 3-oxoadipate and succinyl that goes into the TCA cycle.20 All of the genes coding for proteins involved in the β-ketoadipate ortho degradation pathway, catABC of the catechol branch and pcaBCDG of the protocatechuate branch, have been identified in the P. aeruginosa san ai genome.29 Although it was believed that phthalate and salicylate pathways cannot exist simultaneously, recent study of PYR degradation by Mycobacterium sp. WY10 undoubtedly showed that both pathways simultaneously played roles in degradation.60 In addition, based on identified metabolites, Zhang et al.8 proposed two pathways for FLU degradation by P. aeruginosa DQ8, although key metabolites, phthalate and salicylate, were not detected. Consistent with reports of Sun et al.61 and Zhang et al.,8 and on the basis of metabolites identified in this study, particularly phthalate and salicylate, we propose the simultaneous existence of two FLU degradation pathways, phthalate and salicylate, in P. aeruginosa san ai (Fig. 3).
3, implying an increased accumulation of salicylic acid over time, although metabolites of the salicylate pathway were not identified after 18 h of microbial growth. Substantial secretion and accumulation of intermediates in Pseudomonas growing on aromatic compounds were determined by the limited activity of some of the enzymes of the β-ketoadipate pathway51 – a central pathway to which many different peri-pheral pathways converge.20 Salicylate (X) funnels into catechol (XI), while phthalate (VI) funnels into protocatechuate (VII), both parts of the β-ketoadipate pathway (Fig. 3). Interestingly, catechol formation was identified as the bottleneck in the β-ketoadipate pathway,51 which is in good agreement with the amount of salicylic acid found in our study. The catechol and protocatechuate branches converge at the intermediate, β-ketoadipate enol-lactone, which gives 3-oxoadipate and succinyl that goes into the TCA cycle.20 All of the genes coding for proteins involved in the β-ketoadipate ortho degradation pathway, catABC of the catechol branch and pcaBCDG of the protocatechuate branch, have been identified in the P. aeruginosa san ai genome.29 Although it was believed that phthalate and salicylate pathways cannot exist simultaneously, recent study of PYR degradation by Mycobacterium sp. WY10 undoubtedly showed that both pathways simultaneously played roles in degradation.60 In addition, based on identified metabolites, Zhang et al.8 proposed two pathways for FLU degradation by P. aeruginosa DQ8, although key metabolites, phthalate and salicylate, were not detected. Consistent with reports of Sun et al.61 and Zhang et al.,8 and on the basis of metabolites identified in this study, particularly phthalate and salicylate, we propose the simultaneous existence of two FLU degradation pathways, phthalate and salicylate, in P. aeruginosa san ai (Fig. 3).
Homology search analysis by blastp suite (https://blast.ncbi.nlm.nih.gov/Blast.cgi) of the proteins in Terrabacter DBF63 (ref. 16) involved in FLU biodegradation to phthalate: DbfA (fluorene 9-monooxygenase; 9-fluorenone 4,4a-dioxygenase) for reaction I → II and III → IV; FlnB (9-fluorenol dehydrogenase) for II → III, and; FlnE (2-hydroxy-6-oxo-6-(2′-carboxyphenyl)-hexa-2,4-dienoate hydrolase) for transformation V → VI, showed the existence of similar proteins in P. aeruginosa (ESI Table 1†). Identified metabolites II, III, and VI, in accordance with the KEGG FLU degradation map, imply the transformation catalyzed by the phthalate pathway enzymes. In addition, salicylate (XI) unambiguously documents the existence of the salicylate pathway (Fig. 3).
Furthermore, as salicylate appears as an intermediate, the catechol branch of the β-ketoadipate pathway must be active, and catechol-dioxygenase activity should be detected. In this regard, the catechol-dioxygenase activity of P. aeruginosa san ai biomass grown on FLU, PHE or PYR was measured against catechol and several other aromatic substrates. Our results (Table 2) revealed all examined substrates were preferentially ortho-cleaved. Furthermore, ortho-degradation of catechol by crude enzyme preparation from P. aeruginosa san ai biomass grown on the examined PAHs implies these three PAHs could be ortho-converted by the same enzyme, producing cis,cis-muconic acid. Indeed, the catechol branch of the β-ketoadipate pathway was found to be preferred for several molecules including salicylate.20
| C-source | Substrate | Catechol dioxygenase activity, U mg−1 | |
|---|---|---|---|
| 1,2-Catechol dioxygenase | 2,3-Catechol dioxygenase | ||
| FLU | FLU | 0.275 | 0.039 |
| Catechol | 0.150 | 0.001 | |
| PHE | PHE | 0.285 | 0.010 |
| Catechol | 0.274 | <0.001 | |
| PYR | PYR | 0.151 | <0.001 |
| Catechol | 0.371 | <0.001 | |
4. Conclusion
Polyextremophile P. aeruginosa san ai showed a high degradation capacity for n-alkanes, PAHs and their complex mixture in crude oil hydrocarbon fractions over a period of seven days. PAHs have higher resistance to biodegradation than n-alkanes, and the resistance increases with number of aromatic rings. n-Alkanes were metabolized via the terminal oxidation pathway, while FLU was degraded by both the salicylate and phthalate pathways. Since heavy metals are present in oil spills and alkaline oil refinery effluent, alkaliphilic, metallotolerant, hydrocarbonoclastic P. aeruginosa san ai is an ideal microorganism for remediation of hydrocarbon-contaminated sites, including extreme habitats. Additionally, this bacterium could be considered as an early biomarker of environmental pollution.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Projects III 43004 and 176006), the Japan International Cooperation Agency (JICA) partnership program “Environmental improvement in Pančevo, Serbia through the collaborations among Academia, Government, Industry and Citizens” and the Japan Society for the Promotion of Science (JSPS) Invitation Fellowship Program, ID no. S19013. The authors are grateful to Dr Branka Lončarević for assistance with respiration experiments.References
- Z. Cui, G. Cui, W. Xu, Q. Gao, B. Li, G. Yang and L. Zheng, Int. Biodeterior. Biodegrad., 2014, 91, 45–51 CrossRef CAS.
- V. Beškoski, G. Gojgić-Cvijović, J. Milić, M. Ilić, S. Miletić, T. Šolević and M. Vrvić, Chemosphere, 2011, 83, 34–40 CrossRef PubMed.
- M. Vidali, Pure Appl. Chem., 2001, 73, 1163–1172 CAS.
- J. B. van Beilen, S. Panke, S. Lucchini, A. G. Franchini, M. Rothlisberger and B. Witholt, Microbiology, 2001, 147, 1621–1630 CrossRef CAS PubMed.
- A. Chebbi, D. Hentati, H. Zaghden, N. Baccar, F. Rezgui, M. Chalbi, S. Sayadi and M. Chamkha, Int. Biodeterior. Biodegrad., 2017, 122, 128–140 CrossRef CAS.
- M. Grifoll, S. A. Selifonov and P. J. Chapman, Appl. Environ. Microbiol., 1994, 60, 2438–2449 CrossRef CAS.
- S. M. Resnick and D. T. Gibson, Appl. Environ. Microbiol., 1996, 62, 4073–4080 CrossRef CAS PubMed.
- Z. Zhang, Z. Hou, C. Yang, C. Ma, F. Tao and P. Xu, Bioresour. Technol., 2011, 102, 4111–4116 CrossRef CAS PubMed.
- S. Varjani, Bioresour. Technol., 2017, 223, 277–286 CrossRef CAS PubMed.
- R. Margesin and F. Schinner, Appl. Microbiol. Biotechnol., 2001, 56, 650–663 CrossRef CAS PubMed.
- G. Mehetre, S. Dastager and M. Dharne, Sci. Total Environ., 2019, 679, 52–60 CrossRef CAS PubMed.
- R. J. Watkinson and P. Morgan, Biodegradation, 1990, 1, 79–92 CrossRef CAS PubMed.
- T. Kotani, H. Yurimoto, N. Kato and Y. Sakai, J. Bacteriol., 2007, 189, 886–893 CrossRef CAS PubMed.
- W. Finnerty, in Proceedings of the World Conference on Biotechnology for the Fats and Oil Industry, ed. A. H. Applewhite, 1988, pp. 184–188 Search PubMed.
- J. S. Seo, Y. S. Keum and Q. X. Li, Int. J. Environ. Res. Public Health, 2009, 6, 278–309 CrossRef CAS PubMed.
- H. Habe, J. S. Chung, H. Kato, Y. Ayabe, K. Kasuga, T. Yoshida, H. Nojiri, H. Yamane and T. Omori, J. Bacteriol., 2004, 186, 5938–5944 CrossRef CAS PubMed.
- M. Grifoll, S. A. Selifonov, C. V. Gatlin and P. J. Chapman, Appl. Microbiol. Biotechnol., 1995, 61, 3711–3723 CAS.
- H. Nojiri, H. Habe and T. Omori, J. Gen. Appl. Microbiol., 2001, 47, 279–305 CrossRef CAS PubMed.
- R. A. Kanaly and S. Harayama, J. Bacteriol., 2000, 182, 2059–2067 CrossRef CAS PubMed.
- J. Nogales, J. L. García and E. Díaz, Degradation of aromatic compounds in Pseudomonas: a systems biology view, in Aerobic utilization of hydrocarbons, oils and lipids, Handbook of hydrocarbon and lipid microbiology, ed. F. Rojo, Springer, Cham, 2017, pp. 1–49 Search PubMed.
- S. K. Samanta, O. V. Singh and R. K. Jain, Trends Biotechnol., 2002, 20, 243–248 CrossRef CAS PubMed.
- L. Izrael-Živković, M. Rikalović, G. Gojgić-Cvijović, S. Kazazić, M. Vrvić, I. Brčeski, V. Beškoski, B. Lončarević, K. Gopčević and I. Karadžić, RSC Adv., 2018, 8, 10549–10560 RSC.
- I. Karadzic, A. Masui and N. Fujiwara, J. Biosci. Bioeng., 2004, 98, 145–152 CrossRef CAS PubMed.
- L. Izrael-Živković, V. Beškoski, M. Rikalović, S. Kazazić, N. Shapiro, T. Woyke, G. Gojgić-Cvijović, M. Vrvić, N. Maksimović and I. Karadžić, Extremophiles, 2019, 23, 399–405 CrossRef PubMed.
- M. Rikalović, A. M. Abdel-Mawgoud, E. Déziel, G. Gojgić-Cvijović, Z. Nestorović, M. Vrvić and I. Karadžić, J. Surfactants Deterg., 2013, 16, 673–682 CrossRef.
- A. Dimitrijević, D. Veličković, M. Rikalović, N. Avramović, N. Milosavić, R. Jankov and I. Karadžić, Carbohydr. Polym., 2011, 83, 1397–1401 CrossRef.
- N. Avramović, S. Nikolić-Mandić and I. Karadžić, J. Serb. Chem. Soc., 2013, 78, 639–652 CrossRef.
- T. P. Bastow, B. G. K. van Aarssen and D. Lang, Org. Geochem., 2007, 38, 1235–1250 CrossRef CAS.
- A. Medić, K. Stojanović, L. Izrael-Živković, V. Beškoski, B. Lončarević, S. Kazazić and I. Karadžić, RSC Adv., 2019, 9, 23696–23710 RSC.
- X. Tao, G. Lu, J. Liu, T. Li and L. Yang, Int. J. Environ. Res. Public Health, 2009, 6, 2470–2480 CrossRef CAS PubMed.
- M. Lin, X. Hu, W. Chen, H. Wang and C. Wang, Int. Biodeterior. Biodegrad., 2014, 94, 176–181 CrossRef CAS.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- I. A. Chen, K. Chu, K. Palaniappan, M. Pillay, A. Ratner, J. Huang, M. Huntemann, N. Varghese, J. R. White, R. Seshadri, T. Smirnova, E. Kirton, S. P. Jungbluth, T. Woyke, E. A. Eloe-Fadrosh, N. N. Ivanova and N. C. Kyrpides, Nucleic Acids Res., 2019, 47, D666–D677 CrossRef CAS PubMed.
- M. Kanehisa, Y. Sato, M. Furumichi, K. Morishima and M. Tanabe, Nucleic Acids Res., 2019, 47, D590–D595 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- F. Briganti, E. Pessione, C. Giunta and A. Scozzafava, FEBS Lett., 1997, 416, 61–64 CrossRef CAS PubMed.
- M. Mahiudddin, A. N. M. Fakhruddin and A. A. Mahin, ISRN Microbiol., 2012, 741820 Search PubMed.
- I. Tsirogianni, M. Aivaliotis, M. Karas and G. Tsiotis, Biochim. Biophys. Acta, 2004, 1700, 117–123 CrossRef CAS PubMed.
- N. Jauhari, S. Mishra, B. Kumari and S. N. Singh, Bioresour. Technol., 2014, 170, 62–68 CrossRef CAS PubMed.
- M. C. Romero, M. C. Cazau, S. Giorgieri and A. M. Arambarri, Environ. Pollut., 1998, 101, 355–359 CrossRef CAS.
- I. Amini, A. Tahmourespour and A. Abdollahi, Pollution, 2017, 3, 9–19 CAS.
- E. Z. Ron and E. Rosenberg, Curr. Opin. Biotechnol., 2002, 13, 249–252 CrossRef CAS PubMed.
- S. Sun, Y. Wang, T. Zang, J. Wei, H. Wu., C. Wei, G. Qiu and F. Li, Bioresour. Technol., 2019, 281, 421–428 CrossRef CAS PubMed.
- L. Tao, W. F. Hua, G. L. Ping, L. X. Liang, Y. X. Jin and L. A. Jun, Chemistry, 2012, 55, 1968–1975 Search PubMed.
- H. Liu, J. Xu, R. Liang and J. Liu, PLoS One, 2014, 9, e105506 CrossRef PubMed.
- S. B. Salam, 3 Biotech., 2016, 6, 98 CrossRef PubMed.
- R. S. Kahlon, Biodegradation and bioremediation of organic chemical pollutants by Pseudomonas, in Pseudomonas: Molecular and Applied Biology, ed. R. Kahlon, Springer International Publishing, Switzerland, 2016, pp. 343–417 Search PubMed.
- A. Perfumo, I. M. Banat, F. Canganella and R. Marchant, Appl. Microbiol. Biotechnol., 2006, 72, 132 CrossRef CAS PubMed.
- H. R. Ali, D. A. Ismail and N. S. El-Gendy, Energy Sources, Part A, 2014, 36, 1429–1436 CrossRef CAS.
- P. Censi, S. E. Spoto, F. Saiano, M. Sprovieri, S. Mazzola, G. Nardone and D. Ottonello, Chemosphere, 2006, 64, 1167–1176 CrossRef CAS PubMed.
- K. E. Peters, C. C. Walters and J. M. Moldowan, The biomarker guide: biomarkers and isotopes in the petroleum exploration and earth history, Cambridge University Press, Cambridge, UK, vol. 2, 2005 Search PubMed.
- S. Sudarsan, L. M. Blank, A. Dietrich, O. Vielhauer, R. Takors, A. Schmid and M. Reuss, Metab. Eng. Commun., 2016, 3, 97–110 CrossRef PubMed.
- D. White, The Physiology and Biochemistry of Prokaryotes, Oxford University Press, New York, 2nd edn, 2000, p. 198, 204 Search PubMed.
- C. Park and W. Park, Front. Microbiol., 2018, 9, 1081 CrossRef PubMed.
- J. D. Wang, X. X. Li and C. T. Qu, Curr. Microbiol., 2019, 76, 1270–1277 CrossRef CAS PubMed.
- S. C. George, C. J. Boreham, S. A. Minifie and S. C. Teerman, Org. Geochem., 2002, 33, 1293–1317 CrossRef CAS.
- J. K. Volkman, R. Alexander, R. I. Kagi, S. J. Rowland and P. N. Sheppard, Org. Geochem., 1984, 6, 619–632 CrossRef CAS.
- I. M. Head, D. M. Jones and S. Larter, Nature, 2003, 426, 344–352 CrossRef CAS PubMed.
- L. M. Wenger, C. L. Davis and G. H. Isaksen, SPE Reservoir Eval. Eng., 2002, 5, 375–383 CrossRef CAS.
- K. Stojanović, B. Jovančićević, A. Šajnović, T. Sabo, D. Vitorović, J. Schwarzbauer and A. Golovko, Fuel, 2009, 88, 287–296 CrossRef.
- S. Sun, H. Wang, Y. Chen, J. Lou, L. Wua and J. Xu, J. Hazard. Mater., 2019, 364, 509–518 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra10371f |
| This journal is © The Royal Society of Chemistry 2020 |