Open Access Article
Open Access ArticleIn vitro assessment of the bioactivities of sericin protein extracted from a bacterial silk-like biopolymer
Esamil M. El-Fakharany *a,
Gadallah M. Abu-Elreeshb,
Elbadawy A. Kamouncd,
Sahar Zakib and
Desouky A. Abd-EL-Haleem*b
*a,
Gadallah M. Abu-Elreeshb,
Elbadawy A. Kamouncd,
Sahar Zakib and
Desouky A. Abd-EL-Haleem*b
aProteins Research Dep., Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), New Borg Al-Arab City 21934, Alexandria, Egypt. E-mail: esmailelfakharany@yahoo.co.uk
bEnvironmental Biotechnology Dep., Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), New Borg Al-Arab City 21934, Alexandria, Egypt. E-mail: abdelhaleemm@yahoo.de
cPolymeric Materials Research Dep., Advanced Technology and New Materials Research Institute (ATNMRI), City of Scientific Research and Technological Applications, New Borg Al-Arab City 21934, Alexandria, Egypt
dNanotechnology Research Center (NTRC), The British University in Egypt (BUE), El-Sherouk City, Cairo 11837, Egypt
First published on 31st January 2020
Abstract
Sericin is one of the main components of silk proteins, which has numerous biomedical applications because of its antioxidant, anticancer and antimicrobial properties. We recently isolated and characterized a novel silk-like protein named BNES. It is of non-animal origin and is like a bacterial polymeric silk. Sericin is a very popular protein compound that is effective in treating cancerous tumors. The process of extracting it from natural silk produced by silkworms or spiders is both complex and expensive. From the published scientific literature, it has been shown that sericin has not been previously extracted from a bacterial source. In the present study, sericin was extracted from bacteria capable of producing a biopolymer named BNES whose chemical composition is like that of natural silk and its bio-therapeutic effects were evaluated for the first time. The antioxidant activity of BNES measured by DPPH and ABTS assays showed IC50 values of 0.38 and 0.41 mg mL−1, respectively. BNES displayed satisfactory cytotoxic effect against four cancer cell lines, including Huh-7, Caco-2, MCF-7 and A549 cells, with IC50 values in the ranges of ca. 0.62 ± 0.17, 0.72 ± 0.27, 0.76 ± 0.36 and 0.83 ± 0.31 mg mL−1, respectively, after 24 h of treatment and 0.51 ± 0.22, 0.49 ± 0.19, 0.41 ± 0.25 and 0.55 ± 0.38, respectively, after 48 h of treatment, without affecting normal cells (WI38 cells). The antitumor activity of BNES was established to be an apoptosis-dependent mechanism determined via cellular morphology alterations, cell cycle arrest in the sub-G1 phase and nuclear staining with highly fluorescent fragments. The antimicrobial effects of BNES were examined with yeast and Gram-negative and Gram-positive bacteria. The results confirmed its antimicrobial activity against all tested organisms at concentrations of up to 1.33 mg mL−1. The competitive advantage of the bacterial sericin BNES over sericin extracted from spider or silkworm sources is that it can be produced in very large quantities through large-scale bio-fermenters, which reduces the expected cost of production, in addition to having sustainable bacterial production source.
1. Introduction
Silk is a natural fibrous protein obtained mainly from animal origins. Silk has been playing a significant role in medical applications and economic life, such as in the textile industry from four centuries ago. Silk consists mostly of two types of protein in a core–shell type shape: fibroin comprises the core and is surrounded by the glue-like sericin, which cements itself to the fibroin fibers. Fibroin is composed mainly of three proteins: two proteins linked together, called heavy (H) and light (L) chain proteins, and a small glycoprotein called P25.1 The H-chain protein is hydrophobic, while the L-chain protein demonstrates hydrophilic features and is comparatively elastic with slight or no crystallinity; P25 protein helps retain the combination between silk ingredients through non-covalent connections to the covalent bonds of H-chain and L-chain proteins. The molar ratio of H-chain![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L-chain
L-chain![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P25 proteins in silkworm is 6
P25 proteins in silkworm is 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2,3 Silk fibroin protein can be applied as a biocompatible material in numerous forms, e.g. gels, membranes, sponges, scaffolds and powders.4,5
1.2,3 Silk fibroin protein can be applied as a biocompatible material in numerous forms, e.g. gels, membranes, sponges, scaffolds and powders.4,5
Silk fibroin is characterized by many useful features, including biocompatibility, biodegradability, self-assembly, and strong mechanical properties, which can be improved via various modifications. These chemical modifications can enhance several applications, such as cell or polymer attachments to the silk, the attachment of growth factors, and tissue engineering.6 These applications can also involve immobilization matrices for many enzymes and proteins, burn-wound dressings, nets for use in tissue engineering, vascular prostheses and structural implants.7–9
Sericin is a natural hydrophilic biopolymer with a high content of serine, about one third of the total content, and is composed of 18 types of amino acids, most of which possess strong polar side groups, e.g. carboxyl, hydroxyl and amino groups.10 The glycoprotein sericin possesses several biological properties, including antioxidation, immunomodulation, and inhibition of tyrosinase and elastase activities.11,12 Recently, sericin was found to reduce reactive oxygen species (ROS), protect normal cells from UVB radiation oxidative damage and H2O2, inhibit cancer progression and be an anti-bacterial agent.13,14 The exploitation of purified proteins or their extracts as antioxidant candidates has attracted particular attention. Various proteins, from dairy products such as milk or from other nutriceuticals such as soy, fish, corn and beans, have been demonstrated to have antioxidant properties.14–17 Many reports have revealed that peptides and protein hydrolysate could be replaced and used as potent alternative antioxidant agents.18–21 In addition to its antioxidant effect, sericin protein was found to have potential anticancer activity against many types of cancer cells. Sericin was found to induce the apoptotic effect in SW480 colon cancer cells by enhancing caspase-3 activity and reducing Bcl-2 expression, resulting in cell cycle arrest at the S phase.22 Also, sericin was found to suppress DMBA–TPA of skin tumorigenesis in a mouse model23 and colon tumorigenesis in animal tests.24,25 The antibacterial activity of silk sericin and fibroin proteins against Gram-negative and Gram-positive bacterial strains has been examined in many studies. Senakoon et al.26 demonstrated that the Gram-negative Escherichia coli is more sensitive to degummed eri sericin than the Gram-positive Staphylococcus aureus using the disc diffusion assay and critical-dilution micro assay methods. Basal et al.27 revealed that silk fibroin blend films were found to have antibacterial effect against E. coli and Staphylococcus epidermidis using disc-diffusion and turbidity measurement methods.
BNES silk-like protein is a silk of non-animal origin, as it is produced by a petroleum-originated bacteria called Bacillus sp. strain NE.28 Increased use of bacterial polymeric silk-like proteins is predicted in the near future for many reasons such as quick secretion, low cost-effect and simple product recovery, leading to production of a huge amount of bacterial sericin. In general, production of sericin creates industrial byproducts that are removed in waste products, causing environmental pollution and huge waste of natural resources. So, bacterial sericin will need to be recovered and recycled in adequate manners to provide social and significant economic benefits. There have been several studies investigating the biological activities of isolated sericin of animal origin, but no studies have presented extraction of sericin from a bacterial origin. In the current study, we attempted to investigate the biological activities of a novel bacterial sericin as a beneficial biomaterial to save our resources and protect the environment. The studied biological activities included different effects of the bacterial sericin as an antioxidant against DPPH and ABTS radicals as well as its anticancer effect against different carcinoma cell lines. The antimicrobial activities of BNES sericin were also investigated.
2. Experimental
2.1. Production of BNES
For the production of BNES, Bacillus sp. strain NE was cultured as described previously by Kamoun et al.28 Briefly, after the inoculation of the Bacillus sp. strain NE in a flask containing 100 mL of supplementary nutrient broth (SNB) medium, the bacterial culture was incubated for 24 h at 30 °C and shaken at 150 rpm. About 5 mL of bacterial culture was cultured in another flask containing 100 mL of TSM production medium. Afterward, exo-biopolymer was formed by shaking the culture at ∼150 rpm at 30 °C for 5 days. Then, cell free exo-biopolymer was obtained by centrifugation of the culture for 30 min at 8000 rpm. Bacterial polymeric silk-like supernatant was concentrated to 0.2 volumes using a rotary evaporator and dialyzed against deionized water at 4 °C overnight. After adding 30 mL of cold ethanol to 10 mL of the dialyzed solution, the precipitate was re-suspended in deionized water. Then, 10% cetylpyridinium chloride (CPC) was mixed into the broth with gentle stirring. The broth was left for several hours at room conditions and then centrifuged for 30 min at 5000 rpm to collect the precipitate. After dissolving the precipitate in 0.5 M sodium chloride, three volumes of cold ethanol were added to obtain further solids. Finally, the broth was rinsed three times with 75% ethanol and then lyophilized to obtain purified bacterial BNES.2.2. Preparation of bacterial BNES sericin (BNES)
BNES was obtained from bacterial polymeric BNES as follows: 1 g of lyophilized BNES was chopped, added to 250 mL of Na2CO3 (0.5 M, pH ∼ 11–12), and boiled at 90–95 °C for one hour. The mixture was centrifuged and filtrated to eliminate solid particles and bacterial fibroin. The bacterial filtrate was dialyzed with Milli-Q water for 3 days, then lyophilized and kept at −20 °C for additional use.2.3. Total antioxidant activity of the BNES
The total antioxidant capacity of BNES samples was assessed by phosphomolybdenum procedure, as reported by Prieto et al.29 BNES solutions at concentrations of 0.075, 0.15, 0.3, 0.6 and 1.2 mg mL−1 were added separately in triplicate to 1 mL of phosphomolybdenum reagent solution (4 mM of ammonium molybdate, 28 mM of sodium phosphate and 0.6 M sulphuric acid). All solutions were kept at 95 °C for 90 min and then each sample was spectrophotometrically measured at 635 nm (BMG LabTech, Germany), with ascorbic acid used as the standard. The total antioxidant activity (%) was stated as ascorbic acid equivalents (mg g−1 of sample).2.4. Free radical scavenging activity using DPPH method
In order to examine the antioxidant activity of BNES, the free radical scavenging activity was measured using DPPH method according to the procedure of Blios.30 Sample solutions of BNES at ratios of 0.075, 0.15, 0.3, 0.6 and 1.2 mg mL−1 were added separately in triplicate to 5 mL of 0.1 mM DPPH dissolved in 70% ethanol and then left to stand in the dark in ordinary conditions. After incubation for 20 min, the absorbance of each sample was detected spectrophotometrically at 517 nm. The blank reading was considered the control and the activity of DPPH radical scavenging was calculated by the following formula:| % DPPH free radical scavenging activity = [(X − Y)/X] ×100 | (1) |
2.5. Antioxidant activity using ABTS method
The antioxidant activity of BNES was evaluated according to the procedure of Re et al.31 Both ABTS (7 mM) and ammonium persulfate (APS, 2.45 mM) were diluted in deionized water and mixed in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5. The mixture was kept in dark conditions for 12–16 h at room temperature before being used to produce ABTS˙+ (ABTS radical cation). The ABTS˙+ solution was diluted in 70% ethanol to give an absorbance of 0.70 ± 0.3 at 734 nm. BNES samples at concentrations of 0.075, 0.15, 0.3, 0.6 and 1.2 mg mL−1 were added separately into 1 mL of ABTS˙+ solution in triplicate. Each sample was vortexed for 15 s and incubated in a dark condition at 17 °C for 15 min. After incubation, each sample was spectrophotometrically measured at 734 nm. The scavenging activity of ABTS radicals was calculated according to the following equation:
0.5. The mixture was kept in dark conditions for 12–16 h at room temperature before being used to produce ABTS˙+ (ABTS radical cation). The ABTS˙+ solution was diluted in 70% ethanol to give an absorbance of 0.70 ± 0.3 at 734 nm. BNES samples at concentrations of 0.075, 0.15, 0.3, 0.6 and 1.2 mg mL−1 were added separately into 1 mL of ABTS˙+ solution in triplicate. Each sample was vortexed for 15 s and incubated in a dark condition at 17 °C for 15 min. After incubation, each sample was spectrophotometrically measured at 734 nm. The scavenging activity of ABTS radicals was calculated according to the following equation:| % ABTS˙+ scavenging activity = [1 − (A/B)] × 100 | (2) |
2.6. In vitro cell culture studies
2.7. Antimicrobial activity
The antimicrobial effect of BNES polymer was estimated on Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 15442), Salmonella typhimurium (ATCC 14028), Staphylococcus aureus (ATCC 29213), Bacillus cereus (ATCC 33019), Enterococcus faecalis (ATCC 29212) and Candida albicans (ATCC 10231) by turbidity measurement.35 Sterile Erlenmeyer flasks (250 mL), each containing 30 mL nutrient broth, were inoculated with 0.25 mL of freshly prepared bacterial suspension from each bacterial strain in order to maintain initial bacterial concentrations of 108 CFU mL−1. Subsequently, they were supplemented with different concentrations of BNES polymer (0.08, 0.16, 0.25, 0.33, 0.66, 1.0 and 1.33 mg mL−1) and then incubated in an orbital shaker at 150 rpm at 30.5 ± 2.5 °C for 24 h. Microbial growth was measured as the increase in absorbance at 600 nm as determined by a T60 UV/VIS spectrophotometer. Experiments included a positive control (flask containing polymer and nutrient media, devoid of inoculum) and a negative control (flask contains inoculum and nutrient media, devoid of polymer). The negative controls indicated a microbial growth profile in the absence of the polymer. The absorbance values for positive controls were subtracted from the experimental values (flasks containing nutrient media, inoculum and polymer).2.8. Statistical analysis
All data (n = 3) of cell viability were expressed as the mean ± standard error of the mean (SEM) from three independent experimental measurements. The statistical significance was indicated by student's t-test and McNemar's test. The (p)-values were considered statistically significant at p < 0.05.3. Results and discussion
3.1. Production of bacterial sericin
The bacteria strain Bacillus sp. strain NE isolated from petroleum oil has the ability to form an exo-biopolymer extract named BNES. This extract was identified as a polymeric silk-like protein using exo-polymer physical and chemical characterizations, as discussed in our previously published work.28 Most published studies in this area have focused on the production of biosynthetic spider-silk using genetically engineered bacteria,36–38 whereas the present bacterial silk-like protein is produced by a wild-type bacterial strain. Therefore, it was very important to determine and assess its biological activities for medical applications. In a recent study,28 BNES biopolymer was characterized. It is composed of amino acids with aliphatic and aromatic hydrophobic side chains, with the major repetitive amino acids of the silk-like protein including proline 17%, glycine 14%, alanine 27%, and aspartic acid 7%. BNES can be dissolved in numerous aqueous solutions under very harsh environments by heating and ultrasonic agitation at a very small concentration of protein owing to its high viscosity and amino acid composition. In addition, it has a high stability resistant to thermal degradation conditions as analyzed by TGA, which exhibited that BNES thermally degraded in three thermal degradation regions observed at 140–373 °C. Furthermore, we demonstrated that BNES has a broad smear of proteins on SDS-PAGE related to fibroin (bands at 30, 35 and 400 kDa) and sericin (bands at 6.5, 16, 66 and 95 kDa).283.2. Assessment of the total antioxidant, DPPH and ABTS activities
The total antioxidant activity of BNES was tested using a phosphomolybdenum method; the data is summarized and shown in Table 1. The assay indicated that BNES has a high scavenging activity of 62.42 ± 0.78 mg g−1 ascorbic acid equivalent for 1.2 mg of BNES, which is comparable to the well-known antioxidant standard ascorbic acid.| Sample concentration (mg mL−1) | Total antioxidant (mg g−1) | DPPH radical scavenging activity (%) | ABTS radical scavenging activity (%) |
|---|---|---|---|
| a All values are expressed as mean ± SEM. | |||
| 0.075 | 20.82 ± 0.97 | 21.78 ± 0.42 | 9.33 ± 1.53 |
| 0.15 | 30.63 ± 0.96 | 46.26 ± 0.53 | 19.47 ± 1.21 |
| 0.3 | 39.34 ± 1.05 | 71.28 ± 0.48 | 34.34 ± 1.25 |
| 0.6 | 49.54 ± 1.09 | 73.08 ± 0.57 | 51.96 ± 1.71 |
| 1.2 | 62.42 ± 0.78 | 80.26 ± 0.45 | 72.59 ± 2.34 |
| IC50 (mg mL−1) | — | 0.38 | 0.41 |
The radical scavenging was investigated based on the DPPH assay procedure. Our results indicate that the scavenging capacity (%) on the DPPH radical was clearly improved by increasing the concentration of BNES from 0.075 mg mL−1 to 1.2 mg mL−1 protein. The inhibition of DPPH (%) free radicals ranged from 21.78 ± 0.42% to 80.26 ± 0.45% (in 0.075 mg mL−1 to 1.2 mg mL−1 of BNES) with an IC50 value of 0.38 mg mL−1 (Table 1). The ABTS radical scavenging method is applicable to both hydrophilic and lipophilic samples and is widely used for assessment of antioxidant capacity. The radical scavenging activity was evaluated by ABTS assay. As shown in Table 1, the radical scavenging activity (%) of BNES drastically increased with increase of the concentration of BNES. The inhibition of ABTS (%) free radicals ranged from 9.33 ± 1.53% to 72.59 ± 2.34% (in 0.075 mg mL−1 to 1.2 mg mL−1 of BNES) with an IC50 value of 0.41 mg mL−1. Meanwhile, the extracted sericin of bacterial origin has high antioxidant activity similar to sericin isolated from animal sources. Antioxidant properties, particularly radical scavenging activities, are of importance to remove toxic radicals in food and biological systems. In general, the antioxidant properties of proteins depend on their amino acid conformation and structure. The antioxidant potential of silk sericin is associated with its amino acid composition, especially the –OH groups of threonine and serine and the electron donating groups of the aromatic amino acids (histidine, phenylalanine, tyrosine and tryptophan),28 in addition to the action of secondary metabolites, such as flavonoids and polyphenols, which also contribute.12 Furthermore, alteration in the pH of the protein during the extraction process, as when using a basic or acidic environment, along with the heating process, changes the amino acids' ionizations, thus producing sericin peptides with different lengths that contain many proportions of β-sheets, α-helices, turns and random coils.39 The percentages of random coils and β-sheets respectively reflect the amorphous nature and crystalline structure of sericin peptides.40 The antioxidant mechanism of bacterial sericin is similar to that of animal origin for many reasons, including that the amino acids containing aromatic hydrophobic side chains, such as phenylalanine, histidine and tyrosine, account for about 10 wt% and threonine and serine account for 4.65 wt%. Moreover, BNES was found to contain amino acids with high antioxidant activities, like glycine and proline, as well as aliphatic hydrophobic amino acids like alanine, valine, lucine and isolucine with an overall composition of 32%.28
3.3. Anticancer properties of the bacterial sericin
The viability of WI38, Huh-7, Caco-2, MCF-7 and A549 cells was estimated after treatment with BNES for 24 h and 48 h; the highest SI values refer to the highest safe selectivity of the sample. As shown in Table 2, IC50 values for BNES after treatment for 24 h were found to be 0.62 ± 0.17, 0.72 ± 0.27, 0.76 ± 0.36, and 0.83 ± 0.31 mg mL−1 with SI values of 6.92, 5.88, 5.58 and 5.13 against Huh-7, Caco-2, MCF-7, and A549 cells, respectively. After 48 h of treatment, IC50 values were detected at 0.51 ± 0.22, 0.49 ± 0.19, 0.41 ± 0.25, and 0.55 ± 0.38 with SI values of 8.14, 8.43, 10.25, and 7.62 against Huh-7, Caco-2, MCF-7, and A549 cells, respectively. Notably, MCF-7 cells were found that to be the most sensitive cells to BNES after treatment for 48 h, followed by the Caco-2, Huh-7 and A549 cell lines, respectively.| Cell lines | 24 h | 48 h | ||
|---|---|---|---|---|
| IC50 | SI | IC50 | SI | |
| WI38 | 4.26 ± 0.42 | — | 4.16 ± 0.31 | — |
| Huh-7 | 0.62 ± 0.17 | 6.92 | 0.51 ± 0.22 | 8.14 |
| Caco-2 | 0.72 ± 0.27 | 5.88 | 0.49 ± 0.19 | 8.43 |
| MCF-7 | 0.76 ± 0.36 | 5.58 | 0.41 ± 0.25 | 10.25 |
| A549 | 0.83 ± 0.31 | 5.13 | 0.55 ± 0.38 | 7.62 |
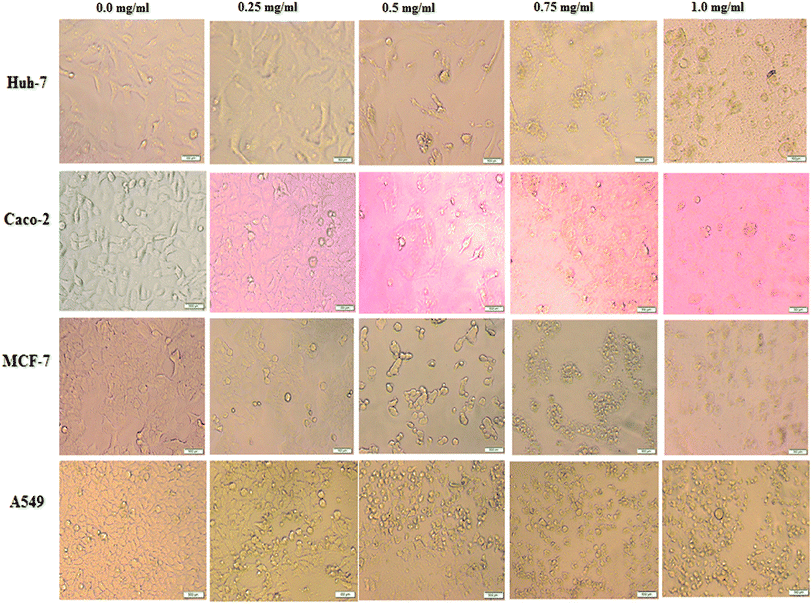
In order to monitor the cellular morphological alterations, all studied human cancer cells were treated with different contents of BNES (0.25–1.0 mg mL−1) for 48 h. Here, all images were taken in live-cell mode via a phase contrast microscope and compared with untreated control cells. The results demonstrate that BNES inflicts obviously selective cell damage and provokes morphological alterations, including blabbing, cell shrinkage and nuclear condensation (Fig. 1), in all treated human cancer cells. Hence, based on these findings, it seems that the bacterial sericin uses the apoptosis mechanism to enhance its anticancer efficiency.41 The apoptosis mechanism is complex due to its involvement in several signaling pathways. It may be generated in extrinsic or intrinsic pathways, resulting in possible alterations of the morphology and biochemistry of the cells. Kaewkorn et al. demonstrated that worm sericin reduced cell viability by stimulating apoptosis of colon tumor cells with reduction of Bcl-2 expression and increased activity of caspase-3.22 Furthermore, the anti-proliferative effect of sericin protein may also contribute to numerous mechanisms other than the apoptosis pathway, such as suppression of cancer progression, inhibition of angiogenesis, and arrest of the tumor cell cycle.22
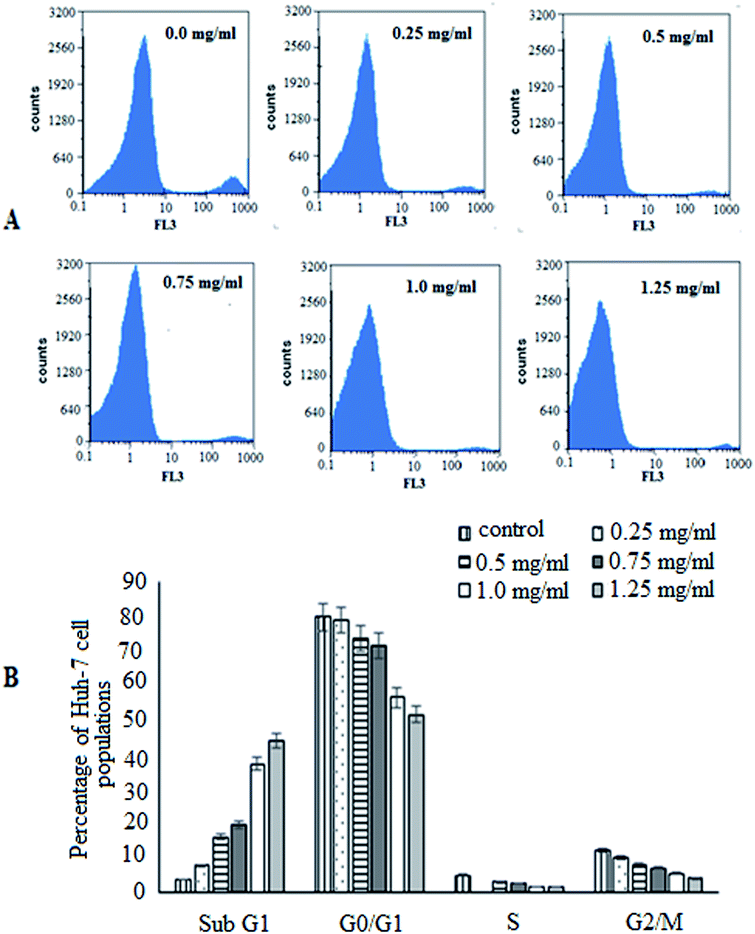
To advance insight into the prospective mechanism displayed by BNES to induce anticancer and anti-proliferative effects, investigations of the cell cycle distribution profiles were conducted. For this purpose, Huh-7 cells were treated with BNES at different concentrations of 0.25–1.25 mg mL−1 for 48 h. As shown in Fig. 2, the bacterial sericin has the capability to stimulate cell cycle distribution captured in the both G0/G1 and G2/M phases (the main checkpoints of cell cycle growth). The presented results show that the percentage of the sub-G1 population increased while S phase decreased in a dose dependent manner. These significant findings indicate that bacterial sericin induced arrest in the cell cycle in the sub-G1 and S phase populations of treated cells as compared with untreated control cells, causing the apoptosis mechanism in a dose dependent manner. In this analysis strategy, results suggest BNES is able to arrest cells in both sub-G1 and S phases, changing the normal cell cycle progression in a markedly dose dependent manner.
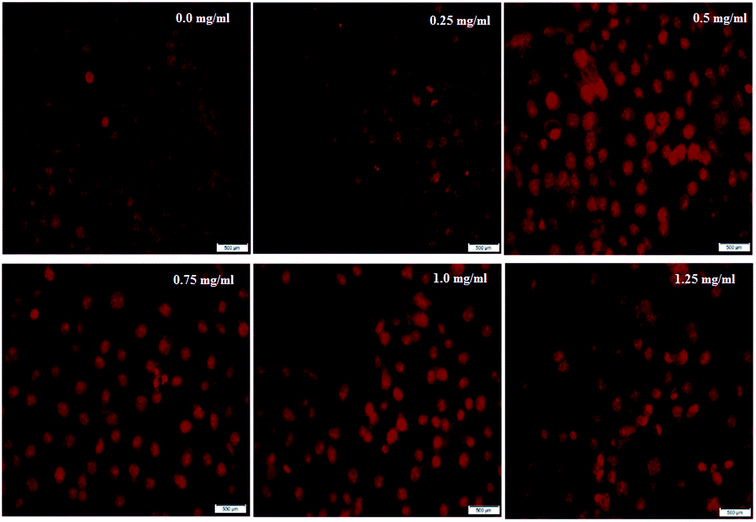
Further support to confirm the potent apoptotic effect and antitumor activity of the bacterial sericin was found by capturing the cellular morphological alternations in Caco-2 cells after treatment through nuclear staining by PI dye. The dye has the ability to bind with fragmented DNA, revealing the apoptotic effect for any tested drug. Although PI dye enters whole cells and binds to plasma membranes, its entry to the nucleus significantly occurs after disruption of cell integrity. After accumulation in the nucleus, PI forms DNA fragment-PI complexes which can be visualized and detected under fluorescence microscopy, as the stained nucleus has extremely brilliant fluorescence. Fig. 3 indicates that the Caco-2 cells appeared to have lost their spindle-shape and converted to a round shape in a dose dependent manner after exposure to BNES for 48 h compared with untreated cells. Results revealed that the nuclei became more brilliantly fluorescent with increasing concentrations of BNES, in addition to their condensation and chromatin fragmentation. Fig. 3A shows the untreated Caco-2 cells with negligible PI positive stained cells, while the maximum effect of BNES was detected at the concentration of 1.25 mg mL−1 (Fig. 3F). Hence, the bacterial sericin seemed to have a high anticancer selectivity against liver, colon, breast and lung tumors while sparing normal WI38 cells. This anticancer selectivity of bacterial sericin seems similar to the silk sericin isolated from silkworms or spiders.22,42
3.4. Antimicrobial activity
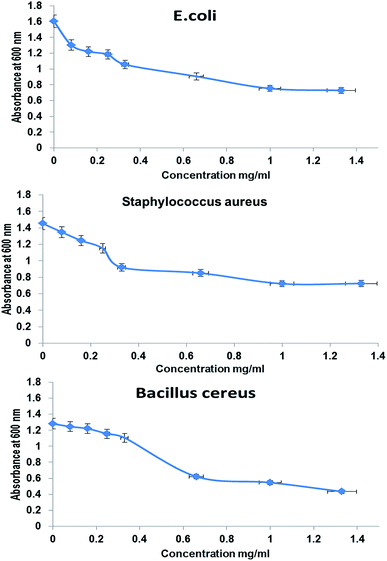
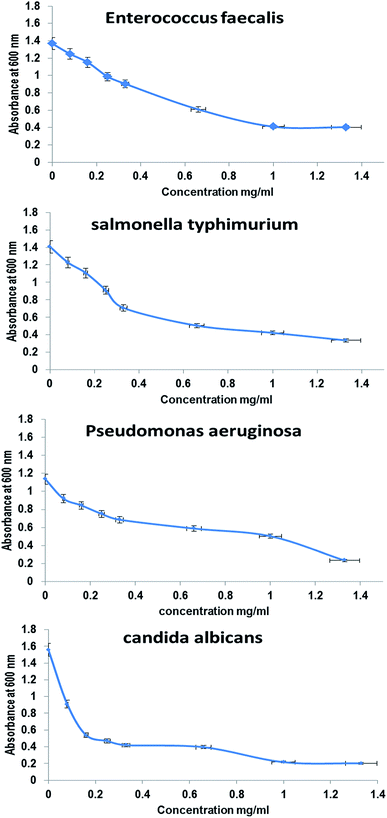
The antimicrobial activities of whole BNES were tested versus Gram-negative and Gram-positive bacteria and yeast by the decrease in the value of optical density (OD600 nm) after exposure to various concentrations of BNES. Fig. 4 and 5 show the OD values of bacteria solutions after treating with BNES. It is clear that a dramatic reduction of OD was observed in the concentration range up to 0.08 mg mL−1 of BNES for all tested organisms, in particular with C. albicans. After that, the reduction of OD values continued, reaching its highest value at 1.33 mg mL−1 BNES.Different studies have reported several types of protein, peptide exotoxins and bacteriocins as antimicrobial agents43–46 and isolated a protein extract from Pseudomonas aeruginosa that has antimicrobial effect against Vibrio sp., E. coli, Klebsiella pneumonia, Salmonella typhi, S. aureus, E. faecalis and Proteus vulgaris. Moreover, Manikandan et al.47 studied the antimicrobial activities of protein extracts from Bacillus bacteria against Staphylococcus aureus and Pseudomonas aeruginosa using an agar well diffusion assay with various concentrations (25, 50, 75, and 100 μL) and the maximum results were zone diameters equal to 18 mm and 15 mm against Staphylococcus aureus and Pseudomonas aeruginosa, respectively. In this respect, Senthilraja et al.47 reported the antimicrobial activity of a protein extracted from Saccharomyces cerevisiae against five bacterial human pathogens: Escherichia coli, Staphylococcus aureus, Vibrio cholera, Salmonella typhi and Shigella flexneri. The modes of action and mechanisms of some proteins and peptides as antimicrobial agents were described by Le et al.48 These include inhibition of nucleic acids, proteins, metabolism, cell wall biosynthesis, lipopolysaccharide-binding peptides, and protein-folding.
Recently, in agreement with the results of the present study, Abd El-Aty et al.49 confirmed the antimicrobial activity of silk fabric extract from neem leaves against different types of microorganisms (bacteria and fungi). In the same context, Ahamad and Vootla50 reported the antimicrobial potential of Antheraea mylitta silk sericin on several pathogenic bacterial and fungal strains. The antibacterial activity of sericin silk proteins from Bombyx mori has been investigated by Saviane et al.,51 who found that it was able to inhibit the growth of Gram-negative bacteria. In contrast, Wright and Goodacre52 found that the web-silk of a native house spider inhibited the growth of the Gram-positive bacterium Bacillus subtilis, while no significant inhibition of growth was observed versus the Gram-negative bacterium Escherichia coli.
4. Conclusions
The present study provided a novel bacterial sericin-like protein (BNES) which shows antioxidant, antimicrobial and anticancer activities. BNES is isolated from a sustainable bacterial source that simplifies its production on the large-scale industrial level. BNES showed effectiveness as an antagonist for a number of tested cancer cells, such as colon, liver, breast and lung cancers. It was also confirmed to be an antimicrobial agent against Gram-positive and -negative bacteria, fungi, and yeasts. BNES has no toxic effects on healthy cells, which make it a candidate for an antagonist with no adverse side effects. BNES can be used as a highly efficient biomaterial to treat many types of tumors and infectious microbial diseases and/or as a mixture with conventional chemical drugs in the treatment of various types of cancer. In vitro studies revealed that BNES showed efficient antioxidant and antibacterial activities and inhibited the proliferation of many types of cancer cells by induction of the apoptosis mechanism. However, in vivo assessment is needed for its use as a potent antioxidant, antitumor and antibacterial candidate in pharmaceutical applications.Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.Conflicts of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.References
- K. Jastrzebska, K. Kucharczyk, A. Florczak, E. Dondajewska, A. Mackiewicz and H. Dams-Kozlowska, Rep. Pract. Oncol. Radiother., 2015, 20, 87–98 CrossRef PubMed.
- C. Z. Zhou, F. Confalonieri, M. Jacquet, R. Perasso, Z. G. Li and J. Janin, Proteins: Struct., Funct., Genet., 2001, 44, 119–122, DOI:10.1002/prot.1078.
- S. Inoue, K. Tanaka, F. Arisaka, S. Kimura, K. Ohtomo and S. Mizuno, J. Biol. Chem., 2000, 275, 40517–40528, DOI:10.1074/jbc.M006897200.
- C. Acharya, B. Hinz and S. C. Kundu, Biomaterials, 2008, 29, 4665–4675, DOI:10.1016/j.biomaterials.2008.08.033.
- L. Uebersax, H. Hagenmüller, S. Hofmann, E. Gruenblatt, R. Müller, G. Vunjak-Novakovic, D. L. Kaplan, H. P. Merkle and L. Meinel, Tissue Eng., 2006, 12, 3417–3429 CrossRef CAS PubMed.
- A. R. Murphy and D. L. Kaplan, J. Mater. Chem., 2009, 19, 6443–6450, 10.1039/b905802h.
- C. Acharya, V. Kumar, R. Sen and S. C. Kundu, Biotechnol. J., 2008, 3, 226–233, DOI:10.1002/biot.200700120.
- L. Meinel, R. Fajardo, S. Hofmann, R. Langer, J. Chen, B. Snyder, G. Vunjak-Novakovic and D. Kaplan, Bone, 2005, 26, 2775–2785, DOI:10.1016/j.bone.2005.06.010.
- I. Dal Pra, G. Freddi, J. Minic, A. Chiarini and U. Armato, Biomaterials, 2005, 26, 1987–1999, DOI:10.1016/j.biomaterials.2004.06.036.
- X. Zhang, M. M. R. Khan, T. Yamamoto, M. Tsukada and H. Morikawa, Int. J. Biol. Macromol., 2012, 50, 337–347, DOI:10.1016/j.ijbiomac.2011.12.006.
- T. Chlapanidas, S. Faragò, G. Lucconi, S. Perteghella, M. Galuzzi, M. Mantelli, M. A. Avanzini, M. C. Tosca, M. Marazzi, D. Vigo, M. L. Torre and M. Faustini, Int. J. Biol. Macromol., 2013, 58, 47–56, DOI:10.1016/j.ijbiomac.2013.03.054.
- J. P. Kumar and B. B. Mandal, Free Radicals Biol. Med., 2018, 108, 803–818, DOI:10.1016/j.freeradbiomed.2017.05.002.
- S. Sarovart, B. Sudatis, P. Meesilpa, B. P. Grady and R. Magaraphan, Rev. Adv. Mater. Sci., 2003, 5, 193–198 CAS.
- M. Chalamaiah, B. Dinesh Kumar, R. Hemalatha and T. Jyothirmayi, Food Chem., 2012, 135, 3020–3038 CrossRef CAS PubMed.
- A. Pihlanto, Int. Dairy J., 2006, 16, 945–960 CrossRef.
- S. Ranamukhaarachchi, L. Meissner and C. Moresoli, J. Membr. Sci., 2013, 429, 81–87, DOI:10.1016/j.memsci.2012.10.040.
- X. J. Wang, X. Q. Zheng, N. K. Kopparapu, W. S. Cong, Y. P. Deng, X. J. Sun and X. L. Liu, Process Biochem., 2015, 44, 842–846, DOI:10.1016/j.procbio.2014.05.014.
- H. Guo, Y. Kouzuma and M. Yonekura, Food Chem., 2009, 113, 238–245, DOI:10.1016/j.foodchem.2008.06.081.
- J. Zhang, H. Zhang, L. Wang, X. Guo, X. Wang and H. Yao, Eur. Food Res. Technol., 2009, 229, 709–719, DOI:10.1007/s00217-009-1103-3.
- B. H. Sarmadi and A. Ismail, Peptides, 2010, 31, 1949–1956 CrossRef CAS PubMed.
- A. M. Ghribi, A. Sila, R. Przybylski, N. Nedjar-Arroume, I. Makhlouf, C. Blecker, H. Attia, P. Dhulster, A. Bougatef and S. Besbes, J. Funct. Foods, 2015, 12, 516–525, DOI:10.1016/j.jff.2014.12.011.
- W. Kaewkorn, N. Limpeanchob, W. Tiyaboonchai, S. Pongcharoen and M. Sutheerawattananonda, Biol. Res., 2012, 45, 45–50, DOI:10.4067/S0716-97602012000100006.
- S. Zhaorigetu, N. Yanaka, M. Sasaki, H. Watanabe and N. Kato, Oncol. Rep., 2003, 10, 537–543 CAS.
- S. Zhaorigetu, M. Sasaki, H. Watanabe and N. Kato, Biosci., Biotechnol., Biochem., 2001, 65, 2181–2186, DOI:10.1271/bbb.65.2181.
- S. Zhaorigetu, M. Sasaki and N. Kato, J. Nutr. Sci. Vitaminol., 2007, 53, 297–300, DOI:10.3177/jnsv.53.297.
- W. Senakoon, S. Nuchadomrong, S. Sirimungkararat, T. Senawong and P. Kitikoon, Asian Journal Of Food & Agro-industry, 2009, 2, S222–S228 Search PubMed.
- G. Basal, D. Altıok and O. Bayraktar, Fibers Polym., 2010, 11, 21–27, DOI:10.1007/s12221-010-0021-0.
- E. A. Kamoun, G. M. Abu-Elreesh, E. M. El-Fakharany and D. Abd-El-Haleem, J. Polym. Environ., 2019, 27, 1629–1641, DOI:10.1007/s10924-019-01459-2.
- P. Prieto, M. Pineda and M. Aguilar, Anal. Biochem., 1999, 269, 337–341, DOI:10.1006/abio.1999.4019.
- M. S. Blios, Nature, 1985, 26, 1199–1200 Search PubMed.
- R. Re, N. Pellegrini, A. Proteggente, A. Pannala, M. Yang and C. Rice-Evans, Free Radicals Biol. Med., 1999, 26, 1231–1237, DOI:10.1016/S0891-5849(98)00315-3.
- M. M. Abu-Serie and E. M. El-Fakharany, Sci. Rep., 2017, 7, 16769, DOI:10.1038/s41598-017-16962-6.
- V. N. Uversky, E. M. El-Fakharany, M. M. Abu-Serie, H. A. Almehdar and E. M. Redwan, Cancer Invest., 2017, 35, 610–623, DOI:10.1080/07357907.2017.1373783.
- E. M. El-Fakharany, M. M. Abu-Serie, E. A. Litus, S. E. Permyakov, E. A. Permyakov, V. N. Uversky and E. M. Redwan, Protein J., 2018, 37, 203–215, DOI:10.1007/s10930-018-9770-1.
- R. C. Goy, S. T. B. Morais and O. B. G. Assis, Brazilian Journal of Pharmacognosy, 2016, 26, 122–127, DOI:10.1016/j.bjp.2015.09.010.
- S. R. Fahnestock, Z. Yao and L. A. Bedzyk, Rev. Mol. Biotechnol., 2000, 74, 105–119 CrossRef CAS PubMed.
- V. A. R. Antony and S. K. Chinnamal, Inter. J. Sci. Tech. Manag., 2015, 4, 458–465 Search PubMed.
- Y. J. Yang, Y. S. Choi, D. Jung, B. R. Park, W. B. Hwang, H. W. Kim and H. J. Cha, NPG Asia Mater., 2013 DOI:10.1038/am.2013.19.
- T. L. Da Silva, A. C. Da Silva, M. Ribani, M. G. A. Vieira, M. L. Gimenes and M. G. C. Da Silva, Chem. Eng. Trans., 2014, 38, 103–108, DOI:10.3303/CET1438018.
- F. R. B. Turbiani, J. Tomadon, F. L. Seixas and M. L. Gimenes, Chem. Eng. Trans., 2011, 24, 1489–1494 Search PubMed.
- G. Pistritto, D. Trisciuoglio, C. Ceci, A. Garufi and G. D'Orazi, Aging, 2016, 8, 603–619 CAS.
- A. Florczak, K. Jastrzebska, A. Mackiewicz and H. Dams-Kozlowska, J. Mater. Chem. B, 2017, 5, 3000–3011, 10.1039/c7tb00233e.
- M. Riley and J. Wertz, Annu. Rev. Microbiol., 2002, 56, 117–137 CrossRef CAS PubMed.
- M. R. Yeaman and N. Y. Yount, Pharmacol. Rev., 2003, 55, 27–55 CrossRef CAS PubMed.
- D. Jayammal and T. Sivakumar, Int. J. Curr. Microbiol. Appl. Sci., 2013, 2, 207–213 Search PubMed.
- P. Manikandan, V. Tamizhazhagan, A. Gnanasekaran and P. Senthikumar, Asian J. Res. Biol. Pharm. Sci., 2017, 5, 75–85 CAS.
- P. Senthilraja, S. K. Sahu and K. Kathiresan, Res. J. Microbiol., 2015, 10, 24–32, DOI:10.3923/jm.2015.24.32.
- C. F. Le, C. M. Fang and S. D. Sekaran, Antimicrob. Agents Chemother., 2017, 61, e02340–16 CAS.
- A. Abd-ElAty, G. El-Bassyouni, N. Abdel-Zaher and O. Guirguis, Fibers Polym., 2018, 19, 1880–1886 CrossRef CAS.
- M. S. I. Ahamad and S. Vootla, International Journal of Recent Scientific Research, 2018, 9, 32019–32022 Search PubMed.
- A. Saviane, O. Romoli, A. Bozzato, G. Freddi, C. Cappelletti, E. Rosini, S. Cappellozza, G. Tettamanti and F. Sandrelli, Transgenic Res., 2018, 27, 87–101, DOI:10.1007/s11248-018-0059-0.
- S. Wright and S. L. Goodacre, BMC Res. Notes, 2012, 5, 326, DOI:10.1186/1756-0500-5-326.
| This journal is © The Royal Society of Chemistry 2020 |