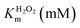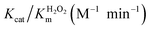Open Access Article
Open Access ArticleA novel nanozyme based on selenopeptide-modified gold nanoparticles with a tunable glutathione peroxidase activity†
Dechen Zhanga,
Na Shena,
Junrong Zhanga,
Jinming Zhub,
Yi Guoa and
Li Xu *a
*a
aKey Laboratory for Molecular Enzymology and Engineering, The Ministry of Education, National Engineering Laboratory for AIDS Vaccine, School of Life Sciences, Jilin University, Changchun 130012, P. R. China. E-mail: xuli@jlu.edu.cn
bChina-Japan Union Hospital of Jilin University, Changchun 130033, P. R. China
First published on 28th February 2020
Abstract
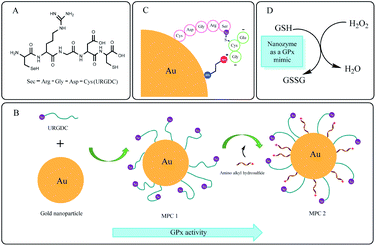
In this study, we successfully prepared a selenium-containing pentapeptide (Sec-Arg-Gly-Asp-Cys)-modified gold nanozyme that exhibited glutathione peroxidase (GPx) activity. The nanozyme catalyzed the reduction of H2O2 by GSH, and its GPx activity was about 14 times that of free selenopeptide. Kinetic analysis indicated that the nanozyme changed the catalytic mechanism from the ping-pong mechanism of the peptide to an ordered mechanism. This indicated that the gold nanoparticle acts as a scaffold that may limit the mobility and constrain the conformation of the peptides, hence exposing the active center to the substrates, and allowing the multivalent selenopeptide to act cooperatively to increase the catalytic rate. Furthermore, upon addition of cysteamine to regulate the surface chemistry of gold nanoparticles and thereby modify the microenvironment of the active center, this nanozyme system could achieve a tunable GPx activity that was about 20 times that of free selenopeptide. Thus, the rational design of the nanozyme greatly improved and amplified the catalytic activity of the ‘active unit’ peptide. This study provides an alternative strategy to establish GPx mimics and provides new insights for the use of gold nanoparticles to develop nanozymes with biological applications.
1. Introduction
Nanozymes are new artificial enzymes in the combined field of nanomaterials and enzymology research that aimed to design novel and efficient enzymatic mimics.1–3 Until now, a variety of nanomaterials have been used to mimic the activity of naturally occurring enzymes,4–8 such as nanocarbon materials,9–11 fullerene,12 carbon nitride,13 Fe3O4 nanoparticles,14 mesoporous silica nanoparticles,15 dopamine nanoparticles,16 melanin nanoparticles,17 and metal–organic complexes.18 Noble metal-based nanomaterials have also shown significant advantages as enzyme mimetics;19,20 gold nanoparticles (AuNPs) possess excellent stability, good biocompatibility, ease of functionalization, and unique optical, and surface properties.21–24 The tailored surface functionalization of gold nanoparticles controls the interface between AuNPs and biomolecules,25–27 and it is crucial for development and design of tunable catalytic activities.28–30Glutathione peroxidase (GPx) is an important selenoenzyme in the cellular antioxidant defense system, which protects cells from oxidative damage by catalyzing the degradation of a variety of hydroperoxides (ROOH) using glutathione (GSH) as the reductant.31–35 GPx contains an essential selenocysteine (Sec) residue at its active site,36–38 and this residue has clear catalytic characteristics.39,40 Considerable effort has been invested in constructing various GPx mimics,41,42 including small molecules,43,44 peptides,45 cyclodextrin derivatives,46,47 abzymes,48 bioimprinting proteins,49,50 and nanozymes.51,52 Small molecular mimics have the advantage of intrinsic structural stability, low immunogenicity, and facile membrane permeability. Ebselen is a well-known GPx mimic, which is a selenium-based organic complex with an anti-inflammatory, anti-oxidant and cytoprotective activity.53,54 However, its catalytic activity is much lower than that of macromolecule protein mimics,41 due to a weaker binding interaction of substrate. The improvement of the catalytic activity of small molecular mimic, while maintaining the above advantageous profile remains a challenge. We hypothesized that multivalent, functional systems anchored on a scaffold, and yet fully soluble, with limited mobility and conformational constraint could be a suitable system to act cooperatively in a catalytic process. Here, a self-assembled selenopeptide-anchored AuNP nanozyme as a GPx mimic was reported.
2. Experimental
Materials and reagents
Auric acid (HAuCl4·4H2O) was purchased from Sinopharm Chemical reagent Co., Ltd. Trisodium citrate (Na3C6H5O7), cysteamine hydrochloride (HSCH2CH2NH2·HCl), glutathione, glutathione reductase, and H2O2 were procured from Sigma-Aldrich. NADPH was purchased from Roche Applied Science. Pentapeptide Ser-Arg-Gly-Asp-Cys (SRGDC) and Sec-Arg-Gly-Asp-Cys (URGDC) were synthesized from GL Biochem (Shanghai) Ltd. Sample stock solution was prepared in Millipore (Milli-Q) water with a resistivity of 18.2 MΩ cm.Apparatus
The kinetic measurements were recorded using a Shimadzu UV2450 spectrophotometer (Japan). TEM analysis was conducted on a JEOL JEM-1200EX model transmission electron microscope. The pH value of the buffer was determined with a Mettler Toledo 320 pH meter. Fourier transform infrared (FTIR) spectral studies were performed on a Nicolet 5700 spectrometer (Thermo Electron Corporation, USA) equipped with an infrared microscope. Spectra were acquired between 4000 and 400 cm−1 with a resolution of 2 cm−1. All freeze-dried powder samples were tiled on the microstat for FTIR measurements.Synthesis of AuNPs
AuNPs were prepared as described earlier.55 Briefly, a 50 mL sample of aqueous HAuCl4 (0.25 mM) was prepared in a 100 mL flask at room temperature. The solution was brought to a boil while being stirred, and the corresponding amount of aqueous sodium citrate with 3.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of citrate to Au3+ was added. The reaction was allowed to run until the solution reached a wine-red colour, which indicated completion of the reaction. These synthesized AuNPs were characterized using UV-vis absorption spectra and dynamic light scattering (DLS) with Zeta Potential Analyser (Brookhaven, USA) coupled to a BI-90Plus digital autocorrelator. According to Liu's method,56 assuming a spherical shape and a uniform fcc structure, the extinction coefficient of the AuNP sample was determined according to the Lambert–Beer law, A = εcl. The AuNP concentration was determined using a Shimadzu UV2450 UV-visible spectrophotometer.
1 molar ratio of citrate to Au3+ was added. The reaction was allowed to run until the solution reached a wine-red colour, which indicated completion of the reaction. These synthesized AuNPs were characterized using UV-vis absorption spectra and dynamic light scattering (DLS) with Zeta Potential Analyser (Brookhaven, USA) coupled to a BI-90Plus digital autocorrelator. According to Liu's method,56 assuming a spherical shape and a uniform fcc structure, the extinction coefficient of the AuNP sample was determined according to the Lambert–Beer law, A = εcl. The AuNP concentration was determined using a Shimadzu UV2450 UV-visible spectrophotometer.
Preparation of nanozyme
Citrate-stabilized colloidal AuNPs were washed by centrifugation at 8000 rpm for 30 min, and re-dissolved in Millipore water. DLS analysis indicated that the hydrodynamic diameter of AuNPs was 20 nm, and the extinction coefficient was calculated to be 6.36 × 108 L mol−1 cm−1. The concentration of AuNPs was adjusted to be 2 nM. For the surface functionalization of AuNPs, the different concentrations of URGDC were added dropwise to the AuNP solution, mixed by violent shaking, and then the homogenate was stored at 4 °C overnight. The resulting Au@SeH with the optimum AuNP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) URGDC molar ratio and maximum enzyme activity was defined as MPC 1. Similar to the preparation of MPC 1, different concentrations of 2-aminoethanethiol was added to the MPC 1 system, mixed by violent shaking, and then the homogenate was stored at 4 °C overnight. The resulting Au@SeH/NH2 with the optimum AuNP
URGDC molar ratio and maximum enzyme activity was defined as MPC 1. Similar to the preparation of MPC 1, different concentrations of 2-aminoethanethiol was added to the MPC 1 system, mixed by violent shaking, and then the homogenate was stored at 4 °C overnight. The resulting Au@SeH/NH2 with the optimum AuNP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cysteamine molar ratio and maximum enzyme activity was termed as MPC 2.
cysteamine molar ratio and maximum enzyme activity was termed as MPC 2.
GPx activity assay
The GPx activity was assayed using H2O2 and GSH as substrates, as previously described.57,58 The reaction was carried out at 37 °C in 700 μL of solution containing 10 mM PBS (pH 7.0), 1 nM EDTA, 1 mM sodium azide, 1 mM GSH, 0.25 mM NADPH, 1 unit of glutathione reductase, and 1–50 μM samples. The mixture was pre-incubated for 5 min and then 0.25 mM NADPH solution was added and incubated for 3 min at 37 °C. The reaction was initiated by the addition of 0.5 mM H2O2. The activity was determined based on the decrease in NADPH absorption at 340 nm. The nonenzymatic reaction affecting the measurement of the initial rate was subtracted to obtain the exact kinetic values.One unit of activity was defined as the amount that utilizes 1 μmol of NADPH per minute. The activity was expressed in U μmol−1.
Enzyme kinetics
The assay for determining enzyme kinetics was similar to that conducted for native GPx.59,60 To investigate the mechanism, assays were carried out under standard reaction conditions as described above by varying concentrations of GSH at a fixed concentration of H2O2 or vice versa. The initial rates were measured by observing the change of NADPH absorption at 340 nm. Data were analyzed using the Origin 8.0 software program. We determined the Kinetics constant by double-reciprocal plots (or Lineweaver–Burk plot) and fit to eqn (1) (ping-pong mechanism) or eqn (2) (ordered mechanism), where V is the reaction velocity, Vmax is the maximal velocity, [ROOH] and [GSH] are the concentrations of substrate H2O2 and GSH, KROOHS is the dissociation constant of enzyme-ROOH, and KROOHm and KGSHm are the Michaelis constants for H2O2 and GSH, respectively.61
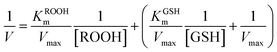 | (1) |
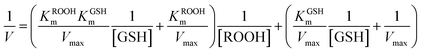 | (2) |
3. Results and discussion
Design of nanozyme
Here, we propose a multivalent catalyst based on selenopeptide (URGDC) and cysteamine co-functionalized AuNPs, to construct a nanozyme with GPx-like activity (Scheme 1). This system provides unique biological functions, coupled with the original properties of AuNPs.Firstly, we designed a selenium-containing pentapeptide (Scheme 1A, URGDC, termed as 5SeP) with a delicate structure, in which the N-terminus was selenocysteine with –SeH (catalytic group of GPx-like activity), and C-terminus was cysteine containing –SH (binding group to AuNPs). Selenocysteine is the active site of natural GPx,62 which enables the pentapeptide GPx-like activity. The high affinity of thiol groups in the cysteine residue for gold atoms enables the pentapeptide binding activity through Au–S bond, resulting in the stable and oriented immobilization of peptides on the AuNPs.63 The RGD motif is the recognition unit used by extracellular matrix proteins (vitronectin, fibrinogen, laminin, and collagen) to bind a variety of integrins that mediates cell attachment. It makes the selenopeptide more potent for identification to facilitate specific binding. Secondly, the selenopeptides were conjugated to AuNP through self-assembly to obtain one mimic (MPC 1). AuNP, as a scaffold, confines the mobility and conformation of randomly oriented URGDC to make most of the –SeH groups exposed in a multivalent environment. Thus, GSH and H2O2 have ready access to the catalytic center, making the catalytic reaction more efficient. Thirdly, the nanozyme–substrate interface offers the opportunity to regulate enzyme activity. Cysteamine was employed to further decorate AuNPs by forming Au–S covalent bond (another mimic defined as MPC 2, Scheme 1B). It can adjust the distance and space structure of URGDC on the AuNP surface. Meanwhile, the electrostatic attraction between anionic GSH substrate and the positively charged monolayer facilitates its transport and binding to the active site (Scheme 1C). MPC 2 mimic provides an electrostatic architecture which, in each reductive step, directs the donor substrate GSH towards the catalytic center in such a way that its sulfhydryl group must react with the selenium moiety. Consequently, the MPC 2 mimic shows a stronger catalysis activity. The catalytic mechanism of the nanozyme is shown in Scheme 1D.
Characterization of the nanozyme
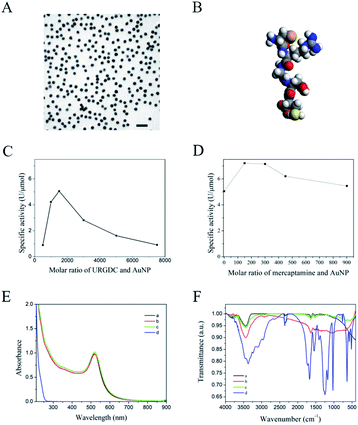
The synthesized AuNPs were investigated by TEM. It is apparent that the AuNPs are monodisperse, spherical in shape, and a narrow particle-size distribution with a mean size of about 20 nm (Fig. 1A). The characteristic surface plasmon resonance (SPR) band at 518 nm is shown in Fig. 1E. The calculated extinction coefficient of AuNP was 6.389 × 108 L mol−1 cm−1.55The molecular simulation of URGDC was obtained by material studio (Fig. 1B), and the calculated cross-sectional diameter was 4 Å. Thus, it was calculated that about 8000 URGDC molecules are needed to completely cover the surface of one 20 nm AuNP.64 Given that, the specific activities of Au@SeH with different Au![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) URGDC molar ratio were tested. It was found that Au@SeH with 1
URGDC molar ratio were tested. It was found that Au@SeH with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1500 molar ratio of Au
1500 molar ratio of Au![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) URGDC (MPC 1) showed the maximum GPx activity of 5.05 U μmol−1 (Fig. 1C, Table 1). Thus, the molar ratio of Au
URGDC (MPC 1) showed the maximum GPx activity of 5.05 U μmol−1 (Fig. 1C, Table 1). Thus, the molar ratio of Au![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) URGDC was fixed to be 1
URGDC was fixed to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1500, and then different proportions of cysteamine were added to obtain Au@SeH/NH2. The tunable specific activity of Au@SeH/NH2 was measured, and we found that Au@SeH/NH2 with 1
1500, and then different proportions of cysteamine were added to obtain Au@SeH/NH2. The tunable specific activity of Au@SeH/NH2 was measured, and we found that Au@SeH/NH2 with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 molar ratio of Au
150 molar ratio of Au![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cysteamine (MPC 2) showed the maximum GPx activity of 7.21 U μmol−1 (Fig. 1D, Table 1). Fig. 1E shows the UV-vis absorption spectra of MPC 1 and MPC 2. The 5SeP and cysteamine modification did not change the optical properties, since the shoulder band towards the longer wavelengths in the range of 600–700 nm did not appear, except that the peak absorbance of MPC 2 was slightly higher. This suggested that the AuNPs did not aggregate, and they are stable in the nanozyme system. It was also confirmed by TEM images (ESI Fig. S1†), there is no obvious change in the morphology of nanozyme. As shown in Fig. 1F, 5SeP exhibited S–H bond stretching vibration falls around 2357 cm−1 for free cysteine, and amide I and II regions showed two peaks located at 1674 and 1635 cm−1, respectively. In the N–H stretching region, 5SeP showed a broader band located at 3373 cm−1. Upon interaction with the surface of AuNP, the whole spectrum of 5SeP was strongly affected: amide I and II bands were weakened, and S–Au vibration appeared at 1644 cm−1.65 Furthermore, the N–H stretching band, which in the free peptide located around 3373 cm−1, became sharp and without any shoulder, probably forbidden by the surface selection rule.66 This result illustrated the conjugation of 5SeP and/or cysteamine over the surface of AuNP.
cysteamine (MPC 2) showed the maximum GPx activity of 7.21 U μmol−1 (Fig. 1D, Table 1). Fig. 1E shows the UV-vis absorption spectra of MPC 1 and MPC 2. The 5SeP and cysteamine modification did not change the optical properties, since the shoulder band towards the longer wavelengths in the range of 600–700 nm did not appear, except that the peak absorbance of MPC 2 was slightly higher. This suggested that the AuNPs did not aggregate, and they are stable in the nanozyme system. It was also confirmed by TEM images (ESI Fig. S1†), there is no obvious change in the morphology of nanozyme. As shown in Fig. 1F, 5SeP exhibited S–H bond stretching vibration falls around 2357 cm−1 for free cysteine, and amide I and II regions showed two peaks located at 1674 and 1635 cm−1, respectively. In the N–H stretching region, 5SeP showed a broader band located at 3373 cm−1. Upon interaction with the surface of AuNP, the whole spectrum of 5SeP was strongly affected: amide I and II bands were weakened, and S–Au vibration appeared at 1644 cm−1.65 Furthermore, the N–H stretching band, which in the free peptide located around 3373 cm−1, became sharp and without any shoulder, probably forbidden by the surface selection rule.66 This result illustrated the conjugation of 5SeP and/or cysteamine over the surface of AuNP.
| Catalyst | Substrate | Activitya (U μmol−1) |
|---|---|---|
| a The GPx activities were measured by a coupled enzymatic assay (as described) in sodium phosphate buffer (10 mM, pH 7.0) at 37 °C. | ||
| Ebselen54 | H2O2, GSH | 0.99 |
| 2-SeCD67 | H2O2, GSH | 7.40 |
| SRGDC | H2O2, GSH | 0 |
| URGDC | H2O2, GSH | 0.36 |
| MPC 1 | H2O2, GSH | 5.05 |
| MPC 2 | H2O2, GSH | 7.21 |
| Native GPx41 | H2O2, GSH | 5780 |
GPx activity and kinetics of 5SeP
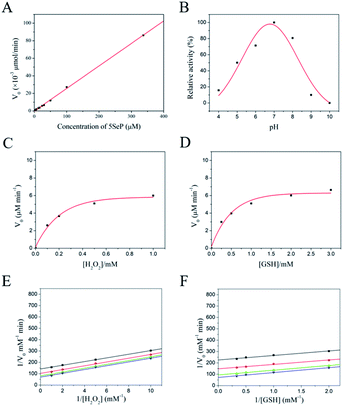
To study the GPx activity of 5SeP, we monitored the initial velocity under several concentrations of 5SeP (Fig. 2A). As a result, a significant liner correlation was observed between them. Analysis of the data revealed that the GPx activity of 5SeP was 0.36 U μmol−1 (Table 1). The SRGDC without –SeH catalytic group did not show GPx activity. Compared with nanozyme system, the GPx activity of MPC 2 was about 20 times that of free selenopeptide and 1.43 times that of MPC 1. Therefore, MPC 2 displayed a higher activity. In a multivalent environment, AuNP scaffold could efficiently facilitate the conjugation between substrate and multi active center. In addition, an electrostatic architecture (cysteamine) improved the reaction process in a more favourable action. For the other small molecular artificial GPx, the GPx activity is close to the same level of 2-SeCD mimic in the absent of hydrophobic environment for binding substrate.67 It means that the multi active center system is more efficient for the artificial enzyme catalytic process.Similar to the crude GPx, the catalytic activity of 5SeP was dependent on pH, GSH, and H2O2 concentration. We measured the GPx-like activity of 5SeP while varying the pH from 4 to 10, the H2O2 concentration from 0.1 to 1 mM, and the GSH concentration from 0.25 to 3 mM. The optimum pH was approximately 7.0 (Fig. 2B). The steady-state rate of catalysis increased towards a saturation plateau with increasing loads of substrate H2O2 or GSH, which was similar to the enzyme catalyzed reaction (Fig. 2C and D). Thus, it was confirmed that 5SeP had GPx-like activity and enzymatic reaction properties.
To investigate the mechanism of GPx activity of 5SeP, we determined the apparent steady-state kinetic parameters for the reaction. The double-reciprocal plots were obtained (Fig. 2E and F), and the parallel lines indicated that the GPx-like reaction of 5SeP comply with a ping-pong mechanism.68
Kinetics study of the nanozyme
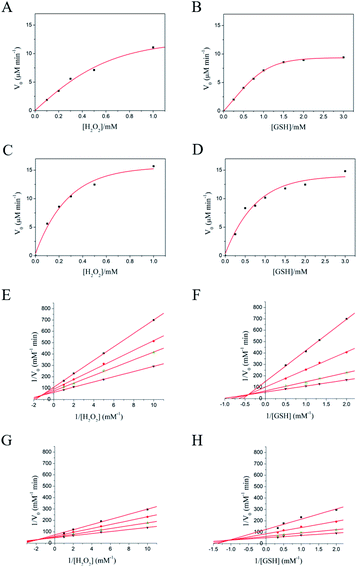
Next, we assessed the first-order kinetics to systematically study the property of nanozyme catalysis. As a result, a kinetic profile of the reaction towards saturation plateau was observed by varying the initial concentration of substrate H2O2 or GSH (Fig. 3A–D). Thus, it was revealed that both MPC 1 and MPC 2 possessed GPx-like activity and enzymatic reaction properties. To further investigate the mechanism of GPx activity of the nanozyme, we determined the apparent steady-state kinetic parameters for the reaction. The double-reciprocal plots were obtained (Fig. 3E–H), and the crossed lines indicated that the GPx-like reaction of MPC 1 and MPC 2 abides by an ordered mechanism.69The kinetic parameters for URGDC, MPC 1, and MPC 2 are listed in Table 2. Their first order constants (Km) for H2O2 diminished in order, and the formal second-order rate constant Kcat/Km of MPC 1 and MPC 2 for H2O2 was 22.7 and 29.2 times higher than that determined under identical conditions for free 5SeP, respectively. The Kcat/Km of MPC 1 and MPC 2 for GSH was 20.0 and 42.1 times higher than that determined under identical conditions for free 5SeP, respectively; this corresponds to the ‘active unit’ on AuNP surface. Thus, by adjusting with cysteamine, the MPC 2 could show more efficient catalysis activity.
Among the three mimics, the Km value of both substrates showed a common profile, with 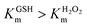 this demonstrated that H2O2 was a priority substrate, likely because H2O2 molecule is smaller than the GSH molecule, making it easier for it to approach the active site of the enzyme. However, while the access of H2O2 to the active site is not specific, the GSH shows specific/selective recognition; this is similar to the characteristics of natural GPx.
this demonstrated that H2O2 was a priority substrate, likely because H2O2 molecule is smaller than the GSH molecule, making it easier for it to approach the active site of the enzyme. However, while the access of H2O2 to the active site is not specific, the GSH shows specific/selective recognition; this is similar to the characteristics of natural GPx.
4. Conclusions
In conclusion, a selenopeptide functionalized AuNP was rationally designed in this study. The nanozyme-catalyzed reaction exhibited saturation kinetics: the reactive rate data complied with the Michaelis–Menten model of enzyme kinetics. The GPx activity of MPC 2 was 7.21 U μmol−1, which is about 20 times of that of free selenopeptide. The kinetics analysis indicated that the catalytic mechanism of free peptide was the ping-pong mechanism, while the nanozymes (MPC 1, MPC 2) showed an ordered mechanism. Furthermore, the catalytic constant Kcat/Km of both GSH and H2O2 were one order of magnitude higher than that of free selenopeptide. Thus, improved properties and synergistic activities could be achieved from an interface-regulating approach, as evident from the measured GPx activity and catalytic characteristics. We obtained not only a peptide-capped nanoparticle (MPC 1) with synergistic enzyme activity, compared with the ‘active unit’, but also a more efficient MPC 2 nanozyme by cysteamine adjustment. Thus, our study successfully demonstrates that AuNPs provide advantages of easily obtainable self-assembled monolayer and specific surface/interface chemistry, displaying a promising platform for improving and establishing delicate and efficient nanozyme catalytic system.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81771965, 31571017, 81571791, 81271697, 21043002), the National “Significant New Drug Creation” Science and Technology Major Program (No. 2012ZX09503001-003), the Project of Science and Technology Department of Jilin Province, China (No. 20170204027NY, 20130206069GX), and the Special Project of Biological Medicine of Jilin Province, China (No. SXGJSF2017-1).Notes and references
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS.
- J. Wu, S. Li and H. Wei, Chem. Commun., 2018, 54, 6520–6530 RSC.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS.
- X. Zheng, Q. Liu, C. Jing, Y. Li, D. Li and W. Luo, et al., Angew. Chem., Int. Ed., 2011, 50, 11994–11998 CrossRef CAS.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang and N. Gu, et al., Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS.
- Y. Tao, E. Ju, J. Ren and X. Qu, Chem. Commun., 2014, 50, 3030–3032 RSC.
- Y. Hu, X. J. Gao, Y. Zhu, F. Muhammad, S. Tan and W. Cao, et al., Chem. Mater., 2018, 30, 6431–6439 CrossRef CAS.
- C. Ge, G. Fang, X. Shen, Y. Chong, W. G. Wamer and X. Gao, et al., ACS Nano, 2016, 10, 10436–10445 CrossRef CAS PubMed.
- K. Fan, J. Xi, L. Fan, P. Wang, C. Zhu and Y. Tang, et al., Nat. Commun., 2018, 9, 1440 CrossRef PubMed.
- L. Huang, J. Chen, L. Gan, J. Wang and S. Dong, Sci. Adv., 2019, 5, eaav5490 CrossRef PubMed.
- D. Zhen, N. Jiang, H. Geng, Y. Qiao, Y. Liu and X. Zhu, et al., Microchim. Acta, 2019, 186, 691 CrossRef PubMed.
- Z. Du, N. Gao, X. Wang, J. Ren and X. Qu, Small, 2018, 14, 1801852 CrossRef PubMed.
- P. Zhang, D. Sun, A. Cho, S. Weon, S. Lee and J. Lee, et al., Nat. Commun., 2019, 10, 940 CrossRef PubMed.
- M. Huo, L. Wang, Y. Chen and J. Shi, Nat. Commun., 2017, 8, 357 CrossRef PubMed.
- J. Kim, H. R. Cho, H. Jeon, D. Kim, C. Song and N. Lee, et al., J. Am. Chem. Soc., 2017, 139, 10992–10995 CrossRef CAS PubMed.
- H. Li, Y. Zhao, Y. Jia, C. Qu and J. Li, Chem. Commun., 2019, 55, 15057–15060 RSC.
- Y. Liu, K. Ai, X. Ji, D. Askhatova, R. Du and L. Lu, et al., J. Am. Chem. Soc., 2017, 139, 856–862 CrossRef CAS PubMed.
- Z. Chen, Z. Wang, J. Ren and X. Qu, Acc. Chem. Res., 2018, 51, 789–799 CrossRef CAS PubMed.
- G. Fang, W. Li, X. Shen, J. M. Perez-Aguilar, Y. Chong and X. Gao, et al., Nat. Commun., 2018, 9, 129 CrossRef PubMed.
- R. Das, A. Dhiman, A. Kapil, V. Bansal and T. K. Sharma, Anal. Bioanal. Chem., 2019, 411, 1229–1238 CrossRef CAS PubMed.
- Y. Hu, H. Cheng, X. Zhao, J. Wu, F. Muhammad and S. Lin, et al., ACS Nano, 2017, 11, 5558–5566 CrossRef CAS PubMed.
- H. Ye, K. Yang, J. Tao, Y. Liu, Q. Zhang and S. Habibi, et al., ACS Nano, 2017, 11, 2052–2059 CrossRef CAS PubMed.
- W.-L. Wan, Y.-J. Lin, H.-L. Chen, C.-C. Huang, P.-C. Shih and Y.-R. Bow, et al., J. Am. Chem. Soc., 2017, 139, 12923–12926 CrossRef CAS PubMed.
- S. Neri, S. G. Martin, C. Pezzato and L. J. Prins, J. Am. Chem. Soc., 2017, 139, 1794–1797 CrossRef CAS PubMed.
- S. Rana, A. Bajaj, R. Mout and V. M. Rotello, Adv. Drug Deliv. Rev., 2012, 64, 200–216 CrossRef CAS PubMed.
- P. M. Kelly, C. Åberg, E. Polo, A. O'connell, J. Cookman and J. Fallon, et al., Nat. Nanotechnol., 2015, 10, 472–479 CrossRef CAS PubMed.
- Y. Chen, Y. Xianyu and X. Jiang, Acc. Chem. Res., 2017, 50, 310–319 CrossRef CAS PubMed.
- Y. Zhao, Y. Huang, H. Zhu, Q. Zhu and Y. Xia, J. Am. Chem. Soc., 2016, 138, 16645–16654 CrossRef CAS PubMed.
- B. Liu and J. Liu, Nano Res., 2017, 10, 1125–1148 CrossRef CAS.
- L. Liu, J. Du, W.-e. Liu, Y. Guo, G. Wu and W. Qi, et al., Anal. Bioanal. Chem., 2019, 411, 2189–2200 CrossRef CAS PubMed.
- A. L. Tappel, Curr. Top. Cell. Regul., 1984, 24, 87–97 CAS.
- I. Ingold, C. Berndt, S. Schmitt, S. Doll, G. Poschmann and K. Buday, et al., Cell, 2018, 172, 409–422 CrossRef CAS PubMed.
- W. S. Hambright, R. S. Fonseca, L. Chen, R. Na and Q. Ran, Redox Biol., 2017, 12, 8–17 CrossRef CAS PubMed.
- Ö. Canli, Y. B. Alankuş, S. Grootjans, N. Vegi, L. Hültner and P. S. Hoppe, et al., Blood, 2016, 127, 139–148 CrossRef PubMed.
- J. Sonet, K. Bierla, A.-L. Bulteau, R. Lobinski and L. Chavatte, Anal. Chim. Acta, 2018, 1011, 11–19 CrossRef CAS PubMed.
- O. Epp, R. Ladenstein and A. Wendel, Eur. J. Biochem., 1983, 133, 51–69 CrossRef CAS PubMed.
- P. Scheerer, A. Borchert, N. Krauss, H. Wessner, C. Gerth and W. Höhne, et al., Biochemistry, 2007, 46, 9041–9049 CrossRef CAS PubMed.
- D. Bhowmick and G. Mugesh, Org. Biomol. Chem., 2015, 13, 10262–10272 RSC.
- K. D. Aumann, N. Bedorf, R. Brigelius-Flohe, D. Schomburg and L. Flohe, Biomed. Environ. Sci., 1997, 10, 136–155 CAS.
- D. E. Fomenko, W. Xing, B. M. Adair, D. J. Thomas and V. N. Gladyshev, Science, 2007, 315, 387–389 CrossRef CAS PubMed.
- X. Huang, X. M. Liu, Q. A. Luo, J. Q. Liu and J. C. Shen, Chem. Soc. Rev., 2011, 40, 1171–1184 RSC.
- F. Yan, Y. Mu, G. L. Yan, J. Q. Liu, J. C. Shen and G. M. Luo, Mini-Rev. Med. Chem., 2010, 10, 342–356 CrossRef CAS PubMed.
- A. Muller, E. Cadenas, P. Graf and H. Sies, Biochem. Pharmacol., 1984, 33, 3235–3239 CrossRef CAS PubMed.
- T. Wirth, Angew. Chem., Int. Ed., 2015, 54, 10074–10076 CrossRef CAS PubMed.
- F. Yan, G. L. Yan, S. W. Lv, N. Shen, Y. Mu and T. Chen, et al., Int. J. Biochem. Cell Biol., 2011, 43, 1802–1811 CrossRef CAS PubMed.
- J. Liu, G. Luo, X. Ren, Y. Mu, Y. Bai and J. Shen, Biochim. Biophys. Acta, 2000, 1481, 222–228 CrossRef CAS.
- M. McNaughton, L. Engman, A. Birmingham, G. Powis and I. A. Cotgreave, J. Med. Chem., 2004, 47, 233–239 CrossRef CAS PubMed.
- X. J. Ren, S. J. Gao, D. L. You, H. L. Huang, Z. Liu and Y. Mu, et al., Biochem. J., 2001, 359, 369–374 CrossRef CAS PubMed.
- L. Liu, S. Z. Mao, X. M. Liu, X. Huang, J. Y. Xu and J. Q. Liu, et al., Biomacromolecules, 2008, 9, 363–368 CrossRef CAS PubMed.
- N. Shen, F. Yan, Y. Guo, S. W. Lue, P. S. Gong and Y. W. Xu, et al., Chem. Res. Chin. Univ., 2011, 27, 258–263 CAS.
- X. Huang, Y. Z. Yin, X. Jiang, Y. Tang, J. Y. Xu and J. Q. Liu, et al., Macromol. Biosci., 2009, 9, 1202–1210 CrossRef CAS PubMed.
- X. Huang, Y. Z. Yin, Y. Tang, X. L. Bai, Z. M. Zhang and J. Y. Xu, et al., Soft Matter, 2009, 5, 1905–1911 RSC.
- M. Parnham and H. Sies, Biochem. Pharmacol., 2013, 86, 1248–1253 CrossRef CAS PubMed.
- G. Mugesh and H. B. Singh, Chem. Soc. Rev., 2000, 29, 347–357 RSC.
- X. Ji, X. Song, J. Li, Y. Bai, W. Yang and X. Peng, J. Am. Chem. Soc., 2007, 129, 13939–13948 CrossRef CAS PubMed.
- X. Liu, M. Atwater, J. Wang and Q. Huo, Colloids Surf., B, 2007, 58, 3–7 CrossRef CAS PubMed.
- F. Yan, G. Yan, S. Lv, N. Shen, Y. Mu and T. Chen, et al., Int. J. Biochem. Cell Biol., 2011, 43, 1802–1811 CrossRef CAS PubMed.
- Y. Sun, T. Y. Li, H. Chen, K. Zhang, K. Y. Zheng and Y. Mu, et al., J. Biol. Chem., 2004, 279, 37235–37240 CrossRef CAS PubMed.
- L. Flohe, G. Loschen, W. A. Gunzler and E. Eichele, Hoppe-Seyler's Z. Physiol. Chem., 1972, 353, 987–1000 CrossRef CAS PubMed.
- C. J. Weydert and J. J. Cullen, Nat. Protoc., 2010, 5, 51–66 CrossRef CAS PubMed.
- S. Toppo, L. Flohe, F. Ursini, S. Vanin and M. Maiorino, Biochim. Biophys. Acta, Gen. Subj., 2009, 1790, 1486–1500 CrossRef CAS PubMed.
- L. Flohe, S. Toppo, G. Cozza and F. Ursini, Antioxid. Redox Signaling, 2011, 15, 763–780 CrossRef CAS PubMed.
- A. M. W. Reed and S. J. Metallo, Langmuir, 2010, 26, 18945–18950 CrossRef CAS PubMed.
- L. Xu, Y. Guo, R. Xie, J. Zhuang, W. Yang and T. Li, Nanotechnology, 2002, 13, 725–728 CrossRef CAS.
- C. K. Yee, A. Ulman, J. D. Ruiz, A. Parikh, H. White and M. Rafailovich, Langmuir, 2003, 19, 9450–9458 CrossRef CAS.
- G. Scari, F. Porta, U. Fascio, S. Avvakumova, V. Dal Santo and M. De Simone, et al., Bioconjugate Chem., 2012, 23, 340–349 CrossRef CAS PubMed.
- J. Q. Liu, S. J. Gao, G. M. Luo, G. L. Yan and J. C. Shen, Biochem. Biophys. Res. Commun., 1998, 247, 397–400 CrossRef CAS PubMed.
- W. W. Cleland, J. Biol. Chem., 1973, 248, 8353–8355 CAS.
- X. Jiang, C. Sun, Y. Guo, G. Nie and L. Xu, Biosens. Bioelectron., 2015, 64, 165–170 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra10262k |
| This journal is © The Royal Society of Chemistry 2020 |