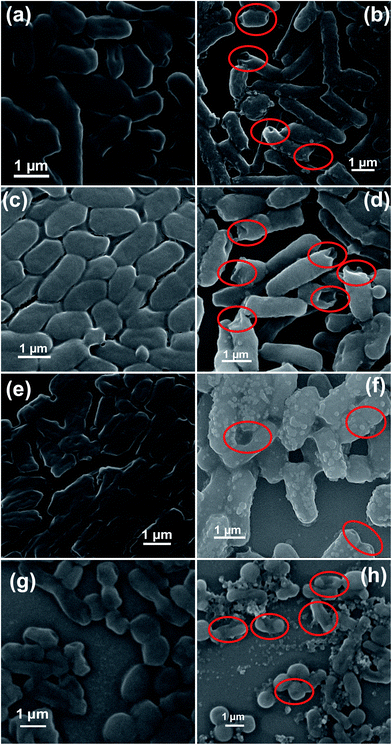
Open Access Article
Open Access ArticleAntibacterial potential of Ni-doped zinc oxide nanostructure: comparatively more effective against Gram-negative bacteria including multi-drug resistant strains
Atanu Naskar†
 ,
Sohee Lee†
,
Sohee Lee† and
Kwang-sun Kim
and
Kwang-sun Kim *
*
Department of Chemistry and Chemistry Institute for Functional Materials, Pusan National University, Busan 46241, South Korea. E-mail: kwangsun.kim@pusan.ac.kr; Tel: +82-51-510-2241
First published on 8th January 2020
Abstract
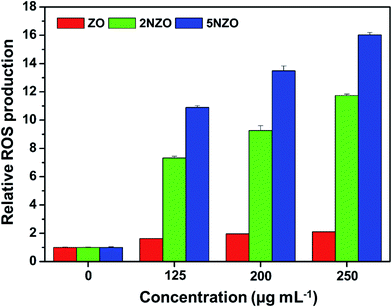
Infections by multidrug-resistant (MDR) bacteria are one of the most threatening concerns for public health. For this purpose, nanomaterials have emerged with great potential for antibacterial activity. In this paper, we report the synthesis of new Ni2+-doped zinc oxide (Ni-ZnO or NZO) nanostructures as targeted antibacterial agents for Gram-negative bacteria. A one-pot low-temperature solution process was used with varying compositions containing 2 or 5% Ni2+ relative to Zn2+, resulting in 2NZO or 5NZO, respectively. X-ray diffractometry, transmission electron microscopy, and X-ray photoelectron spectroscopy were used for material characterization. Further, the antibacterial activity against both Gram-negative [Escherichia coli (E. coli) and Acinetobacter baumannii (A. baumannii) strains including standard, MDR, and clinical isolates associated with mcr-1 gene] and Gram-positive (Staphylococcus aureus and Staphylococcus epidermidis) bacteria were evaluated through analysis of zone of inhibition, minimum inhibitory concentration (MIC), and scanning electron microscopy images. Among the prepared nanostructures, the 5NZO sample showed excellent antibacterial activity against MDR strains of A. baumannii and E. coli. In addition, samples of NZO generated approximately 7 to 16 times more reactive oxygen species (ROS) in E. coli compared to ZnO. Our synthesized nanomaterials have the potential to fight MDR and colistin-resistant Gram-negative bacteria.
1. Introduction
One of the most urgent challenges threatening global public health care systems is infection related to multidrug-resistant (MDR) microorganisms.1 Multidrug resistance is defined as the resistance of microorganisms, and their increasing resistance, against at least two currently administered antimicrobial medications that used to be effective.2 This has a profound impact on public health, as predictions indicate that there will be 10 million annual deaths due to MDR pathogens by 2050, which is higher than that of cancer, unless immediate interventions are developed.3 The grim reality is that human beings will face a time when no antibiotic will act against even a simple bacterial infection if proper action is not taken immediately. At present, some bacterial strains, such as methicillin-resistant Staphylococcus aureus (MRSA) cannot be treated with the last resort antibiotic vancomycin, as a result of the evolution of vancomycin-resistant Staphylococcus aureus (VRSA) strains.4 In addition, the rate of antibiotic development is far less than the attainment of resistance by MDR bacteria.5 Therefore, it is clear that alternatives to traditional antibiotics, such as antibacterial materials, need to be swiftly developed or discovered to fight against MDR bacterial cells.Among many different MDR bacterial cells that require immediate intervention, Gram-negative Acinetobacter baumannii (A. baumannii) is one of 12 families of priority pathogenic bacteria identified by the World Health Organization (WHO) as a major threat to the clinical treatment of infections in human beings.6 This pathogen is mainly found in hospital settings, where critically ill hospitalized patients are exposed through cracks in the skin and respiratory tract.7 In addition, Gram-negative Escherichia coli (E. coli) is the main culprit behind urinary tract infections (UTIs)8 and water borne diseases.9 An epidemiological survey on cystitis, the inflammation of the bladder usually caused by UTIs, showed that more than 10% of E. coli isolates are MDR.10 According to the WHO, unsafe water is reported as the main reason behind 80% of all diseases,11 with E. coli being a member of the fecal coliforms that frequently contaminate drinking water.9 Therefore, alternative antibacterial materials are immediately needed to counter MDR strains of both E. coli and A. baumannii strains.
Alternative approaches to conventional antibiotics to fight bacterial infections include combinations of antibiotics,1 antibacterial antibodies,12 bacteriophages,13 photothermal therapy,14 and nanomaterials.15 Among these alternatives, nanomaterials composed of Ag,16 Au,17 ZnO,18 CuO,19 TiO2,20 graphene,21 and black phosphorus22 are increasingly attracting attention for antibacterial development due to their favorable physiochemical properties. Zinc oxide (ZnO) is one of the most promising nanomaterials as an antibacterial agent due to its effective antibacterial activity and selectivity for bacterial cells, while being minimally toxic to human cells; it is one of the five zinc compounds approved by the U.S. Food and Drug Administration (21CFR182.8991) as a safe antibacterial agent.18 Due to its biocompatible properties, ZnO has already been used as an antibacterial agent in the preparation of various commercial products such as medical equipment, food packaging materials, and cosmetics.23
Moreover, there are several reports describing the particle size-dependent antibacterial activity of ZnO nanoparticles (NPs), where antibacterial activity is inversely correlated with particle size.24 Hence, the particle size of ZnO NPs needs to be reduced to achieve better antibacterial activity. In this regard, doping ZnO with trace amounts of metal ions has shown significant changes in the physical and chemical properties of ZnO NPs, including particle size.25 Several reports are available regarding the doping of ZnO with metal ions of Ni, Co, Fe, Mn, Cr, Al, and Ag to change its properties for photocatalysis, sensors, and antibacterial activity.26–32 Among the above dopants, Ni2+ is favorable to incorporate into the ZnO lattice due to its same valence state (+2) and similar ionic radii (Ni2+, 0.69 Å; Zn2+, 0.74 Å). Moreover, various reports regarding the antibacterial activity of metal ion-doped ZnO, Ag,27,28 Co,29,30 and Cr31,32 have been reported. For instance, the work reported by Dias et al.27 has shown the antibacterial activity of Ag doped ZnO nanoparticles against S. mutans. Similar work was also shown with E. coli by Karunakaran et al.28 Moreover, the work by Vijayalakshmi et al.31 has shown the antibacterial activity of Cr doped ZnO nanorods against E. coli and S. aureus. Meanwhile, the use of Ni2+ as a dopant for ZnO with regard to antibacterial activity against MDR bacteria is sorely lacking.
Despite the excellent antibacterial property possessed by Ni2+-doped ZnO (NZO) NPs/nanostructures, very few reports are available regarding its against MDR pathogens. Therefore, the objective of the present work is to synthesize an effective nanomaterial of NZO NPs and nanostructures as a potential alternative to traditional antibiotics for the fight against MDR bacteria. Herein, we report a one-pot synthesis of NZO NPs prepared using a low-temperature solution process and their antibacterial activity compared to ZO NPs/nanostructures against the Gram-negative bacteria, E. coli and A. baumannii including their MDR strains [E. coli 1368, BAA2452, and 1A626 and A. baumannii (12001)] and colistin-resistant clinical isolates harboring mcr-1 gene.
2. Material and methods
2.1 Synthesis of Ni2+-doped ZnO (NZO) nanostructures
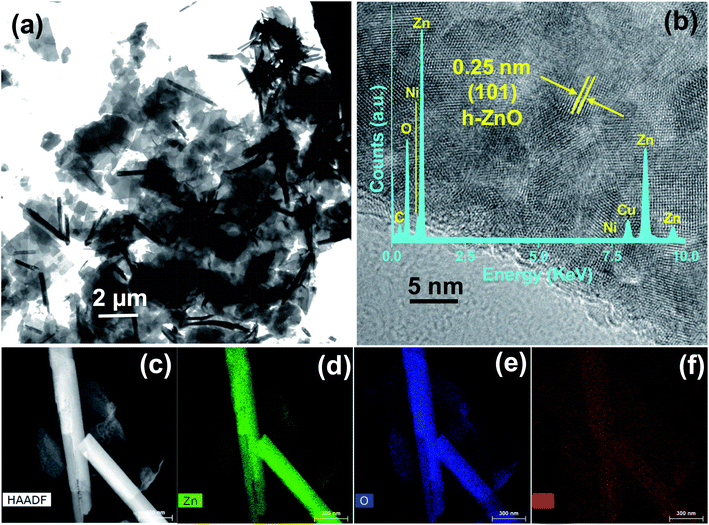
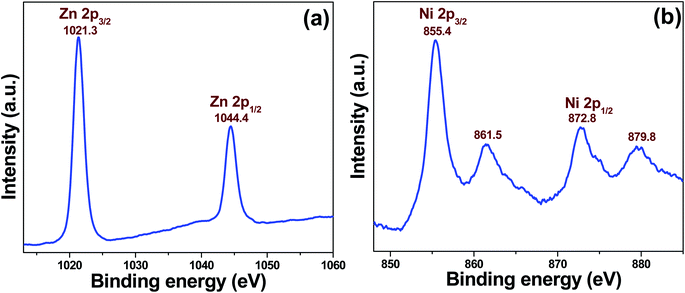
Briefly, 0.1 M of zinc nitrate hexahydrate [Zn(NO3)2·6H2O, Merck] and nickel sulfate hexahydrate (NiSO4·6H2O, Sigma-Aldrich, 99%) are used to prepare 0, 2, and 5 atomic percent (at%) (with respect to Zn2+) ZnO-based NPs. The mixtures were uniformly dispersed in a beaker containing 50 mL of deionized water with continuous stirring for 30 min at room temperature. While continuing to stir, 1 mL of hydrazine hydrate (N2H4·H2O, Merck, 99–100%) was added dropwise to the reaction mixture. After complete addition, the mixture was ultrasonicated for 10 min at 60 °C in a water bath ultrasonicator until a slightly pink precipitate was observed at the bottom of the reaction flask. Further dropwise addition of hydrazine hydrate and ultrasonication was repeated until the pH of the medium reached 8. Afterward, the precipitate was separated by centrifugation. Deionized water and ethanol were then used to wash the product 3–4 times. Finally, the product was dried in an oven at ∼60 °C for 24 h. The synthesized ZnO-based NPs containing 0, 2, and 5 at% Ni are referred to as ZO, 2NZO, and 5NZO, respectively.2.2 Characterizations
3. Results and discussion
3.1 Material properties
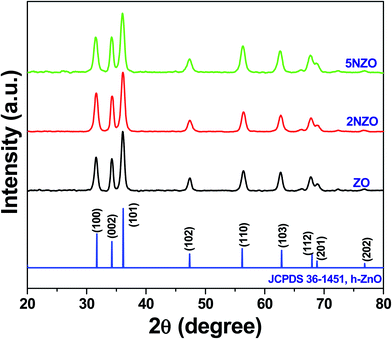
The average crystallite size (D) of ZnO crystallites was measured along the (101) crystal plane using the Debye–Scherrer eqn (1).
D = kλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, θ,
| (1) |
The measured D values of ZO, 2NZO, and 5NZO after solving eqn (1) were approximately 27, 25, and 21 nm, respectively. Therefore, ZnO crystallite size decreases with increasing Ni2+-doping concentration in the precursor solutions. The gradual decrease in D with increasing Ni2+ concentration can be attributed to substitution with Ni2+ ions in the ZnO crystal lattice.26 It appears that the surface area increases due to decreasing ZnO crystallite size, potentially resulting in better antibacterial activity.
3.2 Antibacterial activity
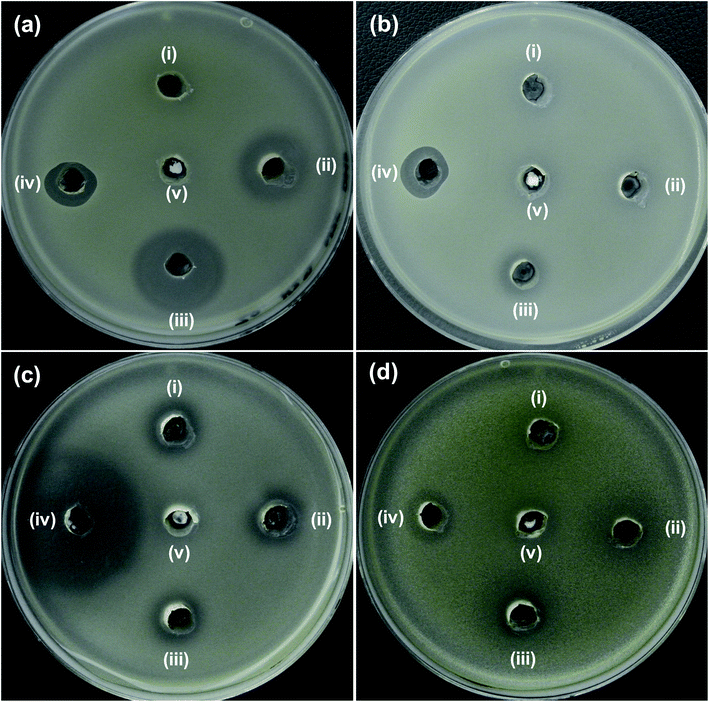 | ||
| Fig. 4 Zone of inhibition (ZOI) of ZnO samples against (a) E. coli, (b) A. baumannii, (c) S. aureus and (d) S. epidermidis. Diameter of ZOI is also displayed in the Table 1 (average from n = 3). | ||
| Bacteria cells | Zone of inhibition (mm) | ||||
|---|---|---|---|---|---|
| (i) ZO | (ii) 2NZO | (iii) 5NZO | (iv) Antibiotics | (v) DMSO | |
| a Antibiotics, aPolymyxin B or bAmpicillin was used as a control for Gram-negative or -positive bacteria, respectively. | |||||
| (a) E. coli ATCC25922 | N.D. | 18 | 24 | 14a | N.D. |
| (b) A. baumannii ATCC19606 | N.D. | N.D. | 12 | 13a | N.D. |
| (c) S. aureus ATCC25923 | 13 | 14 | 14 | 38b | N.D. |
| (d) S. epidermidis ATCC12228 | N.D. | 11 | 14 | 14b | N.D. |
Additionally, the effectiveness of NZO NPs was evaluated for antibacterial activity toward the Gram-positive bacteria, S. aureus (Fig. 4c) and S. epidermidis (Fig. 4d). Although NZO NPs are active against Gram-positive bacteria (ZOI is 14 mm for 5NZO against both S. aureus and S. epidermidis) the activity of such NPs is much less than ampicillin and that for Gram-negative bacteria (ZOI is 24 mm for 5NZO against E. coli) (Table 1). This indicates that Gram-negative bacteria is better target for 2NZO and 5NZO. Therefore, further experiments focused on the MDR strains of the Gram-negative bacteria, E. coli and A. baumannii were performed.
Dependency on the concentration of the synthesized samples was also taken into consideration regarding antibacterial activity. Fig. 5 shows the ZOI of (a) ZO, (b) 2NZO and (c) 5NZO against E. coli at different concentrations. Diameter of ZOI is also displayed in the Table 2. As clearly seen form the Fig. 5 and Table 2 is that 5 mg mL−1 is the most active concentration of 5NZO samples to kill bacterial cells. Therefore, all samples were used with 5 mg mL−1 for future experiments.
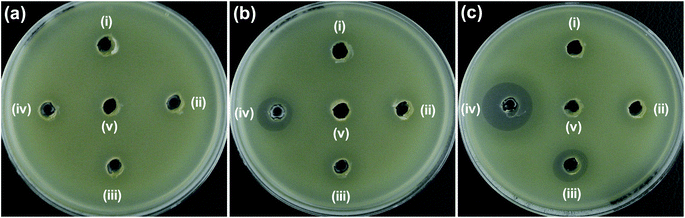 | ||
| Fig. 5 Zone of inhibition (ZOI) of (a) ZO, (b) 2NZO and (c) 5NZO samples against E. coli at different concentrations. Diameter of ZOI is also displayed in the Table 2 (average from n = 3). | ||
| Samples | Zone of inhibition (mm) | |||
|---|---|---|---|---|
| (i) 0.5 mg mL−1 | (ii) 1.25 mg mL−1 | (iii) 2.5 mg mL−1 | (iv) 5 mg mL−1 | |
| a DMSO (v) was used as negative control in all Petri dishes. | ||||
| (a) ZnO | N.D. | N.D. | N.D. | N.D. |
| (b) 2NZO | N.D. | N.D. | N.D. | 15 |
| (c) 5NZO | N.D. | N.D. | 15 | 20 |
The antibacterial activity of the synthesized NPs was investigated with regard to MDR strains of E. coli (Fig. 6a–c) and A. baumannii (Fig. 6d). As seen in Fig. 6, the 5NZO exhibit excellent antibacterial activity against MDR strains of E. coli, as the ZOI for 5NZO are 23, 17, and 21 mm against E. coli 1368, BAA2452, and 1A626, respectively (Table 3). Moreover, 5NZO showed a ZOI of 25 mm for an MDR strain of A. baumannii (12001), indicating that 5NZO is good activity toward Gram-negative bacteria and their MDR strains. Furthermore, 5NZO display comparatively better antibacterial activity than the other NPs (ZO and 2NZO) as the larger ZOI for 5NZO (Fig. 6). This high antibacterial activity of 5ZNO against Gram-negative bacteria and their MDR strains can be attributed to both an increase in Ni2+-doping concentration along with a decrease in crystallite size.24
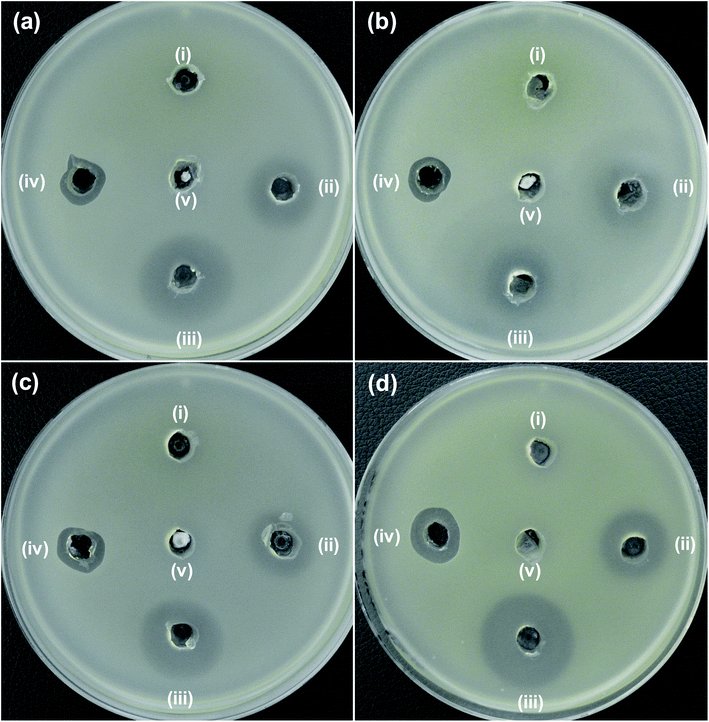 | ||
| Fig. 6 Zone of inhibition (ZOI) of ZnO samples against MDR strains of E. coli (a) 1368, (b) BAA2452, and (c) 1A626 and an MDR strain of A. baumannii (d) 12001. Diameter of ZOI is also displayed in the Table 3 (average from n = 3). | ||
| Bacterial cells | Zone of inhibition (mm) | ||||
|---|---|---|---|---|---|
| (i) ZO | (ii) 2NZO | (iii) 5NZO | (iv) Polymyxin B | (v) DMSO | |
| (a) E. coli 1368 | N.D. | 18 | 23 | 13 | N.D. |
| (b) E. coli BAA2452 | N.D. | 14 | 17 | 12 | N.D. |
| (c) E. coli 1A626 | N.D. | 18 | 21 | 12 | N.D. |
| (d) A. baumannii 12001 | N.D. | 18 | 25 | 15 | N.D. |
Additionally, clinical isolate of E. coli harboring mcr-1 gene was also tested with 5NZO samples to evaluate its efficacy. ZOI (Fig. 7) and its corresponding diameter values (Table 4) showed that 5NZO sample can be an excellent antibacterial agent even against the clinical isolate of E. coli harboring mcr-1 gene with 22 mm and 24 mm of ZOI for NCCP 16283 (Fig. 7a), NCCP 16284 (Fig. 7b) of E. coli, respectively.
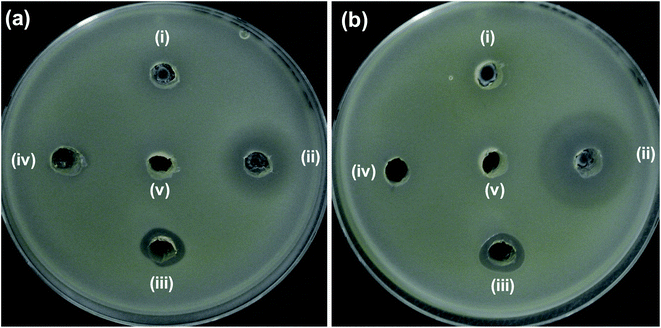 | ||
| Fig. 7 Zone of inhibition (ZOI) of ZnO samples against mcr-1 strains of E. coli (a) NCCP 16283 and (b) NCCP 16284. Diameter of ZOI is also displayed in the Table 4 (average from n = 3). | ||
| Bacterial cells | Zone of inhibition (mm) | ||||
|---|---|---|---|---|---|
| (i) ZO | (ii) 5NZO | (iii) Polymyxin B | (iv) Erythromycin | (v) DMSO | |
| (a) NCCP 16283 | N.D. | 22 | 13 | N.D. | N.D. |
| (b) NCCP 16284 | N.D. | 24 | 13 | N.D. | N.D. |
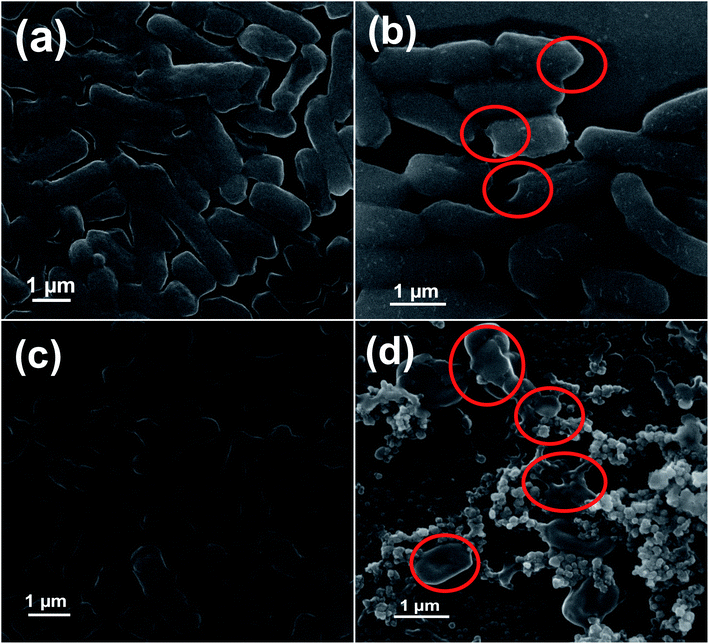
We have also evaluated the MIC value of 5NZO sample for further approval of its antibacterial activity against Gram-negative bacterial cells and showed in Table 5. It can be clearly seen from the table that 5NZO sample not only act on wild type of bacteria[(a) E. coli (ATCC25922) and (b) A. baumannii (ATCC19606)] but also on their MDR strains [(c) E. coli 1368, (d) E. coli BAA2452, (e) E. coli 1A626, and (f) A. baumannii 12001]. Therefore, the synthesized 5NZO nanorods can be an effective nanoweapon against MDR strains of Gram-negative bacterial cells.
| Bacterial cells | Minimum inhibitory concentration (μg mL−1) | ||
|---|---|---|---|
| ZO | 5NZO | ||
| (a) E. coli ATCC25922 | >250 | 200 | |
| (b) A. baumannii ATCC19606 | >250 | 200 | |
| MDR strains | (c) E. coli 1368 | >250 | 200 |
| (d) E. coli BAA2452 | >250 | 200 | |
| (e) E. coli 1A626 | >250 | 200 | |
| (f) A. baumannii 12001 | >250 | 125 | |
4. Conclusions
In summary, this work demonstrates the low-temperature solution synthesis of Ni2+-doped ZnO NPs and the resulting selective antibacterial activity against Gram-negative bacteria, E. coli and A. baumannii including their MDR strains and mcr-1 associated colistin resistant E. coli strains. The 5% Ni2+-doped ZnO (5NZO) NPs even showed comparatively better antibacterial activity at least by zone of inhibition method against Gram-negative bacteria compared to polymyxin B, a last resort peptide antibiotic that is currently used clinically as colistin. Moreover, NZOs produced more ROS compared to ZO. Because the method shown here is a cost-effective and includes simple synthetic strategy, these synthesized NPs can be used as a platform for the development of metal oxide nanomaterials for biomedical applications, in particular the design of nanoweapons against the ever-increasing public health threat of MDR bacteria.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) [grant number 2018R1D1A1B07040941]. We are thankful to Prof. Hyun Deok Yoo at the Department of Chemistry, Pusan National University, for use of the X-ray diffractometer.References
- A. Naskar and K. S. Kim, Microorganisms, 2019, 7, 356 CrossRef PubMed.
- V. A. Kumar and S. Khan, Indian J. Med. Res., 2015, 141, 491–492 CrossRef PubMed.
- J. O'Neill, 2016, available online, https://amr-review.org/sites/default/files/160525_Finalpaper_withcover.pdf, accessed on 2 October 2019.
- W. A. McGuinness, N. Malachowa and F. R. DeLeo, Yale J. Biol. Med., 2017, 90, 269–281 CAS.
- B. Li and T. J. Webster, J. Orthop. Res., 2018, 36, 22–32 Search PubMed.
- World Health Organization, WHO publishes list of bacteria for which new antibiotics are urgently needed, 2017, available online, https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-ofbacteria-for-which-new-antibiotics-are-urgently-needed, accessed on 24 September 2019 Search PubMed.
- V. Tiwari, N. Mishra, K. Gadani, P. S. Solanki, N. A. Shah and M. Tiwari, Front. Microbiol., 2018, 9, 1218 CrossRef.
- N. T. Mutters, A. Mampel, R. Kropidlowski, K. Biehler, F. Günther, I. Bălu, V. Malek and U. Frank, Fitoterapia, 2018, 129, 237–240 CrossRef CAS PubMed.
- S. T. Odonkor and K. K. Addo, Int. J. Microbiol., 2018, 2018, 7204013 Search PubMed.
- G. C. Schito, K. G. Naber, H. Botto, J. Palou, T. Mazzei, L. Gualco and A. Marchese, Int. J. Antimicrob. Agents, 2009, 34, 407–413 CrossRef CAS PubMed.
- H. Ellis and E. Schoenberger, PLoS One, 2017, 12, e0170451 CrossRef PubMed.
- A. DiGiandomenico and B. R. Sellman, Curr. Opin. Microbiol., 2015, 27, 78–85 CrossRef CAS PubMed.
- C. Torres-Barceló, Emerg. Microb. Infect., 2018, 7, 168 Search PubMed.
- J.-W. Xu, K. Yao and Z.-K. Xu, Nanoscale, 2019, 11, 8680–8691 RSC.
- A. Gupta, S. Mumtaz, C.-H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC.
- S. Tang and J. Zheng, Adv. Healthc. Mater., 2018, 7, 1701503 CrossRef PubMed.
- X. Li, S. M. Robinson, A. Gupta, K. Saha, Z. Jiang, D. F. Moyano, A. Sahar, M. A. Riley and V. M. Rotello, ACS Nano, 2014, 8, 10682–10686 CrossRef CAS PubMed.
- A. Naskar, S. Bera, R. Bhattacharya, P. Saha, S. S. Roy, T. Sen and S. Jana, RSC Adv., 2016, 6, 88751–88761 RSC.
- J. Chen, S. Mao, Z. Xu and W. Ding, RSC Adv., 2019, 9, 3788–3799 RSC.
- T. Verdier, M. Coutand, A. Bertron and C. Roques, Coatings, 2014, 4, 670–686 CrossRef.
- A. Naskar, H. Khan, R. Sarkar, S. Kumar, D. Halder and S. Jana, Mater. Sci. Eng. C, 2018, 91, 743–753 CrossRef CAS PubMed.
- Z. Xiong, X. Zhang, S. Zhang, L. Lei, W. Ma, D. Li, W. Wang, Q. Zhao and B. Xing, Ecotoxicol. Environ. Saf., 2018, 161, 507–514 CrossRef CAS PubMed.
- A. Sirelkhatim, S. Mahmud, A. Seeni, N. H. M. Kaus, L. C. Ann, S. K. M. Bakhori, H. Hasan and D. Mohamad, Nano-Micro Lett., 2015, 7, 219–242 CrossRef CAS PubMed.
- K. R. Raghupathi, R. T. Koodali and A. C. Manna, Langmuir, 2011, 27, 4020–4028 CrossRef CAS PubMed.
- S. B. Rana and R. P. P. Singh, J. Mater. Sci.: Mater. Electron., 2016, 27, 9346–9355 CrossRef CAS.
- R. S. Kumar, S. H. S. Dananjaya, M. D. Zoysa and M. Yang, RSC Adv., 2016, 6, 108468–108476 RSC.
- H. B. Dias, M. I. B. Bernardi, V. S. Marangoni, A. C. de Abreu Bernardi, A. N. de Souza Rastelli and A. C. Hernandes, Mater. Sci. Eng. C, 2019, 96, 391–401 CrossRef CAS PubMed.
- C. Karunakaran, V. Rajeswari and P. Gomathisankar, Mater. Sci. Semicond. Process., 2011, 14, 133–138 CrossRef CAS.
- G. Iqbal, S. Faisal, S. Khan, D. F. Shams and A. Nadhman, J. Photochem. Photobiol., B, 2019, 192, 141–146 CrossRef CAS PubMed.
- A. Naskar, H. Khan and S. Jana, Advanced Nano-Bio-Materals and Devices, 2017, 1, 182–190 Search PubMed.
- K. Vijayalakshmi and D. Sivaraj, RSC Adv., 2015, 5, 68461–68469 RSC.
- A. H. Shah, M. B. Ahamed, D. Neena, F. Mohmed and A. Iqbal, J. Alloys Compd., 2014, 606, 164–170 CrossRef CAS.
- K. S. Ong, Y. L. Cheow and S. M. Lee, J. Adv. Res., 2017, 8, 393–398 CrossRef CAS PubMed.
- T. Zhou, M. Hu, J. He, R. Xie, C. An, C. Li and J. Luo, CrystEngComm, 2019, 21, 5526–5532 RSC.
- A. Naskar, S. Bera, R. Bhattacharya, S. S. Roy and S. Jana, J. Alloys Compd., 2018, 734, 66–74 CrossRef CAS.
- G. Vijayaprasath, R. Murugan, S. Palanisamy, N. M. Prabhu, T. Mahalingam, Y. Hayakawa and G. Ravi, Mater. Res. Bull., 2016, 76, 48–61 CrossRef CAS.
- R. S. Ganesh, E. Durgadevi, M. Navaneethan, V. L. Patil, S. Ponnusamy, C. Muthamizhchelvan, S. Kawasaki, P. S. Patil and Y. Hayakawa, Chem. Phys. Lett., 2017, 689, 92–99 CrossRef CAS.
- Z. Y. Dong, M. P. N. Rao, M. Xiao, H. F. Wang, W. N. Hozzein, W. Chen and W. J. Li, Front. Microbiol., 2017, 8, 1090 CrossRef PubMed.
Footnote |
| † Both authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2020 |