Open Access Article
Open Access ArticleThymosin β4 cytoplasmic/nuclear translocation as a new marker of cellular stress. A Caco2 case study
Pierpaolo Conia,
Monica Piras*a,
Anna Mateddua,
Marco Piludub,
Germano Orruc,
Alessandra Scanoc,
Tiziana Cabrasd,
Valentina Pirasd,
Joanna Izabela Lachowicza,
Mariusz Jaremko *e,
Gavino Faaa,
Massimo Castagnolaf and
Giuseppina Pichiria
*e,
Gavino Faaa,
Massimo Castagnolaf and
Giuseppina Pichiria
aDepartment of Medical Sciences and Public Health, University of Cagliari, Cagliari, Italy. E-mail: monipiras@hotmail.com
bDepartment of Biomedical Sciences, University of Cagliari, Cagliari, Italy
cDepartment of Surgical Sciences, OBL, University of Cagliari, Cagliari, Italy
dDepartment of Life and Environmental Sciences, University of Cagliari, Cagliari, Italy
eKing Abdullah University of Science and Technology (KAUST), Biological and Environmental Science & Engineering (BESE), 23955-6900 Thuwal, Saudi Arabia. E-mail: mariusz.jaremko@kaust.edu.sa
fLaboratorio di Proteomica e Metabolomica, IRCCS Fondazione Santa Lucia, Italy
First published on 30th March 2020
Abstract
Biomarkers of cell stress are important for proper diagnosis, and in studies of how cells respond to drug treatment. Biomarkers that respond early to pharmacological treatment could improve therapy by tailoring the treatment to the needs of the patient. Thymosin beta-4 (Tβ4) plays a significant role in many aspects of cellular metabolism because of its actin-sequestering properties. Other physiological functions of Tβ4 have been also reported. Among these, Tβ4 may play a crucial role during cellular stress. We addressed the relevance of Tβ4 in cellular stress conditions by using different treatments (serum starvation, DMSO, and butyrate administration) in a colon adenocarcinoma cell line (CaCo2), a cell line frequently used for in vitro experimental studies of Tβ4. In this study, different stress stimuli were analyzed and the obtained results were compared using immunocytochemistry, and molecular and biochemical methods. Taken together, the data clearly indicate that the Tβ4 peptide is involved in adaptive and defensive cellular mechanisms, and that different stress inducers lead to a similar Tβ4 cytoplasmic/nuclear translocation. The translocation of Tβ4 between the cytoplasm and the nucleus of the cell seems characteristic of a possible molecular response to cellular stress exerted by this peptide.
1. Introduction
Heat shock protein-72 (HSP72), fibroblast growth factor-21 (FGF21), CD133 (prominin-1), vascular endothelial growth factor, and numerous cytokines (e.g. MIP-1 α, IL-8, MIP-1 β, resistin, GDF-15, HGF, CA125, FLRG, VCAM-1, DKK-3, sTNF-R1, CTACK, Acrp30, CXCL-16 and LYVE-1) have been identified as cancer biomarkers expressed in cells under stress conditions.1 Other biomarkers of stress conditions are still under intensive development due to their usefulness in quick diagnosis of the pathology.Thymosin beta-4 (Tβ4) is a 43 amino acid long, ubiquitous intracellular peptide, well known as G-actin sequestering factor in the cytoplasm of mammalian cells.2–4 Various studies suggested that this small peptide has other biological functions related to cell proliferation and differentiation that are necessary to promote and modulate wound healing and inflammatory responses.5
Comparative studies on Tβ4 expression in fetal and adult human tissues showed a preferential expression of Tβ4 during fetal life.6 The role of this peptide in cardiac tissue stress and repair processes, as evidenced by epicardial cell migration, neovascularization and/or revascularization and activation of cardiac progenitor cells in the heart, is already well documented. In human corneal epithelial cells and conjunctival cells, an apoptotic Tβ4 protective effect was reported. This effect took place after stress injury induced by ethanol and hydrogen peroxide (H2O2). Tβ4 also regulates induced-proinflammatory cytokine, while blocking RelA/p65 nuclear translocation.7 Because of this, Tβ4 could be used as a new and early biomarker to diagnose pathology more quickly than cytokines.
Localization of Tβ4 may be easily altered according to different external stimuli. It can cross the nuclear membrane and act as a transcription factor in the nucleus.8 Tβ4 translocation was first described in a paper in which the authors injected this peptide into Xenopus laevis oocytes, where this molecule has reached both nuclear and cytoplasmic compartments.9 In IEC-6 cells, Tβ4 was found predominantly in the nuclei and in the nuclear membrane after cell depletion of polyamines induced by alpha-difluoromethylornithine treatment.10 Thymosin β4 has various functional roles in normal cell biology and its mechanism of action has recently been studied in various tumors. In particular, its role in colorectal cancer invasion and metastasis was suggested in several studies using in vivo and in vitro techniques.
Nuclear localization of Tβ4 in human mammary carcinoma MCF-7 cell line was also reported. The authors, who described this phenomenon, suggested that Tβ4 enters the nucleus through an active transport mechanism, and requires a currently unidentified soluble cytoplasmic factor. The unidentified factor could be the MLH1 protein, a key enzyme in DNA mismatch repair with several additional functions, together with the intranuclear transport of Tβ4.11 A passive, but regulated diffusion mechanism has also been proposed as another way that Tβ4 translocates into the nucleus.12 Recently, the metal binding ability of Tβ4 was suggested as a driving force in the cellular translocation and actin binding activity of this peptide.13 In other experiments, involving stress stimuli induced by the in vitro serum starvation in HepG2 cells, the translocation from the cytoplasm to the nucleus was reported as the characteristic signal of a possible stress regulatory function exerted by this peptide. Altogether, these data underline the ability of Tβ4 to change its subcellular localization in distinct cellular conditions of varying difficulty.
Several questions concerning Tβ4 translocation and its importance in cellular function have yet to be answered, while stress induced events can often trigger a plethora of divergent responses that may interfere with the experimental results, and with the subsequent conclusions.14 For example, short or prolonged incubation in serum-free medium could lead uniform changes in the basal cellular activity, or it could lead to different signaling pathways showing opposing results.
In order to understand better the Tβ4's translocation under stress conditions, we use different stress treatments in Caco2 cell line, a commercial colon cancer cell line frequently used as a normal and human disease model.
2. Material and methods
2.1. Cell culture
Commercial human cell lines (CaCo2, ICLC, Genova) derived from a colon adenocarcinoma were used. CaCo2 cells were maintained in culture with MEM medium (EBSS) containing FBS (fetal bovine serum) at 10%, 100 units per ml penicillin, 100 mg per ml streptomycin, 2 mM L-glutamine, 1% of non-essential amino acids. The confluent cells were detached using trypsin/EDTA as previously described (Pichiri et al., 2013;6 Piludu et al., 2015![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31) in order to obtain different experimental conditions.
31) in order to obtain different experimental conditions.
2.2. Experimental procedures
CaCo2 cells were seeded (2–3 × 104 cells per cm2) in 100 mm culture dish with a cover slides inside and were incubated at 37 °C, 5% CO2 as previously described (Pichiri et al.,6 2013; Piludu et al., 2015![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31).
31).
Different experimental groups were obtained after 24 h of growth with complete medium. One group was incubated with a culture medium without FBS (starvation), while second and third groups were incubated in complete medium in the presence of butyrate (final concentration 1.5 μM) or with DMSO (1% final concentration); the fourth group of cells were incubated in FBS deprived medium and treated with butyrate (final concentration 1.5 μM). A control group was incubated in complete medium without any treatment. After 48 hours all samples were washed with phosphate buffered saline (PBS), fixed in cold acetone for 10 minutes, successively air dried for 30 minutes and used for immunocytochemistry, biochemistry, and RNA analysis.
2.3. Tβ4 immunocytochemistry analysis
Tβ4 immunocytochemistry was performed as previously described in the literature. The cells were briefly rehydrated, and endogenous peroxidase activity was quenched (3 min) by 0.3% hydrogen peroxide in methanol. Cells were incubated with 10% normal goat serum in PBS for 5 min to block non-specific binding and then incubated (30 min) with a polyclonal anti-Tβ4 antibody (Abcam, Anti-Thymosin beta 4 antibody (ab14334); Bachem-Peninsula Lab, San Carlos, CA, USA) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in a blocking solution. Cells were extensively washed with PBS containing 0.01% Triton X-100 and incubated with a secondary reagent (Envision kit) according to the manufacturer's instructions (Dako, Glostrup, Denmark). After additional washes with PBS, the color was developed using AEC reagent (Dako, Glostrup, Denmark); cells were counterstained with hematoxylin and mounted.
100 in a blocking solution. Cells were extensively washed with PBS containing 0.01% Triton X-100 and incubated with a secondary reagent (Envision kit) according to the manufacturer's instructions (Dako, Glostrup, Denmark). After additional washes with PBS, the color was developed using AEC reagent (Dako, Glostrup, Denmark); cells were counterstained with hematoxylin and mounted.
2.4. Biochemistry analysis. Preparation of cytosolic and nuclear proteins/peptides for RP-HPLC-ESIMS analysis
To evaluate the levels of Tβ4 in the cytosol and in the nucleus of the cells under different growing conditions (normal and stressing), subcellular fractions were prepared by the modified method of Galan et al. (Galan et al. 2011![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32). using the commercial kit NE-PER Nucleus and Cytoplasmyc Extraction Reagents (Thermo Scientific-USA, www.thermoscientific.com/pierce).
32). using the commercial kit NE-PER Nucleus and Cytoplasmyc Extraction Reagents (Thermo Scientific-USA, www.thermoscientific.com/pierce).
Cytosolic and nuclear proteins/peptides were fractionated with Amicon centrifugal filters (30 KDa cutoff) (Millipore, Billerica, MA, USA), dialyzed in 25 mM sodium acetate at pH 4.2 and then lyophilized. The powder was dissolved in 100 μl of 0.1% aqueous trifluoroacetic acid (TFA), centrifuged at 8000g for 10 min at 4 °C, and the supernatant analysed by HPLC-ESI-MS. Triplicates of the samples were prepared.
2.5. HPLC-low-resolution ESI-MS analysis and quantification
A Surveyor HPLC system (ThermoFisher, San Jose, CA, USA) connected to an LCQ Advantage mass spectrometer (ThermoFisher) was used for HPLC-low-resolution ESI-MS measurements. The chromatographic column was a Vydac (Hesperia, CA, USA) C8 with 5 mm particle diameter (column dimensions 150 × 2.1 mm). The eluents utilized for the chromatographic separations were: (eluent A) 0.056% aqueous TFA and (eluent B) 0.050% TFA in acetonitrile–water 80/20 (v/v). The applied gradient was linear from 0 to 54% in 39 min (linear) and from 54% to 100% in 10 min (linear), at a flow rate of 0.10 ml min−1. During the first 5 min of separation, eluate was discarded in order to avoid source contamination and instrument damage due to the high salt concentration. Mass spectra were collected every 3 milli-second in the positive ion mode. MS spray voltage was 5.0 kV and capillary temperature was 255 °C.Tβ4 was analysed in the Total Ion Current (TIC) chromatographic profiles of the examined samples by using the extracting ion current (XIC) procedure. The XIC search allowed to extract selectively the ion current peak generated by specific multiply-charged ions (charge) of Tβ4: 993.80 (+5), 1241.90 (+4) and 1655.50 (+3) m/z. A window of ±0.5 Da was used to extract the XIC peak. Experimental average mass value (Mav) was obtained by deconvolution of averaged ESI-MS spectra automatically performed by using MagTran 1.0 software.15 Experimental Mav was compared with the theoretical one available at the UniProt-KB human data-bank (http://us.expasy.org/tools) with the entry P62328. Experimental Mav and elution time were compared also with those obtained by HPLC-ESI-MS of the standard peptide (Bachem, Bubendorf, Switzerland).
The area of XIC peak was used for quantification, since it is proportional to the peptide concentration under constant analytical conditions.16 XIC peak areas were integrated by using the following peak parameters: baseline window 15; area noise factor 50; peak noise factor 50; peak height 15% and tailing factor 1.5. Estimated percentage error of the XIC procedure was <8%. The areas of XIC peaks were correlated to the concentrations expressed in nM, after obtaining a linear regression between the known concentrations of the standard Tβ4 (0.005, 0.025, 0.05, 0.10, 0.25, 0.35, 0.5, 0.75, 1.00, 1.25, 1.50 μM) and the respective XIC peak areas obtained by HPLC-ESI-MS analysis.
Relative abundance (%) in nuclear fraction (NF) with respect to cytosolic fraction (CF) was calculated in the three series of CaCo2 cells as follows:
| % NF = NF XIC peak area × 100/(NF + CF XIC peak areas). |
Statistical comparison of the Tβ4 concentration between the CF and NF samples in the different stress conditions, such as among the Tβ4% in NF, was performed, with 5.0 GraphPad Prism software, by the unpaired t-test with Welch's correction being the samples size equal, small and the variance unequal. The data were analysed also with one-way ANOVA as well as the multicomparison Tukey's test. P values < 0.05 was considered significant.
2.6. HPLC-high-resolution ESI-MS/MS analysis
Characterization of Tβ4 was performed on the intact peptide present in the samples by using an Ultimate 3000 Micro HPLC apparatus (Dionex, Sunnyvale, CA, USA) equipped with a FLM-3000-Flow manager module and coupled to an LTQ Orbitrap XL apparatus (Thermofisher). The injection volume was 20 μl. The column was a Zorbax 300SB-C8 (3.5 μm particle diameter; 1.0 × 150 mm), and the eluents were: A, 0.1% aqueous formic acid solution, and B, 0.1% aqueous formic acid solution in acetonitrile/water 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 v/v. The applied gradient was from 5 to 55% B in 40 min, from 55 to 100% B in 2 min, from 100 to 5% B in 2 min for a total acquisition time of 61 min at a flow rate of 80 l min−1. The MS spectra were acquired in data-dependent mode in the m/z range from 300 to 2000. The three most abundant ions were selected and fragmented by using collision-induced dissociation (CID, 35% normalized collision energy for 30 ms, with an isolation width of 6–10 m/z, activation q of 0.25. MS and MS/MS scans were acquired at a resolution of 60
20 v/v. The applied gradient was from 5 to 55% B in 40 min, from 55 to 100% B in 2 min, from 100 to 5% B in 2 min for a total acquisition time of 61 min at a flow rate of 80 l min−1. The MS spectra were acquired in data-dependent mode in the m/z range from 300 to 2000. The three most abundant ions were selected and fragmented by using collision-induced dissociation (CID, 35% normalized collision energy for 30 ms, with an isolation width of 6–10 m/z, activation q of 0.25. MS and MS/MS scans were acquired at a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). The capillary temperature was set to 250 °C, and the source voltage was set to 4 kV. Data were generated by Xcalibur 2.2 SP1.48 (Thermo Fisher Scientific) using default parameters of the Xtract program for deconvolution. MS/MS data were analyzed by the Proteome Discoverer software (version 1.4.1.14, Thermo Fisher Scientific), based on SEQUEST HT cluster as the search engine against the UniProtKB human database (163, 117 entries, release 2018_02). For peptide matching, high-confidence filter settings were applied: the peptide score threshold was 3.0; the limits for FDR setting were Xcorr scores greater than 1.2 for singly charged ions, and 1.9 and 2.3 for doubly and triply charged ions, respectively. The precursor mass search tolerance was 10 ppm and the fragment mass tolerance were 0.6 Da. Since Tβ4 is N-terminal acetylated, the analysis was performed by using N-terminal acetylation (+42.011 Da) as a dynamic modification.
000). The capillary temperature was set to 250 °C, and the source voltage was set to 4 kV. Data were generated by Xcalibur 2.2 SP1.48 (Thermo Fisher Scientific) using default parameters of the Xtract program for deconvolution. MS/MS data were analyzed by the Proteome Discoverer software (version 1.4.1.14, Thermo Fisher Scientific), based on SEQUEST HT cluster as the search engine against the UniProtKB human database (163, 117 entries, release 2018_02). For peptide matching, high-confidence filter settings were applied: the peptide score threshold was 3.0; the limits for FDR setting were Xcorr scores greater than 1.2 for singly charged ions, and 1.9 and 2.3 for doubly and triply charged ions, respectively. The precursor mass search tolerance was 10 ppm and the fragment mass tolerance were 0.6 Da. Since Tβ4 is N-terminal acetylated, the analysis was performed by using N-terminal acetylation (+42.011 Da) as a dynamic modification.
Peptide sequence was validated by manual inspection of the experimental fragmentation spectra against the theoretical ones generated by MS-Product software available at the ProteinProspector website (http://prospector.ucsf.edu/prospector/mshome.htm).
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://www.ebi.ac.uk/pride) via the PRIDE (Vizcaíno et al., 2016![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33) partner repository with the dataset identifier PXD012161.
33) partner repository with the dataset identifier PXD012161.
Account details for reviewer use only: username: reviwer93637@ebi.ac.uk; password: uNEEGroJ.
2.7. mRNA analysis
To characterize better the different stress stimuli used in this study, the expression of the following genes at the mRNA level was evaluated by real time PCR: PRDX1: this gene encodes a member of the family peroxiredoxin (an antioxidant enzyme); TXN: the protein encoded by this gene is involved in many redox reactions; VCAM: a protein involved in oxidative stress, and as an adhesion molecule in vascular cells; TRND: known as a mitochondrial tRNA useful for regulating the redox state of the cell; Hsp 70: the term HSP70 (Heat Shock Protein 70 kilodaltons) refers to a family of proteins expressed ubiquitously and important for the cellular machinery of protein folding. In fact, the HSP70 proteins help other nascent proteins achieve a definitive spatial structure and help protect the cell against stress factors such as heat; BAX: is a protein of the Bcl-2 family that activates apoptosis; HO-1: is a protein with heme-oxygenase activity in substrates heme and non-heme. We also evaluated a possible change in the expression of Tβ4 induced by these different treatments.The cells were immediately frozen after treatment and stored at −80 °C until RNA extraction. The RNA was extracted with NorDiag ARROW RNA kit using the automated tool of the same company. Actin was used as a reference gene.17
The Real-time reverse-transcriptase PCR was performed using the Roche Light Cycler system and SYBR Green I kit amplification kit (Roche Diagnostics). The following primers (b-actF = 5′-GCATGGGTCAGAAGG-3′, b-actR = 5′-AGGCGTACAGGGATAG-3′, tb4F = 5′-GGCCACTGCGCAGACCAGACT3′, tb4R = 5′CTTGATCCAACCTCTTTGCATCTTACAA-3′) were designed using the sequences of the Tβ4 RNA (GenBank accession no. NM_001101) and the human beta-actin mRNA (GenBank accession no. NM_001101).
The 20 ml final volume contained: 3 mM MgCl2, 0.25 mM of each primer, and 2 ml of RNA extract. Cycling was performed using the following amplification conditions: an initial reverse transcription at 55 °C for 10 min, denaturation at 95 °C for 30 s followed by 35 cycles at 95 °C for 10 s, 53 °C for 10 s, and 72 °C for 8 s with subsequent melting analysis: heating to 95 °C for 20 s, cooling to 45 °C for 10 s and reheating to 95 °C at a rate of 0.2 °C per second.
Fluorescence was detected at the end of the 81 °C segment in PCR step (single mode) and at 45 °C segment in the melting step (continuous mode) in the F1 channel. The relative gene expression was analyzed by using the 2-DDCT method.18
For each analysis, three distinct biological replicas were done, and quantitative data were expressed as a mean. Folding change values in Tβ4 gene expression relative to the beta-actin have been represented as mean + standard error.
3. Results and discussion
Several reports have shown that Tβ4, a multi-functional regenerative peptide,19 could be involved in different critical biological activities, including angiogenesis,20 wound healing,21 inflammatory response,22 cell migration,23 survival, and oxidative stress.24The Tβ4 ability to change conformation and to translocate from the cytoplasm to the nucleus and back, was recently described and associated to stress induced in vitro in the HepG2 cell line by serum starvation.6 The significance of our previous studies was limited by discovering that stress condition like serum starvation, may be characterized by different responses that depend on the experimental conditions.14
In order to clarify better the actual significance of intracellular trafficking of Tβ4, we used different stress stimuli in the Caco2 cell line. The immunocytochemistry, mass spectrometry, and molecular data obtained in this study strongly support the hypothesis that Tβ4 cytoplasmic/nuclear translocation could be a general defensive mechanism against multiple and different adverse environmental changes, and Tβ4 could be used as a new and early biomarker of cellular stress.
Starvation is able to induce stress by serum deprivation in vitro,25 while albumin (the most abundant protein of serum) is the main carrier of essential metal ions and fatty acids for cells.26 Butyrate, a short chain fatty acid produced by commensal bacteria of the human intestine, was already used to induce oxidative stress and apoptosis in CaCo2 cell cultures in Fauser et al.27 Moreover, it was reported that butyrate could act as an epigenetic “switch” of the immune system.28 Butyric acid has been in fact associated with the ability to inhibit histone deacetylases (HDAC), and favors a state of acetylated histones in the cell.29 DMSO is a powerful oxidant that is often used as a solvent in various experimental tests and in NMR analysis, while its signals have low interferences, and it can dissolve a large number of substances. Many embryonic and/or tumor cell lines treated with DMSO in the culture medium are induced to mature, and the hiring of a morphological phenotype more “mature”. Nevertheless, the mechanism of DMSO cellular stress action has not yet been completely deciphered.
Serum starvation, DMSO and butyrate treatment were used in our studies to induce in vitro cellular stress with different pathways.
3.1. Morphological analysis of the cells
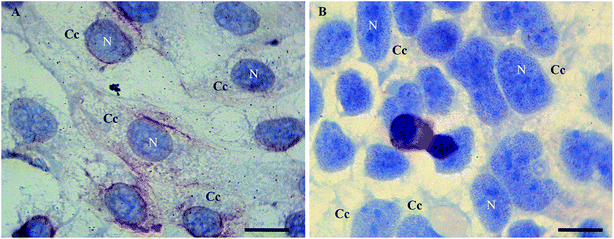
In order to examine morphological changes in the cells, the Caco2 cells were grown under different stress conditions and immunoassayed with a polyclonal anti-Tβ4 antibody. Specific Tβ4 immuno-reactivity was visualized as small roundish spots in all examined samples.In detail, Tβ4 immunocytochemistry was performed in CaCo2 cell lines growing for 48 h with and without fetal bovine serum. Under normal growth conditions, the expression of Tβ4 positivity seemed to be more intense and diffuse in the cytoplasm (Fig. 1A). After 48 h of starvation, a pattern of punctuated nuclear Tβ4 positivity appeared (Fig. 1B).
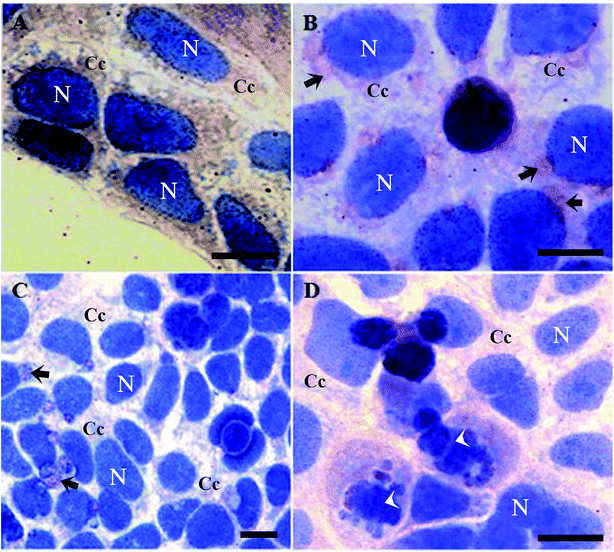
It is worth mentioning that cells treated with butyrate presented a relevant number of Tβ4-positive vesicles (Fig. 2B).
Tβ4 translocation from cytoplasm to the nucleus in CaCo2 cells provoked by DMSO or butyrate treatment confirmed that Tβ4 translocation may be a general signal of cellular stress (Fig. 2A and B). Samples treated with butyrate were characterized by a relevant number of vesicles. Tβ4 translocation and the presence of vesicles are more evident, when CaCo2 cells under starvation were treated with butyrate (Fig. 2C and D).
The number of cellular vesicles increased, when serum starvation and butyrate administration acted together (Fig. 2C and D), suggesting that different stimuli may exert a cumulative effect with increased stress intensity. An increased amount of vesicles could be related to an increased apoptotic activity (more Tβ4 positive vesicles and more cells showing nuclear fragmentation) (Fig. 2B), but the possible presence of Tβ4 inside these vesicles and biological significance of its presence require deeper studies.30
Taken all together, morphological studies show clearly that Tβ4 peptide is involved in adaptive and defensive cellular mechanisms, and that the cytoplasm/nuclear translocation effect, more than simply a different gene/protein expression, is related to a change in its sub cellular localization.
3.2. Expression of Tβ4 and other genes related to stress conditions
Tβ4 RNA expression decreased during all our experimental treatments (Fig. 3), while other genes, related to stress conditions, such as PRDX1, TXN, VCAM, TRND, Hsp 70, BAX, Bcl-2 and HO-1 showed a different pattern related to the changing conditions (Fig. 4, panels A and B).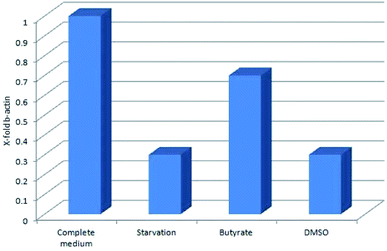 | ||
| Fig. 3 Tβ4 RNA expression in CaCo2 cells exposed to different stress conditions (starvation, butyrate and DMSO). | ||
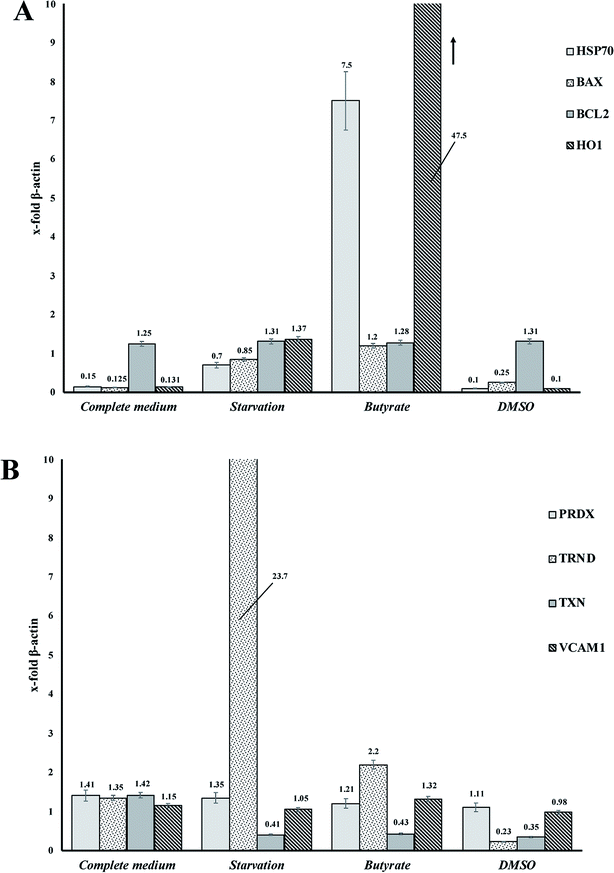 | ||
| Fig. 4 (A and B) RNA expression of several genes related to stress cellular conditions in CaCo2 cells. | ||
In particular, administration of DMSO, characterized by a very low level of Tβ4 RNA, does not seem to corelate with the expression of the other examined genes, whereas butyrate treatment was followed by an increased expression of Hsp70 and HO-1, and starvation conditions induced an increased expression of TRND gene.
In fact, while the Tβ4 RNA expression in CaCo2 cells decreased in all the 3 different experimental stress conditions, a different mRNA pattern of several stress related genes characterized the diversity of these 3 stimuli. It can be assumed that during in vitro cellular stress induced by different stressing stimuli characterized by a different molecular and morphological expression, Tβ4 translocation may be considered a general adaptive response.
3.3. Mass spectrometry analysis
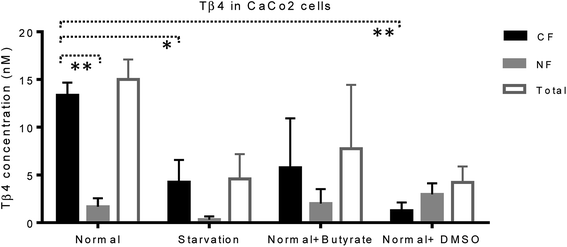
Mass spectrometry analysis was performed in order to evaluate Tβ4 concentration changes in the cytosolic and nuclear fractions of CaCo2 cells, under normal and different stress conditions. Low-resolution liquid chromatography–mass spectrometry (LC-MS) analysis allowed detection of Tβ4 in the nuclear and cytosolic fractions at the elution time of 19.3–19.8 min with an experimental Mav of 4962.8 ± 0.6 (theor. 4963.5 Da); the same elution time and Mav were obtained for the standard peptide analyzed in the same experimental conditions. However, the validation of Mav attribution was performed by high-resolution MS/MS spectra and the MS results were deposited on ProteomeXchange (PXD012161).CaCo2 cells exhibited lower amounts of Tβ4 under starvation, butyrate, and DMSO treatment with respect to the normal conditions (Fig. 5). Despite the limitations due to the low number of samples under study, the statistical comparison performed between the CF and NF samples, by the t-test with Welch's correction, highlighted that Tβ4 concentration in normal cells was significantly higher in cytosolic than in nuclear fractions (p < 0.005). Moreover, cytosolic Tβ4 in starvation conditions and in presence of DMSO significantly decreased with respect to the normal conditions (p < 0.04 and p < 0.003 respectively) (Fig. 5).
The variations of the Tβ4 concentration were comparable to that evidenced by RNA expression, and thus, appeared to be linked to a decreased Tβ4 gene expression. Among stress conditions, the trend of Tβ4 concentrations in the nuclear fraction was interesting: the minimum levels were determined under starvation and the maximum under DMSO treatment, even if not statistically different (Fig. 5). Significant variations were evidenced by ANOVA analysis of the relative abundance of Tβ4 in the nuclear fractions (Fig. 6). The average relative abundance of the peptide in the nucleus decreased from 11% in normal conditions to 7% under starvation and increased to 26% under butyrate treatment, reaching 70% in the presence of DMSO in the growing medium. The percentage of Tβ4 in NF was significantly higher in presence of DMSO with respect to the normal condition (p < 0.01), to the starvation (p < 0.01) and to butyrate presence in the medium (p < 0.05).
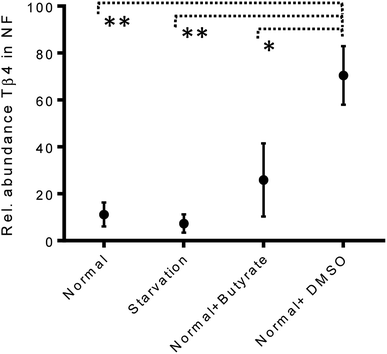 | ||
| Fig. 6 Relative abundance of Tβ4 (%) in nuclear fractions of CaCo2 cells under different growing conditions (from left to right: normal, starvation, normal + butyrate, normal + DMSO). | ||
Interestingly, in the presence of DMSO, the ratio of the cytosol/nucleus fraction of Tβ4 was inverted, as showed in Fig. 5, and since Tβ4 expression at RNA levels in DMSO-treated cells was at the very low level, nuclear Tβ4 protein concentration might be correlated only to a nuclear translocation of this protein from other cellular compartments.
Mass spectrometry analysis of the cytosolic and nuclear protein fractions, under normal and different cell stress conditions, clearly showed the differences in the two cell compartments. Interestingly, the Tβ4 protein levels observed in the nuclear fraction differed according to the varying stimuli, with minimum levels under starvation, and maximum levels after DMSO administration. DMSO represents the only condition in which Tβ4 nuclear concentration is higher than that observed in the cytosolic fraction. Given that in DMSO-treated cells Tβ4 RNA expression was decreased, these differences could be related to a different metabolic response or to a different cellular synchronism induced by the treatment used. In every case, the higher levels of Tβ4 peptide inside the nucleus should be interpreted as a consequence of the Tβ4 translocation from the cytoplasm into the nuclear compartment under the stress conditions to which the cells are exposed.
4. Conclusions
The Tβ4 translocation process from cytoplasm to the nucleus was successfully studied in Caco2 cells under different stress stimuli conditions. In serum starvation conditions, Tβ4 expressed the same cytoplasm/nuclear translocation in Caco2, as it shown in HepG2 cells in the previous studies by Pichiri et al.6 Tβ4 translocation in Caco2 cells can be provoked not only by serum starvation, but also by other stress stimuli as butyrate and DMSO. Contemporary deprivation of serum and addition of butyrate to the cell growing medium lead to the higher rate of Tβ4 translocation from cytoplasmic domains to the nucleus. Overall, stress conditions lead to lower levels of Tβ4 peptide RNA expression. The immunocytochemistry, mass spectrometry, and molecular data obtained in Caco2 cells study, strongly support the hypothesis that Tβ4 cytoplasmic/nuclear translocation could be considered a general defensive mechanism against multiple and different adverse environmental changes. The presented experimental results together with the data that are already available in the literature demonstrate that Tβ4 could be proposed as a new and efficient biomarker of cellular stress at the early stage of pathology.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support of Fondazione Banco di Sardegna, Cagliari, Sardinia, Italy and, the financial support of Regione Autonoma Sardegna RAS-2012, LR 7 2007, CRP-60281. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. M. J. would like to thank King Abdullah University of Science and Technology (KAUST) for the financial support. Authors would like to warmly thank Mawadda Alghrably for arranging references.References
- D. Cai, et al., Extensive serum biomarker analysis in patients with non-small-cell lung carcinoma, Cytokine, 2020, 126, 154868 CrossRef CAS PubMed.
- S. Nemolato, et al., Thymosin β-4 in colorectal cancer is localized predominantly at the invasion front in tumor cells undergoing epithelial mesenchymal transition, Cancer Biol. Ther., 2012, 13, 191–197 CrossRef CAS PubMed.
- M.-C. Tang, et al., Thymosin beta 4 induces colon cancer cell migration and clinical metastasis via enhancing ILK/IQGAP1/Rac1 signal transduction pathway, Cancer Lett., 2011, 308, 162–171 CrossRef CAS PubMed.
- D. Ahmed, et al., Epigenetic and genetic features of 24 colon cancer cell lines, Oncogenesis, 2013, 2, e71 CrossRef CAS PubMed.
- T. Huff, C. S. Müller, A. M. Otto, R. Netzker and E. Hannappel, β-Thymosins, small acidic peptides with multiple functions, Int. J. Biochem. Cell Biol., 2001, 33, 205–220 CrossRef CAS PubMed.
- G. Pichiri, et al., Cellular trafficking of thymosin beta-4 in HEPG2 cells following serum starvation, PLoS One, 2013, 8, e67999 CrossRef CAS PubMed.
- P. Qiu, M. K. Wheater, Y. Qiu and G. Sosne, Thymosin β4 inhibits TNF-α-induced NF-κB activation, IL-8 expression, and the sensitizing effects by its partners PINCH-1 and ILK, FASEB J., 2011, 25, 1815–1826 CrossRef CAS PubMed.
- A. L. Goldstein, E. Hannappel and H. K. Kleinman, Thymosin β4: actin-sequestering protein moonlights to repair injured tissues, Trends Mol. Med., 2005, 11, 421–429 CrossRef CAS PubMed.
- J. D. Watts, P. D. Cary, P. Sautiere and C. Crane Rane-Robinson, Thymosins: both nuclear and cytoplasmic proteins, Eur. J. Biochem., 1990, 192, 643–651 CrossRef CAS PubMed.
- S. A. McCormack, R. M. Ray, P. M. Blanner and L. R. Johnson, Polyamine depletion alters the relationship of F-actin, G-actin, and thymosin β4 in migrating IEC-6 cells, Am. J. Physiol.: Cell Physiol., 1999, 276, C459–C468 CrossRef CAS PubMed.
- A. Brieger, G. Plotz, S. Zeuzem and J. Trojan, Thymosin beta 4 expression and nuclear transport are regulated by hMLH1, Biochem. Biophys. Res. Commun., 2007, 364, 731–736 CrossRef CAS PubMed.
- R. E. Zoubek and E. Hannappel, Subcellular distribution of thymosin β4, Ann. N. Y. Acad. Sci., 2007, 1112, 442–450 CrossRef CAS PubMed.
- J. I. Lachowicz, et al., Metal coordination of thymosin β4: Chemistry and possible implications, Coord. Chem. Rev., 2019, 396, 117–123 CrossRef CAS.
- S. Pirkmajer and A. V. Chibalin, Serum starvation: caveat emptor, Am. J. Physiol.: Cell Physiol., 2011, 301, C272–C279 CrossRef CAS PubMed.
- Z. Zhang and A. G. Marshall, A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra, J. Am. Soc. Mass Spectrom., 1998, 9, 225–233 CrossRef CAS.
- R. Inzitari, et al., HPLC-ESI-MS analysis of oral human fluids reveals that gingival crevicular fluid is the main source of oral thymosins β4 and β10, J. Sep. Sci., 2009, 32, 57–63 CrossRef CAS PubMed.
- J. Durzyńska and E. Barton, IGF expression in HPV-related and HPV-unrelated human cancer cells, Oncol. Rep., 2014, 32, 893–900 CrossRef PubMed.
- K. J. Livak and T. D. Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method, methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- A. L. Goldstein, E. Hannappel, G. Sosne and H. K. Kleinman, Thymosin β4: a multi-functional regenerative peptide. Basic properties and clinical applications, Expert Opin. Biol. Ther., 2012, 12, 37–51 CrossRef CAS PubMed.
- V. Koutrafouri, et al., Effect of thymosin peptides on the chick chorioallantoic membrane angiogenesis model, Biochim. Biophys. Acta, Gen. Subj., 2001, 1568, 60–66 CrossRef CAS.
- K. M. Malinda, et al., Thymosin β4 accelerates wound healing, J. Invest. Dermatol., 1999, 113, 364–368 CrossRef CAS PubMed.
- M. Badamchian, et al., Thymosin β4 reduces lethality and down-regulates inflammatory mediators in endotoxin-induced septic shock, Int. Immunopharmacol., 2003, 3, 1225–1233 CrossRef CAS PubMed.
- M. R. Bubb, Thymosin 4 Interactions, Vitam. Horm., 2003, 66, 297 CrossRef CAS.
- S. Kumar and S. Gupta, Thymosin beta 4 prevents oxidative stress by targeting antioxidant and anti-apoptotic genes in cardiac fibroblasts, PLoS One, 2011, 6, e26912 CrossRef CAS PubMed.
- M. Aghababazadeh and M. A. Kerachian, Cell fasting: Cellular response and application of serum starvation, Journal of Nutrition, Fasting and Health, 2014, 2, 147–150 Search PubMed.
- S. Al-Harthi, J. I. Lachowicz, M. E. Nowakowski, M. Jaremko and Ł. Jaremko, Towards the functional high-resolution coordination chemistry of blood plasma human serum albumin, J. Inorg. Biochem., 2019, 198, 110716 CrossRef CAS PubMed.
- J. Fauser, G. Matthews, A. Cummins and G. Howarth, Induction of apoptosis by the medium-chain length fatty acid lauric acid in colon cancer cells due to induction of oxidative stress, Chemotherapy, 2013, 59, 214–224 CrossRef CAS PubMed.
- G. Serena, et al., Proinflammatory cytokine interferon-γ and microbiome-derived metabolites dictate epigenetic switch between forkhead box protein 3 isoforms in coeliac disease, Clin. Exp. Immunol., 2017, 187, 490–506 CrossRef CAS PubMed.
- S. M. McNabney and T. M. Henagan, Short chain fatty acids in the colon and peripheral tissues: a focus on butyrate, colon cancer, obesity and insulin resistance, Nutrients, 2017, 9, 1348 CrossRef PubMed.
- M. Yáñez-Mó, et al., Biological properties of extracellular vesicles and their physiological functions, J. Extracell. Vesicles, 2015, 4, 27066 CrossRef PubMed.
- M. Piludu, et al., Thymosin Beta 4 May Translocate from the Cytoplasm in to the Nucleus in HepG2 Cells following Serum Starvation. An Ultrastructural Study, PLoS One, 2015, e0119642 CrossRef PubMed.
- J. A. Galan, L. L. Paris, H. Zhang, J. Adler and R. L. Geahlen, et al., Proteomic Studies of Syk-Interacting Proteins Using a Novel Amine-Specific Isotope Tag and GFP Nanotrap, J. Am. Soc. Mass Spectrom., 2011, 22, 319–322 CrossRef CAS PubMed.
- J. A. Vizcaíno, et al., Nucleic Acids Res., 2016, 44(D1), D447–D456 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |