Open Access Article
Open Access ArticleEvaluation of curcumin-mediated photodynamic therapy on the reverse of multidrug resistance in tumor cells†
Ying Li‡
,
Pei Xiao‡,
Zipeng Huang,
Xinru Chen,
Xia Yan,
Junqiu Zhai * and
Yan Ma
* and
Yan Ma *
*
School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, No. 232 Waihuan East Road, Panyu District, Guangzhou, China. E-mail: mayan2006@gzucm.edu.cn; jqzhai@gzucm.edu.cn
First published on 2nd January 2020
Abstract
Curcumin (CUR) possesses photosensitive anti-tumor activity. However, photoactive CUR mainly targets tumor cells sensitive to chemotherapy, whereas the effect on multi-drug resistant cancer cells has not been fully investigated. The study aimed to investigate the anti-tumor activity of CUR on resistant MCF-7/ADM cells and its underlying mechanism providing insights into CUR-mediated PDT and a reference for reversing multidrug resistance. Cell apoptosis and morphological changes were detected by Annexin V-FITC/PI double staining and immunofluorescence, respectively. The apoptosis mechanism of CUR-mediated PDT was investigated by detecting the levels of reactive oxygen species (ROS), mitochondrial membrane potential, and related proteins. MTT and apoptosis results showed that CUR-mediated PDT significantly enhanced cytotoxicity and induced considerable cell apoptosis. After treatment with CUR-mediated PDT, cells became round in shape and shrunk, F-actin was loosely arranged, and the nucleus decreased in size. In addition, the level of ROS increased over time compared to the control and peaked at 6 h. CUR-mediated PDT induced alterations in the mitochondrial membrane potential, increased the release of mitochondrial cytochrome C (Cyt-c), and downregulated caspase-3/7/9, PARP, and P-gp. In conclusion, CUR-PDT induced apoptosis in resistant MCF-7/ADM cells primarily through endogenous mitochondrial apoptosis pathway. Besides apoptosis activation in resistant cells, the reverse of multidrug resistance was ascribed to the downregulation of P-gp expression to a degree.
1. Introduction
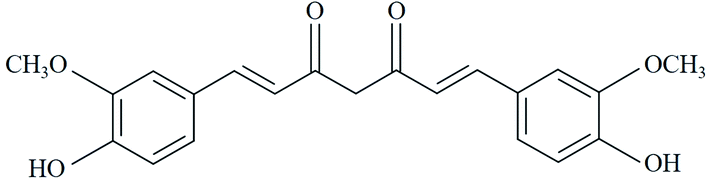
Curcumin (CUR) is the major active component of turmeric and was originally isolated from the plant Curcuma longa.1 CUR possesses a wide range of pharmacologic activities such as antibacterial, antiviral, anti-inflammatory, and antitumor activities.2,3 Fig. 1 showed the chemical structure of CUR. Previous studies had shown that the antitumor activity of CUR could be enhanced by illumination, and low-dose photosensitive CUR exhibited significant antitumor activity compared to non-photosensitive CUR.4,5 As reported, the IC50 of HeLa cells treated with photoactive CUR was 0.1 times that of non-photoactive CUR and the photodynamic antitumor effect of CUR on human epithelial carcinoma A431 cells was significantly enhanced by CUR-mediated photodynamic therapy (PDT) compared with control without light irradiation.6,7PDT is a treatment that uses photosensitizing agents, along with light and oxygen to kill cancer cells by the generation of ROS.8 Photosensitizers as the core component of PDT only work after they have been activated or “turned on” by certain kinds of light.9 Compared with photosensitizers such as hematoporphyrin derivatives, CUR has the characteristics of clear structure, high ROS yield, and low skin phototoxicity. And its applications to PDT has been a recent research topic of interest.10 It has been reported that photosensitized CUR can induce apoptosis of NPC/CNE2,11 HeLa,12 DKMG,13 MCF-7,14 and other tumor cells.15,16
However, the majority of studies have focused on the anti-tumor effects of photosensitive CUR on tumor cells sensitive to chemotherapy, whereas investigations on its effect on multi-drug resistant cells are limited. In the present study, the molecular mechanism by which CUR-PDT induces apoptosis in human breast cancer MCF-7/ADM cells was investigated in order to provide the basis for utilizing PDT to overcome tumor multidrug resistance.
2. Experimental details
2.1. Regents and methods
CUR (>95% purity) was purchased from Sigma-Aldrich Co., Ltd. (Shanghai, China) and dissolved in dimethyl sulphoxide (DMSO) to obtain a 50 mmol L−1 stock solution. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was obtained from Sigma-Aldrich Co., Ltd. (Shanghai, China) and dissolved in PBS to obtain a 2.5 mg mL−1 stock solution. Adriamycin (ADM, 98% purity) was purchased from Beijing Huafeng United Technology Co., Ltd. (Beijing, China). Insulin was obtained from Sigma (St. Louis, MO, USA). Dulbecco's modified eagle medium (DMEM), pancreatin, penicillin–streptomycin, and fetal bovine serum were acquired from Life Technology (USA). The Annexin V-FITC/PI apoptosis detection kit, ROS detection kit, and mitochondrial membrane potential assay kit with JC-1 (5,50,6,60-tetrachloro-1,10,3,30-tetrethyl benzimidalyl carbocyanine iodide) were purchased from Shanghai Best Biotechnology Co., Ltd. (Shanghai, China). Rabbit anti-P glycoprotein antibody was purchased from Abcam (Cambridge, UK). Rabbit caspase-3 antibody, caspase-7 antibody, rabbit caspase-8 antibody, caspase-9, rabbit PARP antibody, rabbit cytochrome c antibody, and anti-rabbit lgG, HRP-linked antibody were obtained from the American Cell Signaling Technology Company (MA, USA). GAPDH antibody and chemiluminescent ECL assay kit were purchased from Affinity Biosciences (USA). TRITC Phalloidin and BCA proteins assay kit were obtained from Dalian Meilun Biotechnology Co., Ltd. (Dalian, China). NCM Universal Antibody Diluent was purchased from New Cell & Molecular Biotech Co., Ltd. (Suzhou, China).2.2. Cell culture
Human breast cancer cell lines MCF-7 and MCF-7/ADM were obtained from the School of Life Sciences and Biopharmaceutics (Guangdong, China). MCF-7 cells were cultured in DMEM medium supplemented with 10% (v/v) FBS, 0.01 mg mL−1 insulin, and 1% penicillin/streptomycin (50 U mL−1 penicillin and 50 μg mL−1 streptomycin) at 37 °C in humidified atmosphere with 5% CO2 and supplied with fresh culture medium every three days. MCF-7/ADM cells were grown in DMEM medium supplemented with 10% FBS, 1.83 μm ADM, and 1% penicillin/streptomycin at 37 °C in 5% CO2. All experiments were conducted with cells in the logarithmic growth phase and without ADM in DMEM medium.2.3. PDT
Cells were seeded into appropriate plates or laser confocal culture dishes at a certain density, and incubated with CUR for 45 min at 37 °C in 5% CO2. Subsequently, cells were supplied with fresh medium (without CUR) after washing twice with PBS, and irradiated by using a set of UV-LED (Shenzhen Hatter Photoelectricity Co., Ltd. Shenzhen, China) at 100 mW cm−2 for 5 min. The wavelength of the UV-LED was set at 450 nm. Light intensity was measured as Lux and inverted to light dose (mW cm−2).2.4. Cell viability assay
To evaluate toxicity induced by CUR-mediated PDT, the viabilities of MCF-7 and MCF-7/ADM cells were evaluated by MTT assay. Cells were seeded into 96-well plates at a density of 8 × 103 cells per well (200 μL in total) and allowed to adherence for 24 h. Then, CUR at different concentrations (2.5, 5.0, 7.5, 10.0, 12.5, 15.0 μM) was subsequently added to designated wells, and DMEM without CUR was set as blank control. One group of cells was treated with PDT as described in Section 2.3, whereas the other was dark treatment (without PDT). After 24 h, MTT solution (2.5 mg mL−1) was added and incubated for another 4 h. Subsequently, the medium containing MTT was completely removed, which was followed by the addition of 150 μL of DMSO, and then the plate was gently vibrated for 15 min. The absorbance was measured at 490 nm using a Bio Tek microplate reader (Thermo Fisher Scientific Inc., Waltham, MA, USA). Cell viability rate was calculated using the following equation:2.5. Cell apoptosis analysis
Annexin V-FITC/PI double staining was used to confirm the apoptotic effect of CUR-mediated PDT on MCF-7 and MCF-7/ADM cells. In short, cells at a density of 1.5 × 105 cells per well (2 mL total volume) were seeded into six-well plates for 24 h and then treated with 7.5 μM CUR-mediated PDT. Approximately 24 h later, cells were harvested and washed twice with 37 °C PBS. Next, cells were suspended in 400 μL binding buffer, followed by staining with 5 μL of Annexin V-FITC for 15 min at 2–8 °C, and then 10 μL of propidium iodide (PI) was added and incubated for 5 min. Cells were immediately analyzed by flow cytometry (Becton, Dickinson and Company, New York, NY, USA).2.6. Assessment of cellular morphology
To observe the morphology of cells, MCF-7 and MCF-7/ADM cells were seeded at a density of 8 × 103 cells well−1 into 96-well plates for 24 h and then treated with 7.5 μm CUR-mediated PDT. Cells were observed using IncuCyte ZOOM (Sintec) 24 h later.To assess changes in F-actin and nuclei,17 MCF-7 and MCF-7/ADM cells were seeded into laser confocal culture dishes (35 mm in diameter) at a density of 5 × 104 cells well−1 for 24 h and processed with 7.5 μM CUR-mediated PDT similarly. Approximately 24 h later, cells were fixed in 4% ice-cold formaldehyde for 10 min, followed by 0.5% Triton X-100 treatment for 10 min at room temperature. Cells were gently washed thrice with PBS and incubated with 200 μL of TRITC-phalloidin in PBS (containing 1% BSA, TRITC-phalloidin: PBS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) for 45 min at room temperature to visualize F-actin. After three washes with PBS, 5 μg mL−1 DAPI was added, and cells were incubated for another 5 min. Finally, cells were observed under laser scanning confocal microscopy (Carl Zeiss AG).
1000) for 45 min at room temperature to visualize F-actin. After three washes with PBS, 5 μg mL−1 DAPI was added, and cells were incubated for another 5 min. Finally, cells were observed under laser scanning confocal microscopy (Carl Zeiss AG).
2.7. Measurement of ROS
ROS levels were measured using a ROS assay kit.18 The probe 2′,7′-dichlorofluorescein-diacetate (DCFH-DA) is easily oxidized to fluorescent dichlorofluorescein (DCF) by intracellular ROS, allowing quantification of ROS levels. Briefly, cells were seeded into a 96-well plate at a density of 1.5 × 104 cells well−1, 5 μM CUR was added to each well, and then cells were incubated for 24 h. After 2, 4, 6, 8, 10, and 12 h of PDT, cells were incubated with 10 μm DCFH-DA in the dark for 30 min at 37 °C and then washed gently with PBS twice. Finally, the intensity of DCF was measured using IncuCyte ZOOM.2.8. Mitochondrial membrane potential (MMP) detection
The fluorescent probe 5,50,6,60-tetrachloro-1,10,3,30-tetrethylbenzimidalyl carbocyanine iodide (JC-1) was used to monitor changes in MMP following CUR-PDT by flow cytometry to verify whether apoptosis induced by CUR-PDT in MCF-7 and MCF-7/ADM cells involved changes in MMP.19 Briefly, MCF-7 and MCF-7/ADM cells were seeded into six-well plates at a density of 1.5 × 105 cells per well. After treated with 7.5 μM CUR-mediated PDT for 24 hours, cells were resuspended and mixed with 500 μL JC-1 staining solution at 37 °C for 20 min. Finally, the JC-1 signals were detected by flow cytometry.2.9. Western blot analysis
Western blot was used to detect the expression of apoptosis-related protein and P-gp. MCF-7 and MCF-7/ADM cells were seeded into six-well plates at a density of 1.5 × 105 cells per well. After treatment of 7.5 μM CUR-mediated PDT for 24 hours, cells were washed with cold PBS and lysed with a 0.1% Triton-X-100 buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, protease, and phosphatase inhibitor (1×) for 15 min. Then, the cellular extract was collected and centrifuged at 12,000g for 15 min at 4 °C. The supernatant was extracted as the total protein fraction. To isolate mitochondrial protein and cytosolic protein separately,20 cells were firstly washed with PBS and lysed with 0.1% Triton-X 100 buffer containing 10 mM Tris (pH 7.5), 1 mM EDTA, and 1 mM phenyl-methylsulfonyl fluoride (PMSF). Secondly, cellular extract was centrifuged at 700g for 5 min (4 °C), and then the supernatant was centrifuged again at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min (4 °C) and collected as the cytosolic protein fraction. The remaining cellular pellet was sequentially washed with Triton-X-100 buffer and centrifuged at 21,000g for 15 min (4 °C). Next, the obtained cellular pellet was further washed with radio-immunoprecipitation assay (RIPA) buffer containing 1 mM PMSF and centrifuged again at 21,000g for 15 min (4 °C). Finally, the supernatant was collected as the mitochondrial protein fraction. Protein concentrations were estimated using the BCA method. An equal volume of a 5× sample buffer was added to the protein samples, followed by denaturation at 100 °C for 5 min. Then, the protein samples were stored at −20 °C until analysis.
000g for 15 min (4 °C) and collected as the cytosolic protein fraction. The remaining cellular pellet was sequentially washed with Triton-X-100 buffer and centrifuged at 21,000g for 15 min (4 °C). Next, the obtained cellular pellet was further washed with radio-immunoprecipitation assay (RIPA) buffer containing 1 mM PMSF and centrifuged again at 21,000g for 15 min (4 °C). Finally, the supernatant was collected as the mitochondrial protein fraction. Protein concentrations were estimated using the BCA method. An equal volume of a 5× sample buffer was added to the protein samples, followed by denaturation at 100 °C for 5 min. Then, the protein samples were stored at −20 °C until analysis.
Approximately 10 μg of protein, 30 μg of mitochondrial protein and/or cytosolic protein were separated using 10–15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis with a 5% stacking gel. Separated proteins were transferred onto a 0.22 μm polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% skim milk or 5% BSA in Tris-buffered saline and 0.1% Tween-20 for 2 h at room temperature. After washing the membrane thrice with TBS-T, the blots were incubated with primary antibodies against P-gp, caspase-3, caspase-7, caspase-9, PARP, Cyt-c, and GAPDH overnight at 4 °C. Next, after washing thrice with TBS-T, the membranes were incubated with the secondary antibodies for 1 h at 37 °C. Then, the proteins were detected using a chemiluminescent ECL assay kit.
2.10. Statistical analysis
All experiments were repeated at least three times, and all results were expressed as the mean ± standard deviation. Comparisons between groups were performed using Student's t test. Statistical significances were presented as *P < 0.05, **P < 0.01, or ***P < 0.001. All statistical analyses were performed using GraphPad Prism 6. Flow cytometry data were analyzed by ModFit LT 4.10 software. Western blots were analyzed by Image J.3. Results and discussion
3.1. Cytotoxicity of photoactive CUR on MCF-7 and MCF-7/ADM cells in vitro
There existed no detectable cytotoxicity when MCF-7 and MCF-7/ADM cells were incubated with a series of concentrations (range from 2.5 μM to 15.0 μM) of CUR with dark treatment (see to S1 and S2†). While the survival rate of cells in the presence of CUR-mediated PDT significantly decreased with light irradiation. However, no difference in cell viability between MCF-7 and MCF-7/ADM cells was observed. The results indicated that CUR-mediated PDT significantly enhanced the cytotoxicity of MCF-7 and MCF-7/ADM cells, and the survival rate was positively correlated with the concentration of CUR. Meanwhile, the cytotoxic effects of CUR-mediated PDT of the two cell lines were basically same under the same concentration range and light conditions. Treatment with 7.5 μM CUR for 45 min and irradiation for 5 min (450 nm, 100 mW cm−2) resulted in the inhibition of about 50% of MCF-7 and MCF-7/ADM cells. Consequently, the treatment of 7.5 μM CUR-mediated PDT was used in the subsequent experiment.3.2. Apoptosis in MCF-7 and MCF-7/ADM cells induced by CUR-PDT treatment
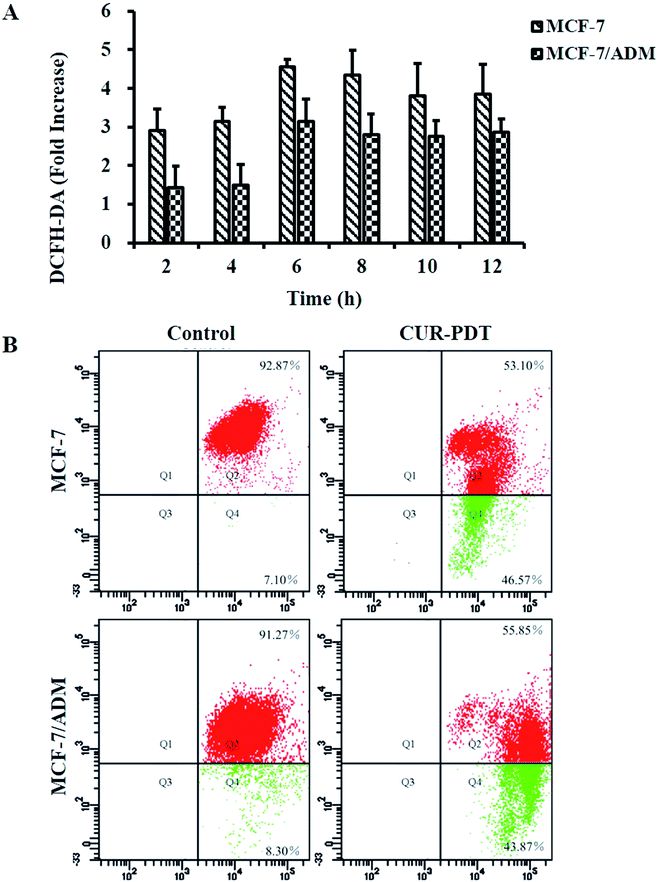
Fig. 2A showed that MCF-7 and MCF-7/ADM cells did not undergo apoptosis when incubated with CUR without light treatment. However, when cells were treated with CUR-PDT, it mainly resulted in early apoptosis. Compared to the control (4.57 ± 0.68%, P < 0.001), the percentage of apoptotic MCF-7 and MCF-7/ADM cells treated with CUR-mediated PDT significantly increased to 56.80 ± 7.64% and 57.07 ± 5.89%, respectively. Both the sensitive and resistant cells showed no significant difference in apoptotic rate after CUR-mediated PDT. Furthermore, the phenotypic characteristics of cells handled with photoactive CUR were also evaluated by IncuCyte ZOOM inspection of overall morphology. In Fig. 2B, cells treated with CUR-mediated PDT showed a marked change in morphology such as rounding and shrinkage.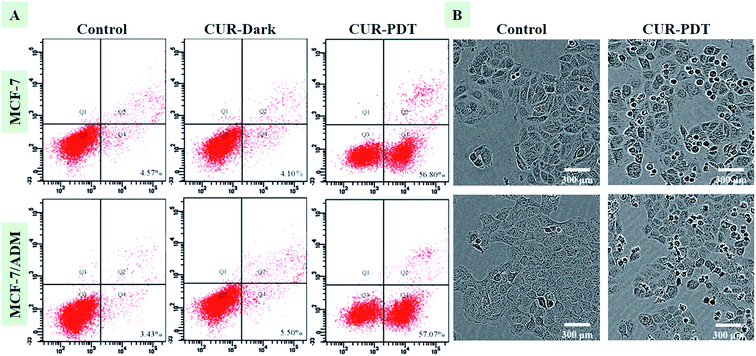 | ||
| Fig. 2 CUR-PDT treatment induces apoptosis (A) and changes in the morphology (B) of MCF-7 and MCF-7/ADM cells after CUR-PDT treatment. | ||
The effect of CUR-mediated PDT on inhibiting the proliferation and promoting apoptosis in MCF-7/ADM cells was similar to MCF-7 cells, indicating that CUR-PDT also had good in vitro anti-tumor activity on drug-resistant cells. Multidrug resistance hinders tumor chemotherapy, and many strategies, including P-gp inhibitors, traditional Chinese medicine reversal agents, RNA interference, nano-medicines, and combinations of delivery strategies have been developed to resolve this problem, but with minimal success.21,22 Here, the results indicated that CUR-mediated PDT may be potentially utilized to overcome multidrug resistance, although the underlying mechanism requires further investigation.
3.3. CUR-PDT treatment disrupts the microfilaments of MCF-7 and MCF-7/ADM cells
CUR-PDT induces a marked change in cellular morphology, as displayed in the rounded morphology of cells. To examine whether the cytoskeleton and nuclei are the targets of CUR-PDT treatment in MCF-7 and MCF-7/ADM cells, TRITC-phalloidin, a specific F-actin fluorescent probe, and DAPI were utilized. Compared to the control, MCF-7 and MCF-7/ADM cells treated CUR-PDT showed a significant change in F-actin microfilaments and nucleus morphology (Fig. 3). F-actin microfilaments displayed a diffused distribution, and the size of the nuclei was smaller than that of control. These findings indicated that CUR-PDT induced apoptosis accompanied by the destruction of F-actin microfilaments and cell nuclei.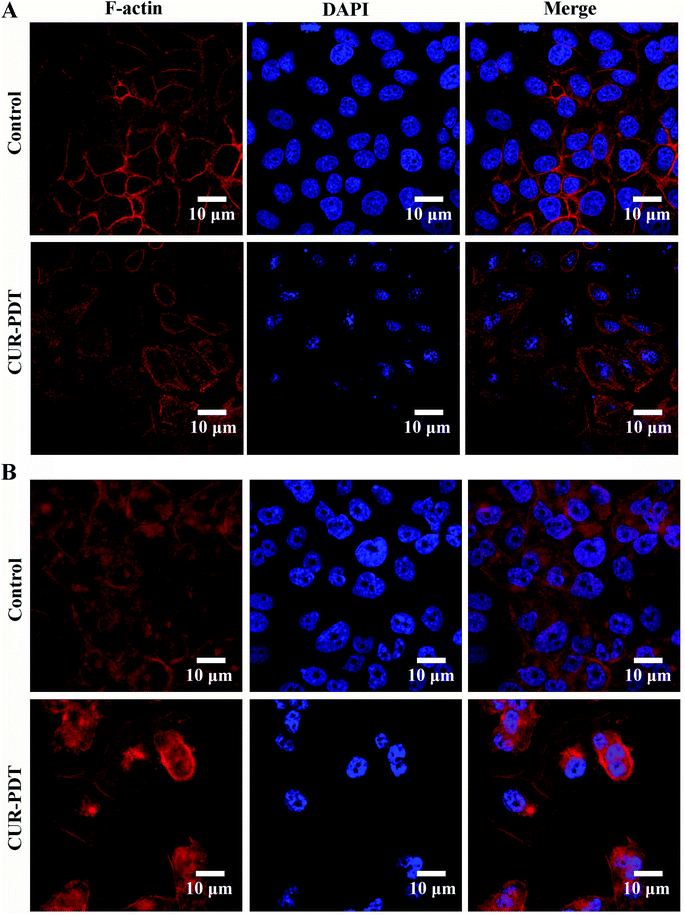 | ||
| Fig. 3 F-actin and cell nuclei structure of MCF-7 and MCF-7/ADM cells. (A) MCF-7 cells. (B) MCF-7/ADM cells. | ||
3.4. Generation of ROS and disruption of MMP in MCF-7 and MCF-7/ADM cells
Intracellular ROS predominantly derived from the mitochondria play an important role in cell survival, proliferation, and death.23,24 PDT induces abundant ROS production that in turn destroys the biomolecular structures of tumor tissue such as nucleic acid, glycoprotein, and phospholipid. Thus, the levels ROS in MCF-7 and MCF-7/ADM cells induced by CUR-mediated PDT were measured (data see to S3†). Fig. 4A shows that ROS generation in MCF-7 and MCF-7/ADM cells significantly increased at a time-dependent manner after treatment with 5 μM CUR and peaked at 6 h. The results indicated that the death of MCF-7 and MCF-7/ADM cells induced by CUR-mediated PDT was ROS-dependent.Variations in MMP are features of cell apoptosis.25 In general, JC-1 agglomerates into J-aggregates emitting red fluorescence in the mitochondrial matrix. When MMP decreases, the J-aggregates decomposes into monomers, and the emitted fluorescence changes from red to green. Thus, the percentage of depolarized cells can be measured through statistical analysis of different fluorescence signals. The ratio of red to green fluorescence normally decreased with the increasingly significant apoptosis, namely, more green fluorescence indicated more apoptosis. Fig. 4B showed that more green fluorescence appeared in both MCF-7 and MCF-7/ADM cells treated with CUR-mediated PDT, and the ratios of red to green fluorescence decreased to 1.14 (13.08 in control) and 1.27 (11.00 in control), respectively. It indicated there existed significant differences in MMP levels between the CUR-mediated PDT group and control (P < 0.001). The results showed that CUR-mediated PDT could significantly reduce the MMP levels of MCF-7 and MCF-7/ADM cells.
3.5. Apoptosis mechanism of multidrug-resistant cells treated with CUR-PDT
Change in the level of MMP often triggers the release of Cyt-c into cytoplasm, subsequently activates the caspase cascade, and ultimately results in apoptosis.26 The release of Cyt-c was consequently examined by western blot analysis. Fig. 5A showed that CUR combined with PDT resulted in a decreased level of Cyt-c in the mitochondria at a dose-dependent manner, while an increased level of Cyt-c in cytoplasm indicated its release from mitochondria into cytosol.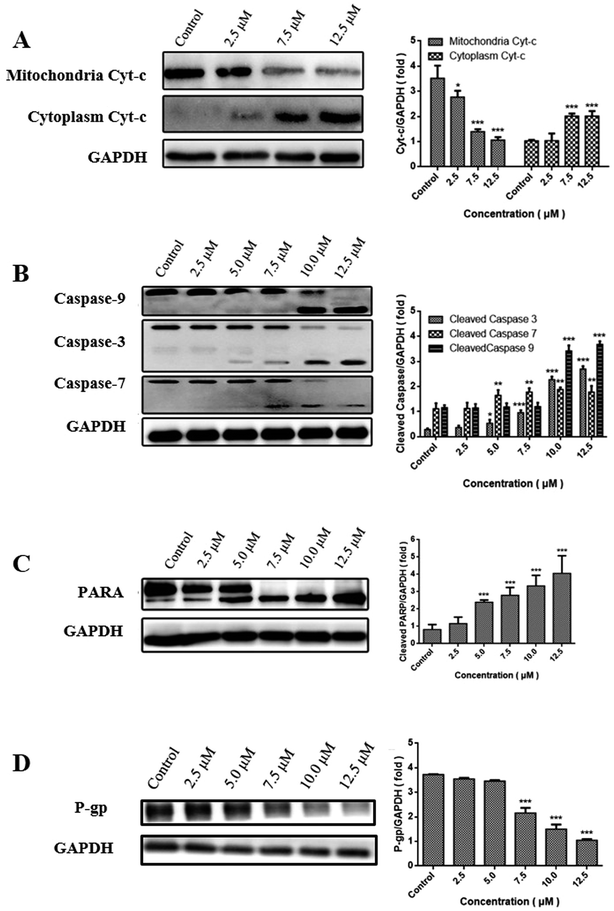 | ||
| Fig. 5 Expression of Cyt-c (A), caspase3/7/9 (B), PARP (C) and (D) in MCF-7/ADM cells in western blot. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. | ||
Caspase family is believed to play a central role in mediating various apoptotic responses.27 In order to confirm whether the apoptotic effects of CUR-PDT are due to the activation of caspase cascades, the protein levels of caspase-3, caspase-7, caspase-9, and PARP were examined. Fig. 5B and C showed that CUR-PDT induced a significant increase in the level of cleaved fragments of caspase-3, caspase-7, caspase-9, and PARP. In brief, these findings indicated that CUR-PDT induced apoptosis by regulating the caspase pathways. (WB results of MCF-7 cells see to S4†).
3.6. Downregulation of P-gp expression reverses multidrug resistance
P-gp is the most studied ABC transporter, which can expel a wide range of anticancer agents from cancer cells, resulting in drug resistance.28 High expression of P-gp can reduce the uptake of API, thus leading to multidrug resistance.29,30To evaluate whether CUR-PDT abolishes multidrug resistance by disrupting the expression of P-gp, the effects of CUR-PDT on P-gp expression in MCF-7/ADM cells were analyzed by western blotting. The results indicated that P-gp expression decreased with increasing CUR-PDT concentrations shown in Fig. 5D. P-gp expression in the control significantly differed (P < 0.001) from those treated with (7.5–12.5 μM) CUR-mediated PDT. The results indicated CUR-mediated PDT could downregulate the expression of P-gp in MCF-7/ADM cells, implying that CUR-mediated PDT might have the potency to overcome multi-drug resistance in tumors by downregulating the expression of P-gp.
4. Conclusions
The studies indicated CUR combined with PDT could effectively produce ROS within tumor cells that in turn induced tumor cell apoptosis and the mechanisms were related to the decreased mitochondrial membrane potential, enhanced membrane permeability, release of Cyt-c from mitochondrial matrix into the cytoplasm, and then the combination of apaf-1 with caspase-9 to form apoptotic complex. The apoptotic complex could further hydrolyze and activate downstream caspase-3/7 that induces cell apoptosis, damages DNA repair enzyme PARP, and causes cell chromatin condensation and nuclear fragmentation, namely accompanied by the disruption of the cytoskeletal structure and other apoptotic characteristics. Besides, CUR-PDT could conquer multidrug resistance in a degree by downregulating the expression of P-gp in MCF-7/ADM cells. In general, CUR-mediated PDT could achieve the reverse of multidrug resistance in chemotherapy by the promotion of endogenous mitochondrial apoptosis and downregulation of P-gp expression. CUR-mediated PDT maybe provide a promising combination with chemotherapeutic drug to overcome the obstacle-multidrug resistance in cancer treatment.Conflicts of interest
The authors had no conflicts of interest to declare.Abbreviations
| CUR | Curcumin |
| PDT | Photodynamic therapy |
| ROS | Reactive oxygen species |
| FITC | Fluorescein isothiocyanate |
| PI | Phosphatidylsilamide |
| Cyt-c | Cytochrome c |
| P-gp | P-glycoprotein |
| MMP | Mitochondrial membrane potential |
| ADM | Adriamycin |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| JC | 1,5,50,6,60-Tetrachloro–1,10,3,30-tetrethyl benzimidalyl carbocyanine iodide |
| DMSO | Dimethyl sulphoxide |
| DMEM | Dulbecco's modified eagle medium |
| PMSF | Phenyl-methylsulfonyl fluoride |
| PVDF | Polyvinylidene difluoride |
Acknowledgements
This work was supported by the National Natural Science Foundation of China (8140140311), Science and Technology Planning Project of Guangdong Province (2016A020217017) and Guangzhou Municipal Science and Technology Project, China (201904010110).References
- H. Mirzaei, A. Shakeri, B. Rashidi, A. Jalili, Z. Banikazemi and A. Sahebkar, Biomed. Pharmacother., 2017, 85, 102–112 CrossRef CAS PubMed.
- S. Ghosh, S. Banerjee and P. C. Sil, Food Chem. Toxicol., 2015, 83, 111–124 CrossRef CAS PubMed.
- P. Anand, C. Sundaram, S. Jhurani, A. B. Kunnumakkara and B. B. Aggarwal, Cancer Lett., 2008, 267, 133–164 CrossRef CAS PubMed.
- A. Bernd, Phytochem. Rev., 2014, 13, 183–189 CrossRef CAS.
- Q. Xiao, J. Wu, X. Pang, Y. Jiang, P. Wang, A. W. Leung, L. Gao, S. Jiang and C. Xu, Curr. Med. Chem.: Anti-Inflammatory Anti-Allergy Agents, 2018, 25, 839–860 CrossRef CAS PubMed.
- U. Bhattacharyya, B. Kumar, A. Garai, A. Bhattacharyya, A. Kumar, S. Banerjee, P. Kondaiah and A. R. Chakravarty, Inorg. Chem., 2017, 56, 12457–12468 CrossRef CAS PubMed.
- J. Dujic, S. Kippenberger, A. Ramirez-Bosca, J. Diaz-Alperi, J. Bereiter-Hahn, R. Kaufmann, A. Bernd and M. Hofmann, Int. J. Cancer, 2009, 124, 1422–1428 CrossRef CAS.
- A. M. Rkein and D. M. Ozog, Dermatol. Clin., 2014, 32, 415–425 CrossRef CAS.
- M. Ethirajan, Y. Chen, P. Joshi and R. K. Pandey, Chem. Soc. Rev., 2011, 40, 340–362 RSC.
- Y. Y. Tian, L. L. Wang and W. Wang, Laser Phys., 2008, 18, 1119–1123 CrossRef CAS.
- H. K. Koon, A. W. N. Leung, K. K. M. Yue and N. K. Mak, J. Environ. Pathol., Toxicol. Oncol., 2006, 25, 205–215 CrossRef CAS.
- S. Banerjee, P. Prasad, A. Hussain, I. Khan, P. Kondaiah and A. R. Chakravarty, Chem. Commun., 2012, 48, 7702–7704 RSC.
- Z. Jamali, S. M. Hejazi, S. M. Ebrahimi, H. Moradi-Sardareh and M. Paknejad, Photodiagn. Photodyn. Ther., 2018, 21, 50–54 CrossRef CAS PubMed.
- A. Garai, I. Pant, S. Banerjee, B. Banik, K. Kondaiah and A. R. Chakravarty, Inorg. Chem., 2016, 55, 6027–6035 CrossRef CAS.
- S. Banerjee, P. Prasad, I. Khan, A. Hussain, P. Kondaiah and A. R. Chakravarty, Z. Anorg. Allg. Chem., 2014, 640, 1195–1204 CrossRef CAS.
- K. Beyer, F. Nikfarjam, M. Butting, M. Meissner, A. König, A. Ramirez Bosca, R. Kaufmann, D. Heidemann, A. Bernd, S. Kippenberger and N. Zölle, J. Cancer, 2017, 8, 1271–1283 CrossRef.
- H. Z. Lee, W. H. Yang, M. J. Hour, C. Y. Wu, W. H. Peng, B. Y. Bao, P. H. Han and D. T. Bau, Eur. J. Pharmacol., 2010, 648, 50–58 CrossRef CAS.
- A. R. Gliga, S. Skoglund, I. Odnevall Wallinder, B. Fadeel and H. L. Karlsson, Part. Fibre Toxicol., 2014, 11, 2–17 CrossRef.
- M. P. Liu, M. Liao, C. Dai, J. F. Chen, C. J. Yang, M. Liu, Z. G. Chen and M. C. Yao, Sci. Rep., 2016, 6, 34245 CrossRef CAS.
- K. Trudeau, T. Muto and S. Roy, Invest. Ophthalmol. Visual Sci., 2012, 53, 6675–6680 CrossRef CAS.
- G. Qi, C. Hongyan, Q. Xianghui, L. Huikai, Y. Peizhi, W. Zhiguo, W. Danqiao and S. Mingyu, Anti-Cancer Agents Med. Chem., 2017, 17, 1466–1476 Search PubMed.
- S. Mollazadeh, A. Sahebkar, F. Hadizadeh, J. Behravan and S. Arabzadeh, Life Sci., 2018, 214, 118–123 CrossRef CAS PubMed.
- C. Gorrini, I. S. Harris and T. W. Mak, Nat. Rev. Drug Discovery, 2013, 12, 931–947 CrossRef CAS.
- D. Trachootham, J. Alexandre and P. Huang, Nat. Rev. Drug Discovery, 2019, 8, 579–591 CrossRef PubMed.
- Y. Ma, J. Zhang, Q. Zhang, P. Chen, J. Song, S. Yu, H. Liu, F. Liu, C. Song and D. Yang, Biochem. Biophys. Res. Commun., 2014, 448, 8–14 CrossRef CAS PubMed.
- S. Elmore, Toxicol. Pathol., 2007, 35, 495–516 CrossRef CAS PubMed.
- O. Julien and J. A. Wells, Cell Death Differ., 2017, 24, 1380–1389 CrossRef CAS PubMed.
- R. J. Kathawala, P. Gupta, C. R. Ashby and Z. S. Chen, Drug Resist. Updates, 2015, 18, 1–17 CrossRef.
- J. Shen, Q. Wang, Q. Hu, Y. Li, G. Tang and P. K. Chu, Biomaterials, 2014, 35, 8621–8634 CrossRef CAS.
- S. Li, M. Gao, Z. Li, S. Li, M. Gao, Z. Li, L. Song, X. Gao, J. Han, F. Wang, Y. Chen, W. Li and J. Yang, Front. Biosci., 2018, 10, 461–468 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09996d |
| ‡ Ying Li and Pei Xiao share first authorship of this article. |
| This journal is © The Royal Society of Chemistry 2020 |