Open Access Article
Open Access ArticleA novel nanoparticle loaded with methyl caffeate and caffeic acid phenethyl ester against Ralstonia solanacearum—a plant pathogenic bacteria†
Jin-Zheng Wang‡
a,
Cheng-Hai Yan‡a,
Xiao-Rui Zhanga,
Qing-Bo Tua,
Yan Xuabcd,
Sheng Shengabcd,
Fu-An Wuabcd and
Jun Wang *abcd
*abcd
aSchool of Biotechnology, Jiangsu University of Science and Technology, Zhenjiang 212018, PR China. E-mail: wangjun@just.edu.cn; Fax: +86-511-85620901; Tel: +86-511-85635867
bSericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang 212018, PR China
cKey Laboratory of Silkworm and Mulberry Genetic Improvement, Ministry of Agriculture and Rural Affairs, Zhenjiang 212018, PR China
dJiangsu Key Laboratory of Sericultural Biology and Biotechnology, Zhenjiang 212018, PR China
First published on 23rd January 2020
Abstract
Developing a novel agent and understanding the interaction model between multipolymer nanoparticles and bacteria could be worthwhile to induce the protection of crops with the prevalence of frequent hazards because of the use of pesticides and chemical resistance. Unlike metal nanoparticles, multipolymer nanoparticles have bacteriostatic properties against Ralstonia solanacearum that can trigger bacterial wilt by infecting the plant. Therefore, a novel poly(lactic-co-glycolic acid) nanoparticle containing caffeic acid phenethyl ester (CAPE) and methyl caffeate (MC) was prepared with the sustained-release property (for 10 d at pH 6.5); here, 50% of the cumulative release rate was achieved. It was observed that the cytomembrane of R. solanacearum was jeopardized by the nanoparticle by the creation of large holes on the bacterial surface. The nanoparticle has an approximate EC50 value of 0.285 mg mL−1 with active pharmaceutical ingredients (APIs), while the drug dosage could be reduced by 2/3. Furthermore, to reveal the possible mechanism of interaction between the multipolymer nanoparticles and bacteria, a formidable inhibition effect was observed; the pathogenicity-related genes, namely, phcA, phcB, pehC, egl, pilT, and polA, of R. solanacearum were downregulated by 1/2, 1/42, 1/13, 1/6, 1/2, and 1/8, respectively, showing significant effects on the major virulence-related genes. Hence, a novel nanoparticle with excellent antibacterial and sustained-release properties has been prepared, possessing the potential to replace chemical pesticides and serve as a new control strategy for mulberry blight disease.
1. Introduction
The benefits of worldwide pesticide utilization have become limited due to their effects on the environment.1 Due to the long-term effect of chemical pesticides, the efficiency of pesticide control has weakened. Many pesticide residues have become ubiquitous in the natural environment and are present ranging from nanogram per liter to low milligram per liter concentrations, which can cause serious microbial species balance,2 soil microenvironment change,3 and effects on human health.4 In particular, during the study on the control strategy of bacterial wilt disease caused by Ralstonia solanacearum, which is one of the most harmful soil-borne diseases in the world due to its wide distribution and host range,5 traditional chemical pesticides and antibiotics have been employed against R. solanacearum despite issues such as limited efficacy, microbial resistance strains, environmental pollution, and potential health risks to human health.6 Therefore, it has become urgent to develop an alternative agent without any collateral damage to replace traditional chemical pesticides and protect economic crops from plant pathogens such as R. solanacearum.In the development of antibacterial agents, polymeric nanoparticles have been considered as novel antibacterial agents with the ability to enhance the antibacterial properties.7 Poly-(lactic-co-glycolic acid) (PLGA), a novel material for preparing polymeric nanoparticles, has been commonly used as the polymer material in drug delivery systems. Generally, PLGA nanoparticles have a sustained-release capacity,8 which has very environmentally friendly properties; this occurs because of the hydrolysis of ester bonds in water to form lactic acid and glycolic acid.9 Further, this release capacity might lead to an incomplete release, which can cause several active pharmaceutical ingredients (APIs) to directly exist in the soil after PLGA degradation. The modification of the nanoparticle surface, such as the application of antibodies,10 might be helpful to resolve this problem. In addition, nanoparticles prepared with PLGA can improve the solubility and dispersibility of nanoparticles in water without affecting their efficacy.11 Furthermore, polymeric nanoparticles have been considered to be the most promising approach against drug resistance of bacteria.12,13 Nanoparticles can be used as alternative tools to handle drug-resistant pathogens.14 Hence, nanoparticles prepared with PLGA have the potential to be an alternative to traditional pesticides.
Due to the ability of R. solanacearum to maintain its activity in soils for a long period of time15 and its complex genetic regulatory system, it is difficult to eradicate this bacterium by means of chemical pesticides and antibiotics, even those that cause serious environmental issues. Nearly 30 provinces in China have suffered from problems related to R. solanacearum, and mulberry trees in Jiangsu and Zhejiang provinces, as well as potatoes in Fujian province, have been afflicted with the most serious damage.16 As early as 2012, R. solanacearum was ranked as the second most harmful plant pathogen in the top 10 bacterial plant pathogens.17 In addition, because R. solanacearum can enter through the plant roots, invade xylem vessels, and rapidly spread to the aerial parts of the plant,15 the swimming motility of R. solanacearum can facilitate its ability to infect plants. Furthermore, the expression of virulence factors also plays a crucial role in the pathogenesis of R. solanacearum. The virulence of filamentous phage against R. solanacearum was evaluated by detecting the expression of pathogenicity-related genes in R. solanacearum (physiological race 1, biochemical type 3), and open reading frame (ORF) 15 of φRSM phage can inhibit the transcriptional regulator phcA, causing pathogenicity in the phenotypic transformation system of R. solanacearum, which can reduce pathogenicity.18 Further, considering the complex regulatory mechanism of R. solanacearum, the combination of multiple APIs might help to reduce the resistance of pathogenic bacteria toward agents.
Since antibiotics can lead to increased resistance of pathogenic bacteria, natural source agents and their derivatives have become the mainstays in the development of antibacterial agents.19 Due to their potential ability to inhibit bacteria, our attention has been focused on caffeic acid—a naturally sourced compound—and its derivatives. Among them, caffeic acid phenethyl ester (CAPE), which is considered to be one of the most popular compounds for the development of new drugs20 and has been the main active ingredient isolated from propolis residues, has shown toxic effects toward R. solanacearum.21 Methyl caffeate (MC) has many pharmacological activities, and it is a synthetic intermediate of many natural drugs and food additives, such as propyl caffeate (PC); CAPE can be prepared by the transesterification of MC with the corresponding alcohol.22 Recently, PLGA particles loaded with CAPE were successfully prepared,20 which demonstrated the feasibility of fabricating PLGA particles loaded with caffeic acid and its derivatives. Many studies have focused on the preparation process as well as the lack of research on the mechanism of bacteriostatic action of the prepared nanoparticles. Therefore, a brief description of the possible bacteriostatic mechanisms has been provided in this study. Further, MC and CAPE were employed as a combination of APIs to develop an alternative agent for controlling plant pathogens, and the preparation of PLGA nanoparticles loaded with MC and CAPE can hopefully become an alternative agent with high efficiency and low toxicity.
In this study, an emulsion solvent evaporation technique was employed to prepare PLGA nanoparticles loaded with MC and CAPE, while the drug loading rate (DL) was selected as the main evaluation standard. Parameters such as polyvinyl alcohol (PVA) concentration, ultrasonic power, ultrasonic time, and ratio of the organic phase to aqueous phase were optimized. The slow-release characteristics of these novel nanoparticles were also investigated. To investigate the antibacterial activity of the nanoparticles on R. solanacearum, RT-PCR was employed to detect the expression levels of pathogenicity-related genes, and scanning electron microscopy (SEM) was employed to observe the treated strains. Susceptibility testing by the disk diffusion method of the nanoparticles was also carried out to preliminarily evaluate the drug resistance after treatment.
2. Experiment method and material
2.1 Experimental materials and strains
Tryptone LP0042 was purchased from Oxoid, UK. Casein hydrolysate, PLGA (lactide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycolide = 50
glycolide = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50; intrinsic viscosity: 0.45–0.60 dL g−1; Mw: ∼38–54 kDa P50/50), and PVA 124 (viscosity: 40 g L−1, 20 °C, 54.0–66.0 (mPa s)−1) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Caffeate derivatives such as MC and CAPE were prepared by our group.21,23 SYBR premix Ex Tap™ II (Tli RNaseH Plus), primer script RT reagent kit with gDNA eraser (Perfect Real Time), DEPC water, and isopropanol were purchased from Takara. The total RNA extraction reagent TRIzol and PCR kit were purchased from Sangon Biotech. RS-5 R. solanacearum strains were used, which were isolated from mulberry roots.21 All the primers were synthesized by Shanghai Biotech; the primer sequences were synthesized as described elsewhere,18 as listed in Table 1.
50; intrinsic viscosity: 0.45–0.60 dL g−1; Mw: ∼38–54 kDa P50/50), and PVA 124 (viscosity: 40 g L−1, 20 °C, 54.0–66.0 (mPa s)−1) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Caffeate derivatives such as MC and CAPE were prepared by our group.21,23 SYBR premix Ex Tap™ II (Tli RNaseH Plus), primer script RT reagent kit with gDNA eraser (Perfect Real Time), DEPC water, and isopropanol were purchased from Takara. The total RNA extraction reagent TRIzol and PCR kit were purchased from Sangon Biotech. RS-5 R. solanacearum strains were used, which were isolated from mulberry roots.21 All the primers were synthesized by Shanghai Biotech; the primer sequences were synthesized as described elsewhere,18 as listed in Table 1.
| Primer name | Nucleotide sequence (5′ → 3′) | Size (bp) |
|---|---|---|
| a F represents the leading chain and R represents the trailing chain. | ||
| Egl3-F | CAGCGCGACCTACTACAAGA | 299 |
| Egl3-R | TCATCAGCCCGAAGATGAC | |
| PhcA(298)-F | GGACATGATCTTCACGGTCAACT | 298 |
| PhcA(298)-R | GACTCATCCTCCTTTTCTGCATC | |
| PhcB(RT)-F | CTACCAGATCGTCGTCAATGAA | 172 |
| PhcB(RT)-R | GTCGAGGTAGTGCTTGATCTTG | |
| HrpB(RT)-F | TTCTCGATGATGTAGCGATAGG | 238 |
| HrpB(RT)-R | GCTGGAATTTTCGACTTCCTCTA | |
| PehC(RT)-F | GTTGTTCGGATTGCTGTACG | 227 |
| PehC(RT)-R | AGTCAAACGATTGCCTGAACTA | |
| PilT(175)-F | AAGAACAAAGCGTCTGATCTGC | 175 |
| PilT(175)-R | CTTCCAGGTTTTCTTCGTAATGCT | |
| polA-238F | GGAATGTCGGAAAGTCAAGAAA | 238 |
| polA-238R | CTTGTAGGCGGGGTACAGTTC | |
| 16SrRNA349-F | CTAGAGTGTGTCAGAGGGAGGTAGA | 349 |
| 16SrRNA349-R | ATGTCAAGGGTAGGTAAGGTTTTTC | |
2.2 Preparation of nanoparticles loaded with CAPE and MC
The preparation of nanoparticles by a single emulsion/solvent evaporation method was based on other studies.11 The effects of PVA concentration, ultrasonic power, ultrasonic time, and ratio of the organic phase to aqueous phase on nanoparticle preparation were investigated. In the first stage, 100 mg PLGA and 55 mg API (MC and CAPE at a mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were dissolved in 1.5 mL dichloromethane and 0.5 mL ethanol, respectively. After complete dissolution, the mixture was shaken for 5 min, completing the preparation of the organic phase. PVA (1%, 2%, 3%, 4%, 5%, and 10% w/w) was heated with deionized water in an ultrasonic water bath at 80 °C until dissolution to prepare the aqueous phase. After the second stage, the organic and aqueous phases were mixed according to various volume ratios (2
1) were dissolved in 1.5 mL dichloromethane and 0.5 mL ethanol, respectively. After complete dissolution, the mixture was shaken for 5 min, completing the preparation of the organic phase. PVA (1%, 2%, 3%, 4%, 5%, and 10% w/w) was heated with deionized water in an ultrasonic water bath at 80 °C until dissolution to prepare the aqueous phase. After the second stage, the organic and aqueous phases were mixed according to various volume ratios (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 2
2, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, and 1
5, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) in an ice bath and then subjected to ultrasonic treatment using a microtip probe sonicator (65, 130, 195, 260, 325, 390, 474.5, and 650 W; amplitude: 80%; 2, 4, 6, 8, and 10 min); the mixture was stirred overnight under magnetic force at room temperature. Thereafter, the obtained white emulsion was centrifuged 3 times at 3000 rpm to remove the precipitate; then, the remaining solution was centrifuged 3 times at 12
3) in an ice bath and then subjected to ultrasonic treatment using a microtip probe sonicator (65, 130, 195, 260, 325, 390, 474.5, and 650 W; amplitude: 80%; 2, 4, 6, 8, and 10 min); the mixture was stirred overnight under magnetic force at room temperature. Thereafter, the obtained white emulsion was centrifuged 3 times at 3000 rpm to remove the precipitate; then, the remaining solution was centrifuged 3 times at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 40 min to obtain the precipitate. The supernatant was discarded. Finally, a small amount of deionized water was added to resuspend the precipitate. The mixture was then lyophilized for 48 h, and the lyophilized nanoparticles were stored in a freezer at −80 °C.
000 rpm for 40 min to obtain the precipitate. The supernatant was discarded. Finally, a small amount of deionized water was added to resuspend the precipitate. The mixture was then lyophilized for 48 h, and the lyophilized nanoparticles were stored in a freezer at −80 °C.
2.3 Determination of DL and encapsulation efficiency (EE) of the nanoparticles
The nanoparticles were dissolved in methanol at a mass-to-volume ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. To determine the MC and CAPE concentrations, the nanoparticle structures were destroyed by ultrasonication at room temperature for 30 min. The total amount of drug substrate in the nanoparticles was determined by high-performance liquid chromatography (HPLC) according to the standard curves of MC and CAPE, and the EE and DL values of the nanoparticles were calculated as follows:
1. To determine the MC and CAPE concentrations, the nanoparticle structures were destroyed by ultrasonication at room temperature for 30 min. The total amount of drug substrate in the nanoparticles was determined by high-performance liquid chromatography (HPLC) according to the standard curves of MC and CAPE, and the EE and DL values of the nanoparticles were calculated as follows:
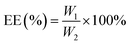 | (1) |
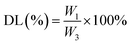 | (2) |
| MC standard curve line: Y = 5.64 × 107 × X − 2.35 × 105, R2 = 0.9914 | (3) |
| CAPE standard curve line: Y = 5.48 × 107 × X − 2.33 × 105, R2 = 0.9908 | (4) |
2.4 FTIR characterization of nanoparticles
Ultraviolet-visible near-infrared absorption spectrometry was employed to reconfirm the presence of MC and CAPE in the nanoparticles.24 Each sample was scanned 10 times, in the range of 4000–750 cm−1 at a resolution of 4 cm−1.2.5 Drug release characteristics of nanoparticles
The determination of sustained-release kinetics was based on an earlier reported method20 with slight modifications. Nanoparticles and phosphate-buffered solution (pH 7.4, 6.5, and 9.5) were mixed at a solid–liquid ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The mixtures were incubated at 37 °C under shaking and sampled at designated time intervals (1, 2, 3, 4, 5, 6, 9, 12, 15, 24, 27, and 30 h after incubation). The release medium was centrifuged at 9000 rpm for 3 min. The supernatant was collected and the solution was replaced with fresh PBS for continued incubation. The collected samples were lyophilized and dissolved in 1 mL methanol solution. Thereafter, HPLC was employed to detect the MC and CAPE concentrations, and the method was performed according to an earlier research.21 The Fickian diffusion model25 was used to evaluate the release process of polymer nanoparticles; the release mechanism was described by a semiempirical eqn (5).
1. The mixtures were incubated at 37 °C under shaking and sampled at designated time intervals (1, 2, 3, 4, 5, 6, 9, 12, 15, 24, 27, and 30 h after incubation). The release medium was centrifuged at 9000 rpm for 3 min. The supernatant was collected and the solution was replaced with fresh PBS for continued incubation. The collected samples were lyophilized and dissolved in 1 mL methanol solution. Thereafter, HPLC was employed to detect the MC and CAPE concentrations, and the method was performed according to an earlier research.21 The Fickian diffusion model25 was used to evaluate the release process of polymer nanoparticles; the release mechanism was described by a semiempirical eqn (5).
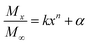 | (5) |
2.6 SEM of R. solanacearum
Antibacterial treatment was carried out according to an earlier research.21 Gemini SEM 300 (ZEISS, Germany) was employed to evaluate the morphology of R. solanacearum, and the inhibitory effect of bacteriostatic agents was evaluated by observing the changes in the cell membrane and cell morphology.2.7 Antibacterial rate of nanoparticles against R. solanacearum
The antibacterial activity of nanoparticles was determined according to an earlier research26 with slight modifications. Different concentrations of nanoparticles (4, 3.2, 1.6, 0.8, 0.4, 0.2, and 0.1 mg mL−1) were added to the wells of a 96-well plate. A microplate reader (SpectraMax i3, Silicon Valley, CA, USA) was used to determine the optical density of the plates at 600 nm at T0 before incubation. The OD600 value was measured as the TF after the plates were incubated at 30 °C overnight under shaking for 5 min. The antibacterial rate of the nanoparticles against R. solanacearum was calculated according to eqn (6), and the EC50 calculation method was used according to the reported methods.27
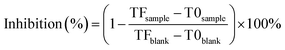 | (6) |
2.8 Pathogenicity-related gene expression of R. solanacearum
The nanoparticles (4 mg mL−1), API (4 mg mL−1), and Triton X-100 were added to the bacterial suspension (OD600 = 0.8–1) at a volume ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and incubated overnight at 30 °C for 24 h. After incubation, this treated bacterial suspension was separately centrifuged at 12
1 and incubated overnight at 30 °C for 24 h. After incubation, this treated bacterial suspension was separately centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min to collect the precipitates of different treatment strains. The total RNA extraction process was carried out according to the protocol modifications.28 Briefly, the Trizol reagent (Invitrogen, USA) was used to extract the total RNA from each treatment sample and DNase I (Promega, Madison, WI, USA) was used to purify the total RNA. The prepared total RNA was stored at −80 °C. The PrimeScript RT Reagent Kit with gDNA Eraser (Takara Biotechnology, Otsu, Japan) was used to synthesize the cDNA, and the product was stored at −80 °C for immediate measurement.
000 rpm for 3 min to collect the precipitates of different treatment strains. The total RNA extraction process was carried out according to the protocol modifications.28 Briefly, the Trizol reagent (Invitrogen, USA) was used to extract the total RNA from each treatment sample and DNase I (Promega, Madison, WI, USA) was used to purify the total RNA. The prepared total RNA was stored at −80 °C. The PrimeScript RT Reagent Kit with gDNA Eraser (Takara Biotechnology, Otsu, Japan) was used to synthesize the cDNA, and the product was stored at −80 °C for immediate measurement.
A LightCycler 96 real-time PCR system (Roche, Switzerland) was used to determine the relative expression of pathogenicity-related genes. Each reaction system (20 μL) contained 10 μL of TB Green, 0.8 μL of the upstream primer, 0.8 μL of the downstream primer, 6.4 μL of RNase-free double-distilled water, and 2 μL of cDNA. The specific primers (Table 1) were synthesized by Sangon in Shanghai according to another research,18 and the 16S rRNA of R. solanacearum was used as the internal control for RT-qPCR to normalize the gene expressions of the pathogenicity-related genes.
2.9 Significance level analysis
Significance level analysis and EC50 calculations were performed using the SPSS 22 software, and the mapping was performed using OriginPro 9.0 software. All the experiments had three sets of repetitions, and the result was reflected in the error bar in the figures. The letters on the column indicate the differences between the different treatment groups, the same letters indicate that the difference is not significant (P > 0.05), and completely different letters indicate significant differences (P < 0.05).3. Results and discussion
3.1 Nanoparticle preparation
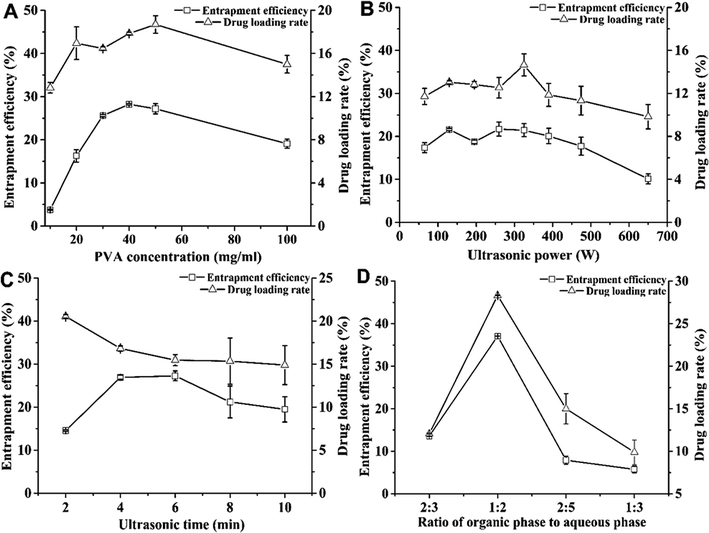
Fig. 1A shows the effects of PVA concentration on the DL and EE of nanoparticles. When the PVA concentration was between 10 and 100 mg mL−1, the EE values of MC and CAPE increased with the concentration, reaching the highest value of 28.25 ± 0.17% at 40 mg mL; further, they decreased from 28.25 ± 0.17% to 19.09 ± 1.04% for PVA concentrations between 40 and 100 mg mL−1. As an emulsifier, the PVA concentration could increase the EE of the drug molecule.29 However, when the emulsifier concentration was considerably increased, the EE and DL of the nanoparticles could reduce.30 When the PVA concentration was 50 mg mL−1, the DL reached the highest value, i.e., 18.70 ± 0.80%. In addition, when the PVA concentration exceeded 40 mg mL−1, the viscosity of the emulsion solvent was too high, making it difficult to separate the nanoparticles; further, at 30 mg mL−1 PVA, the DL (16.46 ± 0.18%) was close to the highest value (18.70 ± 0.80%), and the EE (25.26 ± 0.27%) was relatively high. Therefore, the suitable PVA concentration was 30 mg mL−1.Fig. 1B shows the effect of ultrasonic power on the DL and EE of nanoparticles and the emulsification treatment was carried out in the ultrasonic power range of 65–650 W. When the power was raised from 65 to 325 W, both DL and EE increased. The higher ultrasonic power could facilitate the formation of a uniform stable emulsion droplet.31 When the ultrasonic power was 260 W, the EE reached the maximum value of 21.71 ± 1.64%. Although the DL reached the highest value of 14.65 ± 1.03% under ultrasonic power of 325 W, the EE was 21.44 ± 1.51%. However, increasing the ultrasonic power could enhance cavitation, which could reduce the DL. When the power was increased from 325 to 650 W, both DL and EE decreased. The increase in ultrasonic power could reduce the particle size, while the excessive energy could increase flocculation,32 which could cause this decrease. Therefore, the power condition for obtaining the maximum DL was taken as the optimal condition, and the optimal ultrasonic power condition was 325 W.
Fig. 1C shows the effect of ultrasonic time on the EE and DL values. When the ultrasonic time was 2–10 min, the DL value decreases from 20.54 ± 0.22% to 14.89 ± 2.26%, which indicates that a longer ultrasonic time leads to a decrease in DL. The reason for this decrease might be the long-term cavitation effect caused by ultrasound, which may cause an increase in the droplet sizes33 and the destruction of the formed emulsified droplets, resulting in a decrease in drug content. Furthermore, excessive time has a marginal effect on the formation of emulsion droplets, which could prolong the reaction time.34 This optimal treatment condition with the highest DL was the same as that reported in another research;11 therefore, 2 min was chosen as the suitable ultrasonic time.
Fig. 1D shows the effect of the ratio of the organic phase to aqueous phase on the DL and EE values of the nanoparticles. When this ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the maximum values of EE and DL were achieved, namely, 37.08 ± 0.04% and 28.33 ± 0.03%, respectively. This ratio might not be conducive toward the dispersion of emulsified droplets, while the added PVA concentration was too small. In contrast, the addition of an excessive amount of PVA could result in a decrease in the EE of the nanoparticles due to a reduction in the amount of drugs scattered in the organic phase.35 In addition, this could lead to an increase in the possibility of dissolution of the drug under the action of ultrasound. Therefore, the optimum ratio of the organic phase to aqueous phase was selected to be 1
2, the maximum values of EE and DL were achieved, namely, 37.08 ± 0.04% and 28.33 ± 0.03%, respectively. This ratio might not be conducive toward the dispersion of emulsified droplets, while the added PVA concentration was too small. In contrast, the addition of an excessive amount of PVA could result in a decrease in the EE of the nanoparticles due to a reduction in the amount of drugs scattered in the organic phase.35 In addition, this could lead to an increase in the possibility of dissolution of the drug under the action of ultrasound. Therefore, the optimum ratio of the organic phase to aqueous phase was selected to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, yielding the highest DL of 28.33 ± 0.03%.
2, yielding the highest DL of 28.33 ± 0.03%.
Therefore, the optimum condition was as follows: 3% PVA with the ratio of organic phase to aqueous phase of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 at 325 W under sonication for 2 min. Further, the highest DL (28.33 ± 0.03%) was achieved under this optimum condition.
2 at 325 W under sonication for 2 min. Further, the highest DL (28.33 ± 0.03%) was achieved under this optimum condition.
3.2 Nanoparticle characterization
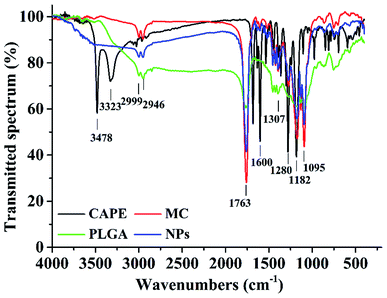
Fig. 2 shows the FTIR spectra of the nanoparticles, CAPE, MC, and PLGA, where 1763 cm−1 is the extension of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond in the ester bond and 1182–1095 cm−1 is the pull of the CO bond. These are the characteristic peaks of the PLGA molecules;36 the main peaks of CAPE at 3478, 3323, and 1600 cm−1 were significantly reduced, indicating that CAPE was successfully embedded in the PLGA.20 The hydroxyl group at 3478 cm−1 was stretched; the 1607, 1535, and 1445 cm−1 peaks denote the C
O bond in the ester bond and 1182–1095 cm−1 is the pull of the CO bond. These are the characteristic peaks of the PLGA molecules;36 the main peaks of CAPE at 3478, 3323, and 1600 cm−1 were significantly reduced, indicating that CAPE was successfully embedded in the PLGA.20 The hydroxyl group at 3478 cm−1 was stretched; the 1607, 1535, and 1445 cm−1 peaks denote the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond stretching of the aromatic compound; and the 1307 cm−1 peak denoted the CO bond of –COOH. These are the characteristic peaks of MC.37 In the nanoparticle spectra, the characteristic peaks, namely, 1281, 1307, 1607, 1535, and 1445 cm−1, were significantly reduced, indicating that MC was successfully embedded in the PLGA. Hence, MC and CAPE were successfully embedded.
C bond stretching of the aromatic compound; and the 1307 cm−1 peak denoted the CO bond of –COOH. These are the characteristic peaks of MC.37 In the nanoparticle spectra, the characteristic peaks, namely, 1281, 1307, 1607, 1535, and 1445 cm−1, were significantly reduced, indicating that MC was successfully embedded in the PLGA. Hence, MC and CAPE were successfully embedded.
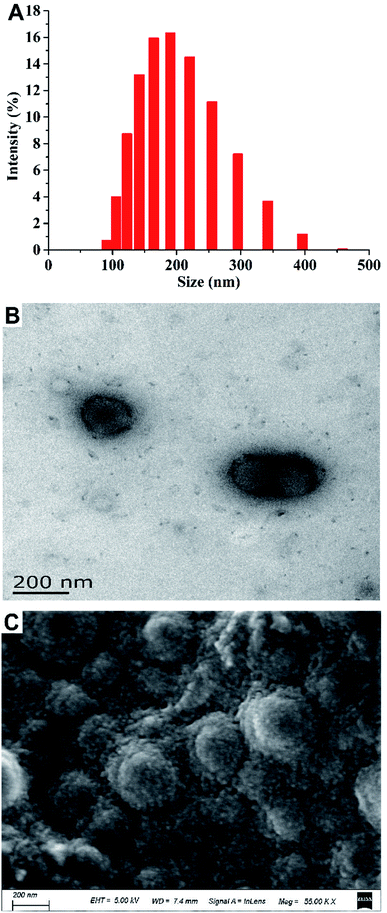
Fig. 3 shows the results of the particle size analysis and electron micrographs of the nanoparticles loaded with MC and CAPE. The particle size distribution is shown in Fig. 3A. Particles with sizes below 200 nm accounted for 58.88%, whereas particles with sizes within 200–458 nm accounted for 37.80% of the total. The hydrated particle size was 188.86 nm. Fig. 3B shows the transmission electron microscopy image of the nanoparticles after reconstitution. The particle size range was 173.78–104.56 nm, and the results were consistent with the particle size analysis results. Fig. 3C shows the SEM image of a nanoparticle sample after lyophilization. The particle size was generally uniform, but the surface of the particle was rough due to lyophilization. In summary, the average particle size of the composite nanoparticles was 188.86 nm, and the particle shape was regular.
3.3 Release kinetics of the nanoparticle
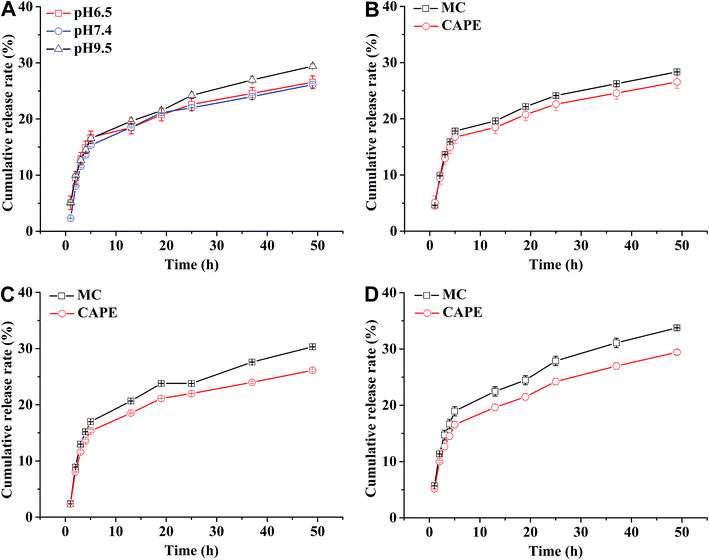
In general, the drug release process of the nanoparticles comprised three stages. Fig. 4A shows that during the first stage (0–5 h) of the release process (called the initial burst release stage11 with the highest rate), the drug molecules were adsorbed and were located adjacent to the surface of the nanoparticles that were first freely diffused into the solvent.20 The cumulative release rate of the drug in the three buffers reached 16.75 ± 1.11% (pH 6.5), 15.34 ± 0.22% (pH 7.4), and 16.54 ± 0.56 (pH 9.5). The second stage (13–50 h) is the release stage in which the diffusion of the drug molecule and PLGA disintegration of the polymer material coexist. During this stage, the fixed drug molecules were released together, and the drug release rate significantly decreased when compared with the first stage. The cumulative release rates of the drug in the three buffers reached 26.56 ± 1.15% (pH 6.5), 26.13 ± 0.17% (pH 7.4), and 29.42 ± 0.42% (pH 9.5). During the third stage, the polymer began to disintegrate, releasing the drug molecules during disintegration; the release rate indefinitely reached zero, eventually entering a nearly linear sustained-release state.11 In theory, there should also be a fourth stage,25 where the remaining PLGA polymer containing the drug molecule rapidly disintegrates and releases the last drug molecule at a higher release rate, but this was not apparent due to the limited time.The Fickian model was employed to formulate the delayed-release kinetic equations of the nanoparticles at pH 6.5, 7.4, and 9.5, which were termed as eqn (7), (8), and (9), respectively. Table 2 lists the results of the theoretical time required to reach 90% release rate and theoretical half-life of the nanoparticles calculated via the semiempirical formula. Under acidic conditions at pH 6.5, the release rate of the agent was lower, the theoretical half-life was approximately 10.0 d, and the theoretical time required for 90% drug release was approximately 44.1 d. The half-life at pH 9.5 and the theoretical time required to achieve a 90% drug release rate were 5.7 and 22.2 d, respectively. The higher pH might lead to the faster degradation of PLGA nanoparticles;9 further, the alkaline environment accelerated the release of drug molecules in the nanoparticles, while the acidic environment slowed down the release of drug molecules. The pH in the mulberry soil was mostly acidic, which was beneficial to the long-term efficacy of nanoparticles in mulberry soil.
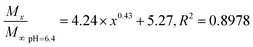 | (7) |
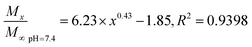 | (8) |
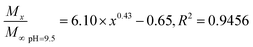 | (9) |
| pH of buffer | Half-life (days) | Time to reach 90% release rate (days) |
|---|---|---|
| 6.5 | 10.0 | 44.1 |
| 7.4 | 5.8 | 21.8 |
| 9.5 | 5.7 | 22.2 |
The nanoparticles in the acidic buffer were found to have a long half-life with a theoretical value of 10.0 d. The theoretical time required to release 90% of the agent under this condition was 44.1 d, but there was a burst in drug release within 0–5 h.11 The drug release rate under these three pH conditions was above 15%: the minimum value was 15.34 ± 0.22% (pH 7.4). This phenomenon was mainly caused by the excessive adsorption of drug molecules on the surface of the nanoparticles and adjacent surfaces. Therefore, the nanoparticles exhibited excellent sustained-release properties, and the half-life reached up to 10.0 d at pH 6.5.
Fig. 4B–D show the release curves of MC and CAPE at different pH values in the nanoparticles. The maximum cumulative release rates of MC (18.95 ± 0.82%) and CAPE (15.63 ± 2.27%) were achieved during the first stage under pH 9.5 and 6.5, respectively. After 13 h, the degradation stage of the PLGA particles was accompanied by the slow dissolution of MC and CAPE. At pH 9.5 and a release time of 50 h, the cumulative release rate of MC was 33.76 ± 0.01%, and the cumulative release rate of CAPE was 24.73 ± 0.64%. The cumulative release rates of MC were larger than those of CAPE at pH 6.5, 7.4, and 9.5. This phenomenon could be attributed to the different distributions of MC and CAPE during emulsification. The shorter alkyl chain of MC has a stronger affinity toward water than CAPE, which might result in different distributions during emulsification. The MC molecule adsorbed and embedded itself on the surface of PLGA nanoparticles more than that for CAPE. The release of the drug molecules adsorbed and embedded on the surface of the PLGA particles is faster,38 resulting in the larger cumulative release rate of MC within the same period of time during the first two stages of drug release. Therefore, combined with the multiple actions of nanoparticles, the inhibitory effect of nanoparticles containing MC and CAPE on R. solanacearum not only complicates inducing drug resistance, but also has the effect of batch release of drug molecules on the cell membrane of R. solanacearum.
3.4 Electron micrographs of R. solanacearum after nanoparticle treatment
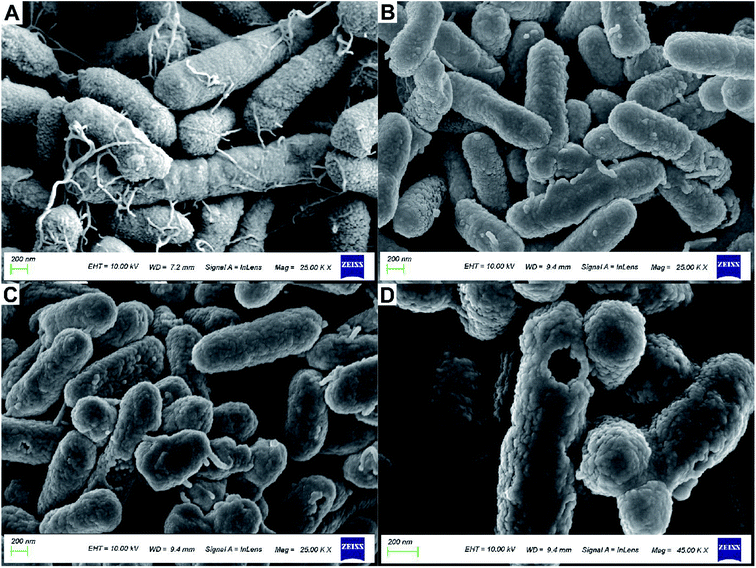
Fig. 5 shows the treatment of R. solanacearum with API and nanoparticles; morphological changes were observed by using SEM. Fig. 5A shows the morphology of R. solanacearum. It was found that the flagellar structure of the strains treated with API and nanoparticles was destroyed, while the untreated strains were unharmed and the flagella structure was clearly visible. Fig. 5B shows the observation of R. solanacearum strains treated with API alone. The rod shape of the bacterium was deformed, the shape of the strains was distorted, and the flagella were also damaged to some extent, but the cell surface was not obviously damaged. However, Fig. 5C shows many strains with damaged cell membranes and incomplete flagella. Further, a large circular cavity appeared on the surface of R. solanacearum, as shown in Fig. 5D, and the cell membrane structure was seriously damaged, which may be related to the perforation effect of nanoparticles on the cell membrane.39 Furthermore, the size of this cavity was similar to the hydrated particle size (188.86 nm), which confirmed that the particle might be involved. It was reported that PLGA-embedded drug particles might have certain adhesion to the surface of Candida albicans,40 which can explain the huge pores appearing on the surface of R. solanacearum, possibly due to the attachment of nanoparticles to the cell surface. The PLGA-embedded drug particles disintegrate and release a large number of drug molecules, which causes the formation of pores on the cell membrane. Therefore, the nanoparticle has a certain perforation effect on the membranes of R. solanacearum.3.5 Antibacterial activity of nanoparticles against R. solanacearum
Table 3 shows that under optimal conditions, the DL values of MC and CAPE in the nanoparticles were 14.50 ± 0.03% and 13.61 ± 0.27%, respectively. According to the literature,27 the EC50 value of the nanoparticles was calculated to be 0.285 mg mL−1. When a 50% inhibition rate was reached, the masses of MC and CAPE contained in 1 mL suspension containing the nanoparticle were 41.33 and 38.79 μg, respectively. Under the same bacteriostatic effect, the qualities of MC or CAPE alone were 7.5 and 4.3 times the masses of MC and CAPE in the nanoparticle, respectively, and the quality of the APIs was required to be 3 times that of the nanoparticle. Considering that the free bulk drug could facilitate the absorption of bacteria, its excellent efficacy needs to be based on the use of a higher amount of APIs, and the nanoparticle prepared in this study could considerably reduce the use of APIs, indicating its worthwhile technical economy. Furthermore, Fig. S1† shows the image of the susceptibility testing by the disk diffusion method. Evidently, the strains treated with the nanoparticles could not form visible plaques when compared with the other plates after incubation for 7 d. This phenomenon indicated that the novel nanoparticles loaded with MC and CAPE might possess the ability against the resistance of R. solanacearum, which could be supported by the data from recent studies according to which polymeric nanosystems might be a new prospect for treating multidrug-resistant bacteria.13 Therefore, nanoparticles can effectively improve the antibacterial properties of caffeic acid esters in the aqueous phase with less API.| Antibacterial agents | EC50 (mg mL−1) |
|---|---|
| MC | 0.310 |
| CAPE | 0.165 |
| APIs | 0.248 |
| Nanoparticles | 0.285 |
3.6 Expressions of pathogenicity-related genes during the early stage of infection after treatment
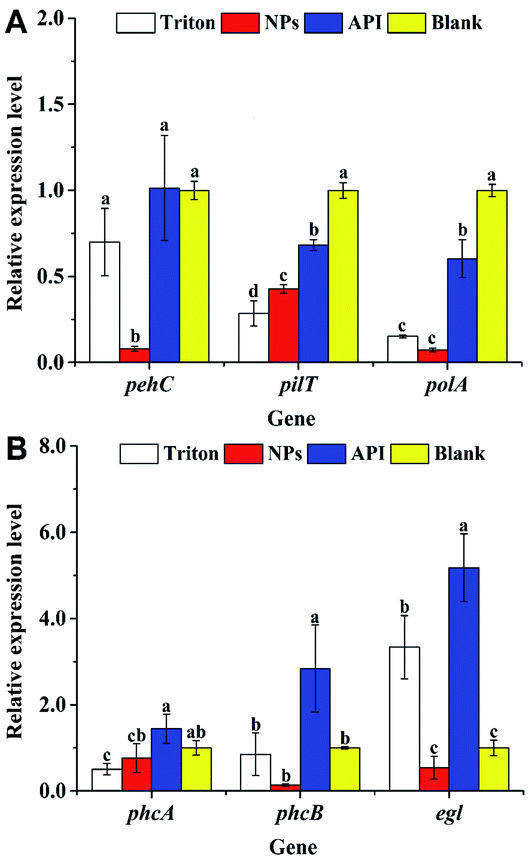
Fig. 6A shows the expression levels of pehC, pilT, and polA during the early stage of R. solanacearum infection. The expression levels of the polygalacturonase gene pehC41 in the API and blank groups were 12.8 and 12.7 times than those in the nanoparticle group, respectively. The expression levels in the nanoparticle group were significantly lower than those in the other groups (P < 0.05). The expression of the pilT gene, which is a twitching motility-related gene,42 also appeared to be inhibited, as shown in Fig. 6A; electron microscopy data confirmed that the number of bacterial flagella in the nanoparticle treatment group was significantly reduced. There was a significant difference in the expression levels in the nanoparticle and blank groups (P < 0.05). The expression levels of the blank and API groups were 2.3 and 1.6 times those of the nanoparticle group, respectively. In addition, the expression level of the DNA-polymerase-related gene polA43 is shown in Fig. 6A. There was a significant difference among the four treatment groups (P < 0.05), where the nanoparticle group had the lowest expression level. The expression levels of the blank and API groups were 13.7 and 8.3 times those of the nanoparticle group, respectively. Therefore, the nanoparticles could inhibit the expressions of pehC, pilT, and polA.3.7 Expressions of pathogenicity-related genes during the late infection stage after treatment
Fig. 6B shows the expression levels of phcA, phcB, and egl; phcA is the core regulatory gene of the phenotypic transformation system, which is the core system regulating the virulence and pathogenicity of the R. solanacearum infection44 and regulates the expression of virulence factors such as EPS, plant cell wall-degrading enzymes, motility, and other regulatory elements.45 As shown in Fig. 6B, the expression levels of phcA in the blank and API groups were 1.3 and 1.9 times those of the nanoparticle group, respectively. CAPE has a certain inhibitory effect on the various transcription factors (such as interleukin-6 and cyclooxygenase-2) and transcriptional activators in cells,46 which may lead to the decreased expression of phcA. Further, the decreased expression of phcA might be one of the reasons for the decreased motility and the formation of a biofilm.43 Furthermore, the phcB gene, which is involved in group signaling, was also inhibited, as shown in Fig. 6B. The expression of the phcB gene was significantly lower than those of the others (P < 0.05), resulting in the attenuation of the activation of the phcA expression,47 which led to the downregulation of phcA. The expression of the phcA gene has a regulatory effect on the endoglucanase-related gene egl, confirming the results of the decrease in the expression level of egl (Fig. 6B). Fig. 6B shows that in the presence of nanoparticles, the expression level of egl was at least 0.54 ± 0.3, which was 1/2 that of the blank group; however, there was no significant difference between the blank and nanoparticle groups (P > 0.05). The above results reveal that the nanoparticles loaded with MC and CAPE have a certain inhibitory effect on the egl, pehC, phcA, phcB, pilT, and polA pathogenicity genes of R. solanacearum. The expression levels of egl, pehC, phcA, phcB, pilT, and polA were downregulated to 1/6, 1/13, 1/2, 1/42, 1/2, and 1/8 of that of the drug treatment group, respectively.Fig. 7 shows the schematic diagram of the regulation of nanoparticles on some pathogenicity-related genes of R. solanacearum. When the nanoparticle was added to the bacterial suspension, the cell membrane and flagellum structure were severely damaged due to the multiple actions of the nanoparticles, which could be confirmed with the data shown in Fig. 5. The nanoparticles containing CAPE and MC continuously released CAPE and MC into the cells of R. solanacearum. The expression of phcA, an important regulatory factor in the phenotype transformation system, was inhibited; further, the expressions of the EGL and EPS phytotoxic factors were downregulated. In addition, in the detection of the pathogenicity-related genes of R. solanacearum before infecting the plants, it was found that the expression level of the transcriptional activator hrpB in the T3SS secretion system of R. solanacearum was extremely low to be used to calculate the relative expression. However, it has been reported that the expression of hrpB can be induced when R. solanacearum is cocultured with the plant suspension,48 which might be the reason for the low expression of hrpB. In summary, the nanoparticles loaded with MC and CAPE have a certain inhibitory effect on the phenotypic transformation system and the mobilization of R. solanacearum, and it is expected to develop into a new type of antibacterial agent.
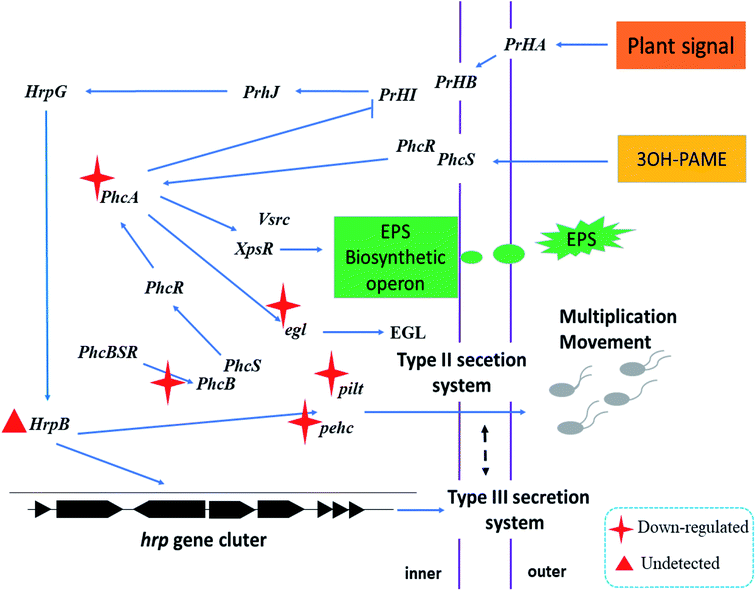 | ||
| Fig. 7 Schematic of the regulation of nanoparticles loaded with MC and CAPE on some pathogenicity-related genes of R. solanacearum. | ||
4. Conclusions
In this paper, PLGA nanoparticles loaded with MC and CAPE were successfully prepared by a single emulsion/solvent evaporation method. The optimum conditions for obtaining the highest DL (28.33 ± 0.03%) were as follows: 3% PVA with a ratio of the organic phase to aqueous phase of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 at 325 W under sonication for 2 min. Using the Fickian diffusion model to fit the sustained-release curve, the half-lives at pH 6.5, 7.4, and 9.5 were calculated to be 10.0, 5.8, and 5.7 d, respectively, indicating worthwhile sustained-release properties. The nanoparticles loaded with MC and CAPE were used for the antibacterial experiment of R. solanacearum, and the EC50 value was 0.285 mg mL−1. Under the same antibacterial effect, the amount of nanoparticles used was 1/3 of the drug dose of API. The expression levels of the pathogenicity-related genes, namely, egl, pehC, phcA, phcB, pilT, and polA, of R. solanacearum were downregulated by RT-PCR to 1/6, 1/13, 1/2, 1/42, 1/2, and 1/8 of the drug-treated group, respectively, which have significant inhibitory effects on the major transcription factor phcA in the phenotype switching system. Therefore, the nanoparticles loaded with MC and CAPE show good sustained-release properties and good antibacterial effects, as observed by the disruption of the cytomembrane and suppression of pathogenicity-related gene expressions in R. solanacearum, which has the potential to be considered as a novel alternative agent for replacing traditional chemical pesticides.
2 at 325 W under sonication for 2 min. Using the Fickian diffusion model to fit the sustained-release curve, the half-lives at pH 6.5, 7.4, and 9.5 were calculated to be 10.0, 5.8, and 5.7 d, respectively, indicating worthwhile sustained-release properties. The nanoparticles loaded with MC and CAPE were used for the antibacterial experiment of R. solanacearum, and the EC50 value was 0.285 mg mL−1. Under the same antibacterial effect, the amount of nanoparticles used was 1/3 of the drug dose of API. The expression levels of the pathogenicity-related genes, namely, egl, pehC, phcA, phcB, pilT, and polA, of R. solanacearum were downregulated by RT-PCR to 1/6, 1/13, 1/2, 1/42, 1/2, and 1/8 of the drug-treated group, respectively, which have significant inhibitory effects on the major transcription factor phcA in the phenotype switching system. Therefore, the nanoparticles loaded with MC and CAPE show good sustained-release properties and good antibacterial effects, as observed by the disruption of the cytomembrane and suppression of pathogenicity-related gene expressions in R. solanacearum, which has the potential to be considered as a novel alternative agent for replacing traditional chemical pesticides.
Conflicts of interest
The authors have declared no conflicts of interest.Acknowledgements
This study was financially supported by the Key Research and Development Program (Modern Agriculture) of Zhenjiang City (NY2017010), the National Modern Agricultural Industry Technology System of China (CARS-18-ZJ0305), the 333 High-level Talent Training Project of Jiangsu Province (BRA2019281), the Qing Lan Project of Jiangsu Province (Year of 2019), and the Shen Lan Young scholars program of Jiangsu University of Science and Technology (Year 2015).References
- K. Fenner, S. Canonica, L. P. Wackett and M. Elsner, Evaluating pesticide degradation in the environment: Blind spots and emerging opportunities, Science, 2013, 341, 752–758 CrossRef CAS PubMed.
- M. E. Delorenzo, G. I. Scott and P. E. Ross, Toxicity of pesticides to aquatic microorganisms: A review, Environ. Toxicol. Chem., 2001, 20, 84–89 CrossRef CAS.
- T. T. Iyaniwura, Non-target and environmental hazards of pesticides, Rev. Environ. Health, 1991, 9, 161–176 CAS.
- A. F. Hernandez, T. Parron, A. M. Tsatsakis, M. Requena, R. Alarcon and O. Lopez-Guarnido, Toxic effects of pesticide mixtures at a molecular level: Their relevance to human health, Toxicology, 2013, 307, 136–145 CrossRef CAS PubMed.
- B. Kim, E. French, D. Caldwell, E. J. Harrington and A. S. Iyer-Pascuzzi, Bacterial wilt disease: Host resistance and pathogen virulence mechanisms, Physiol. Mol. Plant Pathol., 2016, 95, 37–43 CrossRef.
- J. Chen, Y. Yu, S. Li and W. Ding, Resveratrol and coumarin: novel agricultural antibacterial agent against Ralstonia solanacearum in vitro and in vivo, Molecules, 2016, 21, 1–18 CrossRef PubMed.
- J. Chen, F. Wang, Q. Liu and J. Du, Antibacterial polymeric nanostructures for biomedical applications, Chem. Commun., 2014, 50, 14482–14493 RSC.
- F. Danhier, E. Ansorena, J. M. Silva, R. Coco, A. Le Breton and V. Préat, PLGA-based nanoparticles: An overview of biomedical applications, J. Controlled Release, 2012, 161, 505–522 CrossRef CAS PubMed.
- V. Singh, S. Singh, S. Das, A. Kumar, W. T. Self and S. Seal, A facile synthesis of PLGA encapsulated cerium oxide nanoparticles: release kinetics and biological activity, Nanoscale, 2012, 4, 2597–2605 RSC.
- C. Yang and D. Merlin, Can naturally occurring nanoparticle-based targeted drug delivery effectively treat inflammatory bowel disease?, Expert Opin. Drug Delivery, 2019, 1–4 Search PubMed.
- T. Arasoglu, S. Derman and B. Mansuroglu, Comparative evaluation of antibacterial activity of caffeic acid phenethyl ester and PLGA nanoparticle formulation by different methods, Nanotechnology, 2016, 27, 25103 CrossRef PubMed.
- R. Y. Pelgrift and A. J. Friedman, Nanotechnology as a therapeutic tool to combat microbial resistance, Adv. Drug Delivery Rev., 2013, 65, 1803–1815 CrossRef CAS PubMed.
- X. Ding, A. Wang, W. Tong and F. Xu, Biodegradable antibacterial polymeric nanosystems: A New hope to cope with multidrug-resistant bacteria, Small, 2019, 15, 1900999 CrossRef PubMed.
- B. Syed, M. N. N. Prasad, M. K. Kumar and S. Satish, Bioconjugated nano-bactericidal complex for potent activity against human and phytopathogens with concern of global drug resistant crisis, Sci. Total Environ., 2018, 637, 274–281 CrossRef PubMed.
- G. Huet, Breeding for resistances to Ralstonia solanacearum, Front. Plant Sci., 2014, 5, 1–5 Search PubMed.
- G. Jiang, Z. Wei, J. Xu, H. Chen, Y. Zhang, X. She, A. P. Macho, W. Ding and B. Liao, Bacterial wilt in China: History, current status, and future perspectives, Front. Plant Sci., 2017, 8, 1–10 Search PubMed.
- J. Mansfield, S. Genin, S. Magori, V. Citovsky, M. Sriariyanum, P. Ronald, M. Dow, V. Verdier, S. V. Beer, M. A. Machado, I. Toth, G. Salmond and G. D. Foster, Top 10 plant pathogenic bacteria in molecular plant pathology, Mol. Plant Pathol., 2012, 13, 614–629 CrossRef PubMed.
- H. S. Addy, A. Ahmed, K. Takeru, F. Makoto and Y. Takashi, Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by φRSM filamentous phages, Phytopathology, 2012, 102, 469–477 CrossRef CAS PubMed.
- G. D. Wright, Opportunities for natural products in 21st century antibiotic discovery, Nat. Prod. Rep., 2017, 34, 694–701 RSC.
- S. Derman, Caffeic acid phenethyl ester loaded PLGA nanoparticles: Effect of various process parameters on reaction yield, encapsulation efficiency, and particle size, J. Nanomater., 2015, 2015, 1–12 CrossRef.
- Y. Xu, S. Sheng, X. Liu, C. Wang, W. Xiao, J. Wang and F. A. Wu, Cooperative reinforcement of ionic liquid and reactive solvent on enzymatic synthesis of caffeic acid phenethyl ester as an in vitro inhibitor of plant pathogenic bacteria, Molecules, 2017, 22, 72 CrossRef PubMed.
- J. Wang, S. S. Gu, H. S. Cui, X. Y. Wu and F. A. Wu, A novel continuous flow biosynthesis of caffeic acid phenethyl ester from alkyl caffeate and phenethanol in a packed bed microreactor, Bioresour. Technol., 2014, 158, 39–47 CrossRef CAS PubMed.
- S. S. Wang, Z. J. Li, S. Sheng, F. A. Wu and J. Wang, Microfluidic biocatalysis enhances the esterification of caffeic acid and methanol under continuous-flow conditions, J. Chem. Technol. Biotechnol., 2016, 91, 555–562 CrossRef CAS.
- S. Derman, Z. A. Mustafaeva, E. S. Abamor, M. Bagirova and A. Allahverdiyev, Preparation, characterization and immunological evaluation: canine parvovirus synthetic peptide loaded PLGA nanoparticles, J. Biomed. Sci., 2015, 22, 89 CrossRef PubMed.
- A. Budhian, S. J. Siegel and K. I. Winey, Controlling the in vitro release profiles for a system of haloperidol-loaded PLGA nanoparticles, Int. J. Pharm., 2008, 346, 151–159 CrossRef CAS PubMed.
- C. F. Yang, Y. Zhou, Y. Zheng, C. L. Li, S. Sheng, J. Wang and F. A. Wu, Enzymatic modification of chitosan by cinnamic acids: Antibacterial activity against Ralstonia solanacearum, Int. J. Biol. Macromol., 2016, 87, 577–585 CrossRef CAS PubMed.
- K. A. Nath, A. J. Croatt, S. Likely, T. W. Behrens and D. Warden, Renal oxidant injury and oxidant response induced by mercury, Kidney Int., 1996, 50, 1032–1043 CrossRef CAS PubMed.
- S. Sheng, C. W. Liao, Y. Zheng, Y. Zhou, Y. Xu, W. M. Song, P. He, J. Zhang and F. A. Wu, Candidate chemosensory genes identified in the endoparasitoid Meteorus pulchricornis (Hymenoptera: Braconidae) by antennal transcriptome analysis, Comp. Biochem. Physiol., Part D: Genomics Proteomics, 2017, 22, 20–31 CAS.
- A. Sahin, F. Spiroux, I. Guedon, F. B. Arslan, E. T. Sarcan, T. Ozkan, N. Colak, S. Yuksel, S. Ozdemir and B. Ozdemir, Using PVA and TPGS as combined emulsifier in nanoprecipitation method improves characteristics and anticancer activity of ibuprofen loaded PLGA nanoparticles, Pharmazie, 2017, 72, 525–528 CAS.
- G. Gaucher, M. Poreba, F. Ravenelle and J. C. Leroux, Poly(N-vinyl-pyrrolidone)-block-poly(D,L-lactide) as polymeric emulsifier for the preparation of biodegradable nanoparticles, J. Pharm. Sci., 2007, 96, 1763–1775 CrossRef CAS PubMed.
- S. Freitas, G. Hielscher, H. P. Merkle and B. Gander, Continuous contact- and contamination-free ultrasonic emulsification—a useful tool for pharmaceutical development and production, Ultrason. Sonochem., 2006, 13, 76–85 CrossRef CAS PubMed.
- K. C. Guedes Silva and A. C. Kawazoe Sato, Sonication technique to produce emulsions: The impact of ultrasonic power and gelatin concentration, Ultrason. Sonochem., 2019, 52, 286–293 CrossRef PubMed.
- S. M. M. Modarres-Gheisari, R. Gavagsaz-Ghoachani, M. Malaki, P. Safarpour and M. Zandi, Ultrasonic nano-emulsification – A review, Ultrason. Sonochem., 2019, 52, 88–105 CrossRef CAS PubMed.
- X. Luo, J. Cao, H. Yan, H. Gong, H. Yin and L. He, Study on separation characteristics of water in oil (W/O) emulsion under ultrasonic standing wave field, Chem. Eng. Process., 2018, 123, 214–220 CrossRef CAS.
- X. Song, Y. Zhao, S. Hou, F. Xu, R. Zhao, J. He, Z. Cai, Y. Li and Q. Chen, Dual agents loaded PLGA nanoparticles: Systematic study of particle size and drug entrapment efficiency, Eur. J. Pharm. Biopharm., 2008, 69, 445–453 CrossRef CAS PubMed.
- W. Hai, Z. Ying, W. Yan, H. Yu-Lin, N. Kaihui, N. Guangjun and C. Hao, Enhanced anti-tumor efficacy by co-delivery of doxorubicin and paclitaxel with amphiphilic methoxy PEG-PLGA copolymer nanoparticles, Biomaterials, 2011, 32, 8281–8290 CrossRef PubMed.
- M. K. Pyo, Y. Lee and H. S. Yun-Choi, Anti-platelet effect of the constituents isolated from the barks and fruits of Magnolia obovata, Arch. Pharmacal Res., 2002, 25, 325–328 CrossRef CAS PubMed.
- P. Rafiei and A. Haddadi, Docetaxel-loaded PLGA and PLGA-PEG nanoparticles for intravenous application: Pharmacokinetics and biodistribution profile, Int. J. Nanomed., 2017, 12, 935–947 CrossRef CAS PubMed.
- Y. J. Tang, J. M. Ashcroft, D. Chen, G. Min, C. Kim, B. Murkhejee, C. Larabell, J. D. Keasling and F. F. Chen, Charge-associated effects of fullerene derivatives on microbial structural integrity and central metabolism, Nano Lett., 2007, 7, 754–760 CrossRef CAS PubMed.
- L. Roque, N. Duarte, M. R. Bronze, C. Garcia, J. Alopaeus, J. Molpeceres, E. Hagesaether, I. Tho, P. Rijo and C. Reis, Development of a bioadhesive nanoformulation with Glycyrrhiza glabra L. extract against Candida albicans, Biofouling, 2018, 34, 880–892 CrossRef CAS PubMed.
- E. T. Gonzalez and C. Allen, Characterization of a Ralstonia solanacearum operon required for polygalacturonate degradation and uptake of galacturonic acid, Mol. Plant-Microbe Interact., 2003, 16, 536–544 CrossRef CAS PubMed.
- N. Singh, T. Phukan, P. L. Sharma, K. Kabyashree, A. Barman, R. Kumar, R. V. Sonti, S. Genin and S. K. Ray, An innovative root inoculation method to study Ralstonia solanacearum pathogenicity in tomato seedlings, Phytopathology, 2018, 108, 436–442 CrossRef CAS PubMed.
- W. Raza, J. Wang, Y. Wu, N. Ling, Z. Wei, Q. Huang and Q. Shen, Effects of volatile organic compounds produced by Bacillus amyloliquefaciens on the growth and virulence traits of tomato bacterial wilt pathogen Ralstonia solanacearum, Appl. Microbiol. Biotechnol., 2016, 100, 7639–7650 CrossRef CAS PubMed.
- Y. Hikichi, Y. Mori, S. Ishikawa, K. Hayashi, K. Ohnishi, A. Kiba and K. Kai, Regulation Involved in colonization of intercellular spaces of host plants in Ralstonia solanacearum, Front. Plant Sci., 2017, 8 DOI:10.3389/fpls.2017.00967.
- R. P. Garg, J. Huang, W. Yindeeyoungyeon, T. P. Denny and M. A. Schell, Multicomponent transcriptional regulation at the complex promoter of the exopolysaccharide I biosynthetic operon of Ralstonia solanacearum, J. Bacteriol., 2000, 182, 6659–6666 CrossRef CAS PubMed.
- G. Murtaza, A. Sajjad, Z. Mehmood, S. H. Shah and A. R. Siddiqi, Possible molecular targets for therapeutic applications of caffeic acid phenethyl ester in inflammation and cancer, J. Food Drug Anal., 2015, 23, 11–18 CrossRef CAS PubMed.
- S. M. Brumbley, B. F. Carney and T. P. Denny, Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional regulator, J. Bacteriol., 1993, 175, 5477–5487 CrossRef CAS PubMed.
- D. Aldon, B. Brito, C. Boucher and S. Genin, A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes, EMBO J., 2014, 19, 2304–2314 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09441e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |