Open Access Article
Open Access ArticleQSAR modeling of the toxicity classification of superparamagnetic iron oxide nanoparticles (SPIONs) in stem-cell monitoring applications: an integrated study from data curation to model development†
Marianna I. Kotzabasaki *,
Iason Sotiropoulos
*,
Iason Sotiropoulos and
Haralambos Sarimveis
and
Haralambos Sarimveis *
*
School of Chemical Engineering, National Technical University of Athens, 9 Heroon Polytechneiou Street, Zografou Campus, 15780, Athens, Greece. E-mail: mariannako@chemeng.ntua.gr; jasonsoti1@gmail.com; hsarimv@central.ntua.gr; Fax: +302107723138; Tel: +302107723236 Tel: +306936396688 Tel: +302107723237
First published on 3rd February 2020
Abstract
The use of in silico approaches for the prediction of biomedical properties of nano-biomaterials (NBMs) can play a significant role in guiding and reducing wetlab experiments. Computational methods, such as data mining and machine learning techniques, can increase the efficiency and reduce the time and cost required for hazard and risk assesment and for designing new safer NBMs. A major obstacle in developing accurate and well-validated in silico models such as Nano Quantitative Structure–Activity Relationships (Nano-QSARs) is that although the volume of data published in the literature is increasing, the data are fragmented in many different publications and are not sufficiently curated for modelling purposes. Moreover, NBMs exhibit high complexity and heterogeneity in their structures, making data collection and curation and QSAR model development more challenging compared to traditional small molecules. The aim of this study was to construct and fully validate a Nano-QSAR model for the prediction of toxicological properties of superparamagnetic iron oxide nanoparticles (SPIONs), focusing on their application as Magnetic Resonance Imaging (MRI) contrast agents for non-invasive stem cell labelling and tracking. To achieve this goal, we first performed an extensive search through the literature for collecting and curating relevant data and we developed a dataset containing both physicochemical and toxicological properties of SPIONs. The data were analysed next, using Automated machine learning (Auto-ML) approaches for optimising the development and validation of nanotoxicity classification QSAR models of SPIONs. Further analysis of relative attribute importances revealed that physicochemical properties such as the size and the magnetic core are the dominant attributes correlated to the toxicity of SPIONs. Our results suggest that as more systematic information from NBM experimental tests becomes available, computational tools could play an important role in supporting the safety-by-design (SbD) concept in regenerative medicine and disease therapeutics.
Introduction
Stem-cell therapy is the use of stem cells to treat or prevent a disease for which traditional medical therapies are insufficient, bringing new hope in regenerative medicine.1 The most widely used stem-cell therapy is the transplantation of blood stem cells to treat diseases of the blood and immune system. According to the US National Marrow Donor Program (NMDP), there is a full list of diseases that are treated by hematopoietic stem cell transplantation (HSCT). In Europe, more than 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 patients are treated with blood stem cells every year.2 The six classes of stem cells, which are, embryonic stem cells (ESCs), bone marrow stem cells (BMSCs), mesenchymal stem cells (MSCs), induced pluripotent stem cells (iPSCs), umbilical cord stem cells (UCSCs) and tissue specific progenitor stem cells (TSPSCs), are widely used in regenerative medicine and disease therapeutics.3 The main areas of ongoing or completed clinical trials concern macular degeneration, neurological conditions, such as Parkinson's disease, Huntington's disease and Motor Neuron Disease (MND), diabetes, spinal cord injury, and myocardial infarction.2
000 patients are treated with blood stem cells every year.2 The six classes of stem cells, which are, embryonic stem cells (ESCs), bone marrow stem cells (BMSCs), mesenchymal stem cells (MSCs), induced pluripotent stem cells (iPSCs), umbilical cord stem cells (UCSCs) and tissue specific progenitor stem cells (TSPSCs), are widely used in regenerative medicine and disease therapeutics.3 The main areas of ongoing or completed clinical trials concern macular degeneration, neurological conditions, such as Parkinson's disease, Huntington's disease and Motor Neuron Disease (MND), diabetes, spinal cord injury, and myocardial infarction.2
Over the last few decades nano-biomaterials (NBMs) have very wide usage in regenerative medicine and disease therapeutics. NBMs in order to be used in regenerative medicine must fulfil some important requirements. They must be biodegradable, biocompatible – nontoxic to the cells, effective at therapeutic doses and chemically stable in physiological conditions. Ideally, they should maintain their physical properties after surface functionalization, to have regulatory approval and not to affect stem cells characteristics.4
Among all NBMs, magnetic nanoparticles (MNPs) are the most promising candidates as molecular contrast nanoprobes in magnetic resonance imaging (MRI), due to their excellent biocompatibility and low toxicity.5
The use of MNPs based on iron oxide for MRI is receiving significant attention in regenerative medicine for stem cells labelling, tracking and activation in vitro and in vivo.4,5 Iron oxide based nanoprobes for use in MRI are desired to exhibit superparamagnetic properties.4 Superparamagnetic iron oxide nanoparticles (SPIONs) are composed of either maghemite (γ-Fe2O3) or magnetite (Fe3O4) core ranging in size from 10 to 100 nm. Their surface can be modified by organic and/or inorganic coating material.6 Owing to their high sensitivity to external magnetic fields, SPIONs have been regarded as novel T2-weighted MRI contrast agents in the last decade, leading to a rapid progress in the field of stem cell labeling and tracking.7 In essence, stem cells internalize SPIONs, thus magnetic properties are transferred to the cells. Labelled cells are then implanted within the body and visualized by MRI. In this way location, migration and the number of stem cells are monitored over time.4
MRI with the use of contrast nanoprobes is regarded as the safest non-invasive, non-ionizing method for stem cell studies. However, commercially available SPIONs (e.g. Feridex® and Revosit®) combined with potentially toxic transfection agents (TAs) (e.g. Superfect, poly(L-lysine) (PLL)) may cause severe effects to living bodies.7 Therefore, there is the need of designing novel SPIONs with specific surface coatings or functional groups for the effective cellular internalization without the use of TAs, thus reducing the potential nanotoxicity.
SPIONs' toxicity depends on many factors, such as, size, surface chemistry, charge, dose and exposure duration.8 Developing MNPs with suitable coatings make the particles less toxic, however a variety of in vitro and long-term in vivo studies have to be conducted to evaluate the toxicological profiles of SPIONs in living organs. These evaluations are very expensive and time-consuming.
Computational modelling constitutes an alternative approach relying on the systematic analysis of nanotoxicity related data, which can guide the experimental studies and reduce their costs. Nano Quantitative Structure Activity Relationships (nano-QSARs),9 are in silico prediction models, which are based on the idea that the structure of a substance affects its activity and thus similar structures exhibit similar activities. These tools can assist researchers to effectively screen the NBMs with the desired properties before their synthetic route, and perform experiments only on the most promising candidates. Researchers have already developed efficient mathematical (QSAR) models in order to accurately assess the toxicity endpoint of new or untested nanomaterials.10–20 For instance, Byun et al.,11 developed a generalized QSAR model for oxide nanomaterials that is able to determine toxicity based on different biological conditions. In another study, Kim and co-workers20 proposed a Quasi-SMILES-Based nano-QSAR model to predict the cytotoxicity of multiwalled carbonnanotubes to human lung cells. Medintz's group15 meta-analysed a range of published studies focusing on the cellular toxicity of cadmium-containing semiconductor quantum dots. In addition, Jiang et al.,16 have successfully developed robust nano-QSAR models for predicting the cytotoxicity of MeOx NPs to both E. coli and HaCaT cells.
However, to the best of our knowledge, no studies have been published yet for SPIONs-based MRI contrast in stem cells monitoring applications. In this work, we developed first a novel nanosafety dataset of SPIONs, by collecting, assembling and curating data about their physicochemical and toxicological properties as MRI contrast agents for non-invasive stem cell labelling and tracking. Secondly, we developed and validated a nanotoxicity classification QSAR model for predicting cell viability of SPIONs based on physicochemical characteristics, using automated machine learning (Auto-ML).21 This approach allowed us to compare multiple machine learning algorithms in order to produce the best performing model. Finally, we analyzed the most important attributes affecting the toxicity of SPIONs and defined the domain of applicability (DOA)22 of the model. The model was implemented as a ready-to-use web application in the Jaqpot computational platform and is available to the community through the BIORIMA virtual organisation.
Materials and methods
Workflow of model development
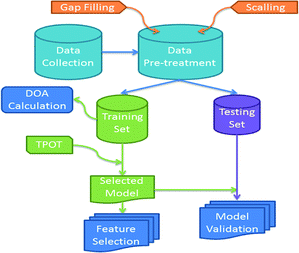
The study followed the workflow depicted below in Fig. 1.The physicochemical properties included in our data set relative to the nanotoxicity of SPION-based MRI contrast agents in stem-cell based therapies are shown in Table 1. The endpoint to be predicted was cell viability. Two classes were defined for the output variable by defining a cut-off value.
| Data attributes | Explanation | Value |
|---|---|---|
| Magnetic core | Chemical formula maghemite | γ-Fe2O3/Fe3O4 |
| Zeta potential (mV) | Electrokinetic potential in colloidal dispersions | (−42) to (+40) |
| Size (nm) | Particle size (nm) | 2–150 |
| B0 (T) | Magnetic field strength | 0.47–4.7 |
| Fe/cell (pg) | Iron concentration per cell (pg) = the amount of SPIONs internalized inside the cells | 2.15–1459 |
| Relaxivity, r2 (s−1 mM−1) | Relaxation rate as a function of concentration | 27.6–549 |
Data were normalized by subtracting from each feature the lowest value and dividing it with the min–max difference (min–max scaling). A hierarchical clustering approach was used to fill missing values of one attribute. The full dataset comprising 16 observations was then split into training (11 samples) and validation (5 samples) sets using the Kennard–Stone algorithm.23 Subsequently, the training set was used to develop multiple nano-QSAR classification models using the TPOT24 Auto-ML library.21 In the process of model development, 3-fold cross-validation was applied on the training set and was used as the model performance metric for internal validation. The validation set was not involved in the training procedure and was used only for the final external performance evaluation of the model.25
The domain of applicability (DOA)22 was calculated according to the distance-based leverage method. When the model is used to predict the toxicity of a SPION, the leverage value indicates if the sample is within the DOA of the model and only in this case the prediction is considered reliable. Finally, the attribute importance was analysed to identify the most important features in predicting the nanotoxicity related endpoint.
Nano-related data curation
The data that were used in this study were collected from an extensive literature search on in vitro and in vivo studies of SPIONs-based MRI contrasts agents used for stem cell labelling detection. From the Scopus database26 we initially extracted the toxicological characteristics of many novel SPIONs, by performing manual data mining on 71 published articles with the keywords “toxicity studies”, “iron oxide NPS (SPIONs)”, “MRI”, “stem cell therapy” and “regenerative medicine”. However, in most of these articles, physicochemical attributes were missing. Only 12 of these studies contained physicochemical data.27–38 Subsequently, we extracted information on physicochemical and in vitro/in vivo toxicological properties of SPIONs from these 12 studies. Only one literature source was found and used for each material, but some sources contained information for more than one SPION. The produced curated nano-related dataset consists of 16 novel SPIONs along with their physicochemical properties and their toxicological profiles. The physicochemical characteristics (features) were: “the magnetic core”, “the zeta potential”, “the particle size”, “the field strength”, “the concentration of iron per cell” and the “relaxivity”. These attributes are presented in Table 1 along with the respective minimum and maximum values. “Cell viability” was the toxicological property-the endpoint-in our predictive model. The dataset is presented in Table S1 in the ESI.†Data pre-processing
Firstly, the numerical attributes of Table S1† (the full dataset) were scaled between 0 and 1 (min–max scaling). For the “magnetic core” attribute the value was set to 1 for maghemite and 0 for magnetite. Missing values of the “relaxivity” feature were filled by applying agglomerative hierarchical clustering (Fig. 2) on the rest of the variables. More specifically, the agglomerative clustering method was applied from the scikit-learn Python library, which merges recursively pairs of clusters. The method minimally increases a distance, which in our application was selected as the Euclidean maximum distance between all observations of two clusters. The dendrogram produced by the method is depicted in Fig. 2. Three clusters of SPIONs were defined: {Cluster 1: SPIONs with IDs 12, 9, 7, 6, 14, 5, 15, 8}, {Cluster 2: SPIONs with IDs 13, 0, 11, 4, 3, 1, 2}, {Cluster 3: SPION with ID 10}. Missing “relaxivity” values were computed as the average “relaxivity” of the rest of the samples that were included in the same cluster.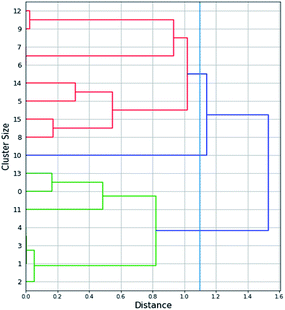 | ||
| Fig. 2 Dendrogram of the agglomerative hierarchical clustering that was applied in order to fill the “relaxivity” missing values. | ||
Regarding the endpoint, two classes, namely “Toxic” and “Non-Toxic” were defined for the classification model development. A material was characterized as “Toxic” if the cell viability percent was less than 75%, otherwise it was characterized as “Non-toxic”. “Cell viability” in bibliography is very well-defined as the quantification of the number of live cells and is usually expressed as a percentage of the control.33
We excluded “zeta potential” from the rest of this study, because zeta potential values were missing for 50% of the SPIONs in the dataset and data gaps were not balanced between toxic and nontoxic SPIONs.
Nanotoxicity QSAR model development and validation
A Jupyter notebook was developed in Python (version 3.6)39 which optimizes the procedure for developing a predictive nanoQSAR model that describes the correlation of cell viability and some physicochemical properties. The dataset produced after the pre-processing stage was partitioned into training and test sets, using the Kennard–Stone algorithm.23 Then the TPOT24 auto-ML tool was used to train the model.AutoML21 is an automatic process of model algorithm selection, hyper-parameter tuning, iterative modelling, and model assessment. The TPOT tool uses binary expression trees to represent ML pipelines with optimization provided by genetic programming and other stochastic search methods. The TPOT classifier iterated for 10 minutes through several algorithms, using as a validation metric a 3-fold cross validation test, and concluded into a logistic regression model and the following hyperparameters:
• C = 20.0. The parameter C represents the inverse of regularization strength.
• Penalty = “l2”. Penalty is used to specify the norm that is used in the penalization.
The exported logistic regression model was tested with several validation metrics and the results are presented in the results and discussion section.
Model explanation & information extraction
Last step of our analysis was to define the DOA of the model and the features with the highest predictive importance. DOA describes the physicochemical space on which the developed model is trained, and thus can be applied to make predictions. In this study, DOA was calculated according to the Leverage method, to which a threshold, is defined from the size of the training dataset and the number of features according to the following formula.
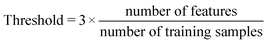 | (1) |
A nanoQSAR model prediction is associated with the calculation of the leverage value hi
| hi = xiT(XTX)−1xi | (2) |
The feature importances were exported from sklearn logistic regression results. This attribute displays the logistic regression model coefficients. A table with the leverage values and a diagram, presenting the absolute value of these coefficients, are presented in the results and discussion section.
Results and discussion
Data curation
A novel nanotoxicity dataset of SPIONs was extracted manually from literature data mining in 12 studies. The full curated dataset used for modelling is given in Table S1.† As shown in Table S1,† physicochemical and toxicological characteristics of the presented materials were obtained from several different literature sources and curated as a dataset of 16 iron oxide SPIONs. The toxicological end-point was clearly defined according to OECD principles;40 specifically two cell viability classes were derived from the toxicological characteristics: if the viability percent was more than 75% the material was considered as “Non-toxic” and the end-point value was set at 0, otherwise it was considered as “Toxic” and the end-point value was set at 1. For the material with ID 13, the cell viability was not clearly defined in the literature so we considered these materials as non-toxic, from the presented experimental studies.35,36Nano-QSAR model performances
The performance of the model on the training and validation sets are evaluated in Table 3 using the metrics defined in eqn (3)–(5). True positives (TP) are toxic SPIONs correctly classified as positives, true negatives (TN) are nontoxic SPIONs correctly classified as negatives, while false positives (FP) are nontoxic SPIONs incorrectly classified as toxic and false negatives (FP) are toxic SPIONs incorrectly classified as nontoxic.
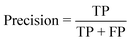 | (3) |
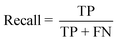 | (4) |
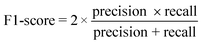 | (5) |
All metrics agree that the produced model predicts accurately the correct class of SPIONs regarding cell viability. The model failed in only one training sample and predicted the correct class for all validation samples, thus the testing accuracy (both precision and recall) is 100%.
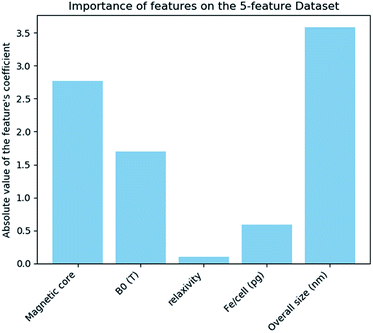
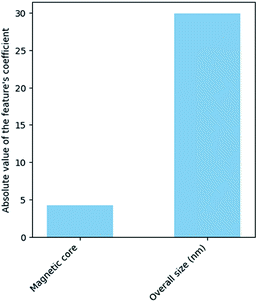
The results of the analysis for the most important features for the prediction of the class are displayed in Fig. 3. “Magnetic core” and “size” are the most important features in predicting the cell viability of SPIONs.
The results from the above analysis indicated that features “Bo”, “Relaxivity” and “Fe/cell” play an insignificant role in the decision of the model, since their contribution to the exponential of the logistic regression model is very small. In order to reduce the dimensionality, we examined if a nanoQSAR model trained and tested on the same samples, with “Bo”, “Relaxivity”, “Fe/cell” excluded from the set of independent variables produces similarly accurate results with the original model. The new dataset contains only two independent features, namely the magnetic core and the size (Fig. 4).
We used Auto-ML again to optimise the modelling procedure after excluding the less accurate features. We obtained again a logistic regression model with hyperparameters C = 40 and penalty function = l1. The nanoQSAR model predictions on both the training and the validation sets are shown in Table 2. The last two columns in Table 2 show the probabilities computed by the reduced logistic regression model of belonging to classes 0 and 1. The performance metrics of the reduced model are shown in Table 4. It is clear that the model is successful and provides more accurate results compared to the full model as all SPIONs have been classified correctly. This confirms that the rest of the features considered in the original nanoQSAR model are not important.
| Material ID | Material | Training (T) validation (V) | True class 1: toxic, 0: nontoxic | Predicted class 1: toxic, 0: nontoxic | Probability of class 0 by the reduced model | Probability of class 1 by the reduced model |
|---|---|---|---|---|---|---|
| 0 | Fe2O3-PLL27 | T | 0 | 0 | 0.52 | 0.48 |
| 1 | Uncoated γ-Fe2O3 (ref. 29) | T | 1 | 1 | 0.28 | 0.72 |
| 2 | D-Mannose-coated-γ-Fe2O3 (ref. 29) | T | 1 | 1 | 0.07 | 0.93 |
| 3 | Fe2O3-PLL29 | V | 1 | 1 | 0.28 | 0.72 |
| 4 | PDMAAm-coated-γ-Fe2O3-PLL29 | V | 1 | 1 | 0.28 | 0.72 |
| 5 | N-Dodecyl-PEI2k/SPIO30 | T | 0 | 0 | 1 | 0 |
| 6 | Iron oxide-loaded cationic nanovesicle31 | T | 0 | 0 | 1 | 0 |
| 7 | Iron oxide-loaded cationic nanovesicle31 | V | 0 | 0 | 1 | 0 |
| 8 | CMCS-SPIONs32 | V | 0 | 0 | 1 | 0 |
| 9 | ED-pullulan coating SPIO33 | T | 0 | 0 | 1 | 0 |
| 10 | IONP-6PEG-HA34 | T | 0 | 0 | 1 | 0 |
| 11 | PDMAAm-coated-γ-Fe2O3-PLL35 | T | 0 | 0 | 1 | 0 |
| 12 | Citrate SPION36 | V | 0 | 0 | 1 | 0 |
| 13 | D-Mannose-coated SPIONs29 | T | 1 | 1 | 0.15 | 0.85 |
| 14 | SPIO@SiO2–NH2 (ref. 37) | T | 0 | 0 | 0.95 | 0.05 |
| 15 | TAT-CLIO38 | T | 0 | 0 | 1 | 0 |
The importance of the two features remained in the final nanoQSAR model are displayed in Fig. 4 and they are both significant. The leverage values were computed for all validation samples (Table 5). All leverage values are below the threshold value of 0.5, which means that all the predictions are considered to be inside the DOA of the model.
| ID | Name | Leverage value | Reliability |
|---|---|---|---|
| 3 | Fe2O3-PLL | 0.2 | Reliable |
| 4 | PDMAAm-coated-g-Fe2O3-PLL | 0.2 | Reliable |
| 7 | Iron oxide-loaded cationic nanovesicle | 0.47 | Reliable |
| 8 | CMCS-SPIONs | 0.06 | Reliable |
| 12 | Citrate SPION | 0.17 | Reliable |
Due to the simplicity of the final model, we were able to arrive to some simple rules concerning the toxicity of SPIONs in stem-cell therapy. SPIONs with magnetite cores are non-toxic while SPIONs with maghemite cores are toxic for small sizes and nontoxic for larger sizes, with the cut-off value around 15 nm. Clearly additional experimental information is needed to define more accurately this cut-off value.
The logit function of the LR model that calculates the probability of belonging to the toxicity class is shown next:
 | (6) |
Web implementation of the model
The source code for developing the model is available at: https://github.com/ntua-unit-of-control-and-informatics/SPIONs. The model has been implemented as a web service in the Jaqpot 5 modelling platform (https://app.jaqpot.org/) and is available in the following URL: https://app.jaqpot.org/model/DcWnWFp9GESI16R4o2av under the BIORIMA organisation. In the overview tab, more details about the model are presented including a Predictive MarkUp Language (PMML) representation which contains the scaling coefficients and the logit function (eqn (6)). For accessing the model, the interested user should first register in Jaqpot 5 and then become a member of the BIORIMA organisation by sending an e-mail to: E-mail: hsarimv@central.ntua.gr.Conclusions
Addressing the toxicity of a potential NBMs in stem-cell therapy, requires unwrapping the key physicochemical nanomaterial properties that lead to toxicity at the cellular level. Herein, we present an integrated study from data curation to model development, handling a set of nano-related toxicological and physicochemical data for SPIONs-based MRI agents, used in stem-cell labelling and monitoring. Specifically, in our meta-analysis more than 70 publications were mined manually, generating a nanotoxicity dataset composed of 16 novel SPIONs and 5 categorical/numerical attributes applicable in regenerative medicine.The applicability of our curated dataset in developing predictive models for SPION toxicity was examined by applying Auto-ML techniques. In fact, we compared multiple ML models for developing a high-performance nanotoxicity classification QSAR model for iron oxide NPs. The most robust model in our study with the highest performance (100% train accuracy and 100% test accuracy) was built with the logistic regression (LR) algorithm. Attribute significance, evaluated in conjunction with LR model development, indicated that SPIONs-induced toxicity response correlated primarily with key intrinsic NPs properties. Thus, our analysis revealed that physicochemical properties such as “magnetic core” and “the size”, are the predominant attributes to the toxicity of magnetic NPs, the “iron concentration per cell is following, whereas other attributes such as “relaxivity”, and “magnetic field” are of low measurable significance in correlating toxicity. Finding that SPIONs “core” and “size” are the most important correlating attributes for SPIONs toxicity, was conclusively demonstrated by our analysis of the curated literature data.
Overall, this study suggests that as more systematic information from NBMs becomes available in the literature, QSAR modelling could actually provide guidance regarding key NBM attributes (for example, physicochemical properties) that should be experimentally characterized and reported in NBM toxicity studies. Identifying the dominant features to SPIONs nanosafety, as well as the dependant attribute–toxicity relationship, will support the development of NBMs that are safe-by-design. This will bring many promises in regenerative medicine and disease therapeutics.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
H. Sarimveis and M. Kotzabasaki acknowledge financial support by BIORIMA (Grant Agreement 760928), a project funded by the European Commission under the Horizon 2020 Programme.Notes and references
- M. Maumus, D. Guérit, K. Toupet, C. Jorgensen and D. Noël, Stem Cell Res. Ther., 2011, 2(2), 14 CrossRef PubMed.
- https://www.eurostemcell.org/.
- R. S. Mahla, Int. J. Cell Biol., 2016, 2016(7), 1–24 CrossRef PubMed.
- H. Markides, M. Rotherham and A. J. El Haj, J. Nanomater., 2012, 2012, 1–11 CrossRef.
- T. Lam, P. Pouliot, P. K. Avti, F. Lesage and A. K. Kakkar, Adv. Colloid Interface Sci., 2013, 199–200, 95–113 CrossRef CAS PubMed.
- Y. Liu, M. Li, F. Yang and N. Gu, Sci. China Mater., 2017, 60(6), 471–486 CrossRef CAS.
- L. Li, W. Jiang, K. Luo, H. Song, F. Lan, Y. Wu and Z. Gu, Theranostics, 2013, 3(8), 595–615 CrossRef PubMed.
- N. Lewinski, V. Colvin and R. Drezek, Small, 2008, 4(1), 26–49 CrossRef CAS PubMed.
- S. C. Peter, J. K. Dhanjal, V. Malik, N. Radhakrishnan, M. Jayakanthan and D. Sundar, Encyclopedia of Bioinformatics and Computational Biology, 2019, vol. 2, pp. 661–676 Search PubMed.
- T. X. Trinh, M. K. Ha, J. S. Choi, H. G. Byun and T. H. Yoon, Environ. Sci.: Nano, 2018, 5, 1902–1910 RSC.
- M. K. Ha, T. X. Trinh, J. S. Choi, D. Maulina, H. G. Byun and T. H. Yoon, Sci. Rep., 2018, 8(1), 3141 CrossRef PubMed.
- J. M. Gernand and E. A. Casman, Risk Anal., 2014, 34(3), 583–597 CrossRef PubMed.
- R. Liu, R. Rallo, S. George, Z. Ji, S. Nair, A. E. Nel and Y. Cohen, Small, 2011, 7, 1118–1126 CrossRef CAS PubMed.
- R. Liu, R. Rallo, R. Weissleder, C. Tassa, S. Shaw and Y. Cohen, Small, 2013, 9, 1842–1852 CrossRef CAS PubMed.
- E. Oh, R. Liu, A. Nel, K. B. Gemill, M. Bilal, Y. Cohen and I. L. Medintz, Nat. Nanotechnol., 2016, 11(5), 479–486 CrossRef CAS PubMed.
- Y. Pan, T. Li, J. Cheng, D. Telesca, J. I. Zink and J. Jianga, RSC Adv., 2016, 6, 25766–25775 RSC.
- T. Puzyn, D. Leszczynska and J. Leszczynski, Small, 2009, 5, 2494–2509 CrossRef CAS PubMed.
- T. Puzyn, B. Rasulev, A. Gajewicz, X. Hu, T. P. Dasari, A. Michalkova, H.-M. Hwang, A. Toropov, D. Leszczynska and J. Leszczynski, Nat. Nanotechnol., 2019, 6(3), 175–178 CrossRef PubMed.
- K. P. Singh and S. Gupta, RSC Adv., 2014, 4, 13215–13230 RSC.
- T. X. Trinh, J.-S. Choi, H. Jeon, H.-G. Byun, T.-H. Yoon and J. Kim, Chem. Res. Toxicol., 2018, 31(3), 183–190 Search PubMed.
- M. Feurer, A. Klein, K. Eggensperger, J. T. Springenberg, M. Blum and F. Hutter, Adv. Neural Inf. Process. Syst., 2015, 2962–2970 Search PubMed.
- K. Roy, S. Kar and P. Ambure, Chemom. Intell. Lab. Syst., 2015, 145, 22–29 CrossRef CAS.
- P. Taylor, R. W. Kennard and L. A. Stone, Technometric, 1969, 11, 137–148 CrossRef.
- R. S. Olson, R. J. Urbanowicz, P. C. Andrews, N. A. Lavender, L. C. Kidd and J. H. Moore, Lect. Notes Comput. Sci., 2016, 9597, 123–137 Search PubMed.
- M. Stone, J. Roy. Stat. Soc. B, 1974, 36(2), 111–147 Search PubMed.
- https://www.scopus.com/.
- S. Ju, G. Teng, Y. Zhang, M. Ma, F. Chen and Y. Ni, Magn. Reson. Imaging, 2006, 24, 611–617 CrossRef PubMed.
- M. Babič, D. Horák, M. Trchová, P. Jendelová, K. Glogarová, P. Lesný, V. Herynek, M. Hájek and E. Syková, Bioconjugate Chem., 2008, 19, 740–750 CrossRef PubMed.
- D. Horák, M. Babič, P. Jendelová, V. Herynek, M. Trchová, K. Likavčanová, M. Kapcalová, M. Hájek and E. Syková, J. Magn. Magn. Mater., 2009, 321, 1539–1547 CrossRef.
- G. Liu, Z. Wang, J. Lu, C. Xia, F. Gao, Q. Gong, B. Song, X. Zhao, X. Shuai, X. Chen, H. Ai and Z. Gu, Biomaterials, 2011, 32(2), 528–537 CrossRef CAS PubMed.
- R. M. Guo, N. Cao, F. Zhang, Y. R. Wang, X. H. Wen, J. Shen and X. T. Shuai, Eur. J. Radiol., 2012, 22, 2328–2337 CrossRef PubMed.
- Z. Shi, K. G. Neoh, E. T. Kang, B. Shuter, S. C. Wang, C. Poh and W. Wang, ACS Appl. Mater. Interfaces, 2009, 1, 328–335 CrossRef CAS PubMed.
- J. ichiro Jo, I. Aoki and Y. Tabata, J. Controlled Release, 2010, 142, 465–473 CrossRef PubMed.
- H. J. Chung, H. Lee, K. H. Bae, Y. Lee, J. Park, S. W. Cho, J. Y. Hwang, H. Park, R. Langer, D. Anderson and T. G. Park, ACS Nano, 2011, 5(6), 4329–4336 CrossRef CAS PubMed.
- M. Babič, D. Horák, M. Trchová, P. Jendelová, K. Glogarová, P. Lesný, V. Herynek, M. Hájek and E. Syková, Bioconjugate Chem., 2008, 19, 740–750 CrossRef PubMed.
- K. Andreas, R. Georgieva, M. Ladwig, S. Mueller, M. Notter, M. Sittinger and J. Ringe, Biomaterials, 2012, 33, 4515–4525 CrossRef CAS PubMed.
- H. H. Wang, Y. X. J. Wanu, K. C. F. Leung, D. W. T. Au, S. Xuan, C. P. Chak, S. K. M. Lee, H. Sheng, G. Zhang, L. Qin, J. F. Griffith and A. T. Ahuja, Chem.–Eur. J., 2009, 15, 12417–12425 CrossRef CAS PubMed.
- M. Song, W. K. Moon, Y. Kim, D. Lim, I. C. Song and B. W. Yoon, Korean J. Radiol., 2007, 8(5), 365–371 CrossRef PubMed.
- Python Software Foundation, Python Language Reference, version 3.6. available at http://www.python.org Search PubMed.
- OECD, Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models, OECD Series on Testing and Assessment, No. 69, OECD Publishing, Paris, 2014, DOI:10.1787/9789264085442-en.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09475j |
| This journal is © The Royal Society of Chemistry 2020 |