Open Access Article
Open Access ArticleHarmane ameliorates obesity though inhibiting lipid accumulation and inducing adipocyte browning
Yanwen Lia,
Chanjuan Lib,
Jiaqiang Wua,
Wenfeng Liu a,
Dongli Li
a,
Dongli Li a and
Jun Xu
a and
Jun Xu *ab
*ab
aSchool of Biotechnology and Health Sciences, Wuyi University, Jiangmen 529000, China. E-mail: junxu@biochemomes.com
bSchool of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou 510006, China
First published on 27th January 2020
Abstract
Harmane, a compound derived from Peganum harmala L., was discovered as a lipid accumulation inhibitor with our triglyceride (TG) assay. To investigate the molecular mechanism underlying the role of harmane in the prevention of lipid accumulation, a number of biological experiments have been designed to determine if harmane reduces lipid accumulation by suppressing adipogenesis, lipogenesis, and regulating a specific signal transduction pathway. Our experimental data show that harmane inhibits TG accumulation though down-regulating the expression of adipogenic and lipogenic factors, up-regulating adipocyte browning markers and activating the SIRT1-LKB1-AMPK pathway. Therefore, harmane ameliorates obesity though inhibiting lipid accumulation and inducing adipocyte browning.
1. Introduction
Obesity is caused by acuminated white adipose tissues, which store excess energy in the form of triglyceride (TG).1 The prevalence of overweight and obesity is rising among children and adolescents.2 Obesity is a major cause of metabolic disorders such as cardiovascular disease, type 2 diabetes (T2D), and nonalcoholic fatty liver disease. The primary cause of T2D is obesity-driven insulin resistance in white adipose tissue (WAT), and insufficient secretion of insulin.3 These diseases reduce life quality as well as life span.4The differentiation of preadipocytes and the subsequent lipid accumulation in mature adipocytes are well-organized process accompanied by coordinated genetic changes.5 At the early stage of preadipocyte differentiation, the transient expression of CCAAT/enhancer binding protein-β (C/EBPβ) induce the expression of C/EBPα and peroxisome proliferator activated receptor γ (PPARγ), which promote the formation of mature adipocytes and lipid production.6,7 In addition, acetyl-CoA carboxylase 1 (ACC1), fatty acid synthase (FAS), and stearyl coenzyme A desaturated enzyme 1 (SCD-1) are rate-limited metabolic enzymes involved in lipogenesis.8,9
Obesity results from greater energy intake than energy expenditure, leading to excessive deposition of WAT in the body.10 WAT can be induced into brown fat-like adipocytes (BAT), promoting energy expenditure.11 This process is mediated by uncoupling protein 1 (UCP1, the only specific brown adipocyte marker) and activation of oxidative metabolism. BAT is characterized by multilocular lipid droplet morphology, high mitochondrial content and the expression of a core set of brown fat-specific genes such as UCP1, PPARα and PGC1α. These genes and pathways provide a variety of promising therapeutic targets for obesity treatment.12
The FDA approved anti-obesity drugs are mainly appetite inhibitors and gastrointestinal lipase inhibitor (orlistat). However, these drugs are not adequate therapies on their own in the long-term, as they have adverse effects in mood changes and gastrointestinal or cardiovascular complications.13 Therefore, searching for novel anti-obesity agents is compelling.
Harmane is a compound derived from Peganum harmala L. Previous reports demonstrated that harmane had activities against inflammatory, diabetes, and depression.14,15 However, the mechanisms of these activities were not articulated.
Recently, we revealed that harmane inhibited lipid accumulation, but also induced adipocyte browning in 3T3-L1 pre-adipocytes differentiation model.
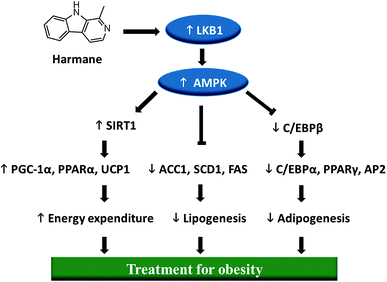
In order to determine if harmane can be developed into an agent against obesity, we have to investigate the mechanism of action underlying the role of harmane in the prevention of lipid accumulation. Based on the previous reports5,9,16 and our experimental data, we hypothesize that harmane might regulate the SIRT1-LKB1-AMPK pathway (Fig. 1). Biological experiments were designed to detect liver kinase B1 (LKB1) and all downstream genes have been regulated by harmane for the obesity treatment.
2. Experimental methods
2.1 3T3-L1 cell differentiation and compound preparation
3T3-L1 pre-adipocytes was cultured in DMEM supplemented with 10% fetal bovine serum (Hyclone, USA), 1% penicillin and streptomycin (Gibco, USA) at 37 °C in 5% CO2. 3T3-L1 pre-adipocytes was differentiated as previously reported.17 Two days after confluence (day 0), the medium was changed to a differentiation cocktail mixture containing 0.5 mM 3-isobutyl-1-methylxanthine (Sigma, USA), 100 ng mL−1 dexamethasone (MP, USA), 2 μg mL−1 insulin (Sigma, USA) and 10 ng mL−1 biotin in DMEM with 10% FBS and 1% penicillin and streptomycin for 3 days (day 3). Then, the medium was replaced by 10% FBS/DMEM containing 2 μg mL−1 insulin for another 3 days (day 6). Then, the differentiation medium was changed to culture medium. Harmane was purchased from TargetMol, dissolved in dimethyl sulfoxide (DMSO), and supplemented at indicated concentrations during indicated time course of differentiation. The equal amount of DMSO was used as vehicle.2.2 TG content assay
Cells were washed three times with PBS (pH 7.4) and then ultrasonically lysed. TG contents were measured using commercial TG assay kit (Jiancheng Bioengineering Institution, Nanjing, China) according to the manufacture's protocol. Results were presented as the relative TG content compared to the vehicle group.2.3 Oil Red O (ORO) staining
Cells were fixed with 4% formaldehyde (v/v) for 1 h at room temperature and stained with freshly prepared ORO working solution for 1 h at room temperature and washed three times with distilled water. The plates were scanned using an OLYMPUS CKX41 microscope and camera (OLYMPUS). The ORO working solution was prepared by mixing 6 mL of 0.5% ORO (w/v, Sigma) in isopropanol with 4 mL of distilled water followed by filtration through a 0.22 μM filter (Millipore).2.4 Cytotoxicity analysis
3T3-L1 and RAW264.7 cells were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). 3T3-L1 and RAW264.7 cells were seeded at the density of 5000 per well into 96-well plates to grow overnight. Then the cells were treated with various concentrations of compounds for 72 hours. Same volume of DMSO is added as vehicle control. 0.5 mg mL−1 MTT solution was added to each well and incubated for 4 h, after which 150 μL DMSO was added to each well. The absorbance of wells was recorded at 492 nm after 10 min of shaking. The vitality percentage was calculated as (Acompound − Ablank)/(Avehicle − Ablank) × 100%. The viability of vehicle control is defined as 100%.2.5 Determination of mitochondrial DNA copy numbers
Mitochondrial DNA (mtDNA) copy numbers was achieved by qPCR. Briefly, DNA was extracted using Sangon Biotech kit. The copy numbers of nuclear DNA (nDNA) and mtDNA were assessed by qPCR using primers targeted toward the cytochrome B gene (for mtDNA) and 18S rRNA (for nDNA).2.6 Real-time qPCR
Total RNA was isolated using RNAiso plus (Takara, Dalian, China) according to the manufacturer's protocol. Total RNA (1 μg) was converted to cDNA using ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan) according to the manufacturer's protocol. cDNA and SYBR Green were used to amplify specific target genes by Real-time PCR Master Mix (Toyobo, Osaka, Japan). The sequence of primers was described in this table.| Gene | Forward primer (5′–3′) | Reversed primer (5′–3′) |
|---|---|---|
| PPARγ | TGCTGTTATGGGTGAAACTCTG | GAAATCAACTGTGGTAAAGGGC |
| C/EBPα | AGTCGGTGGACAAGAACAGCAAC | CGGTCATTGTCACTGGTCAACTC |
| C/EBPβ | ACCGGGTTTCGGGACTTGA | CCCGCAGGAACATCTTTAAGTGA |
| FAS | ATTGGCTCCACCAAATCCAAC | CCCATGCTCCAGGGATAACAG |
| SCD1 | TGTTCGTTGCCACTTTCTTG | GCTAATGTTCTTGTCATAAGGAC |
| ACC1 | AGGATTTGCTGTTTCTCAGAGCTT | CAGGATCTACCCAGGCCACAT |
| AP2 | GTCACCATCCGGTCAGAGAGTAC | TCGTCTGCGGTGATTTCATC |
| Cyto b | GGCTACGTCCTTCCATGAGG | TGGGATGGCTGATAGGAGGT |
| 18S rRNA | AACTTTCGATGGTAGTCGC | TTCCTTGGATGTGGTAGCC |
| β-Actin | TGGAATCCTGTGGCATCCATGAAA | TAAAACGCAGCTCAGTAACAGTCC |
2.7 Western blotting
The cell lysates were prepared with RIPA lysis buffer containing protease inhibitor cocktail (Beyotime, Shanghai, China) and phosphatase inhibitor (Beyotime, Shanghai, China). Protein concentrations were determined using Pierce™ Rapid Gold BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, USA). Protein samples were denatured in SDS buffer and then subjected to SDS-PAGE and blotted onto polyvinylidene difluoride (PVDF) membrane (Millipore, Darmstadt, Germany). The membranes were blocked using 5% (w/v) skim milk in TBST (Tris-buffered saline containing 0.1% (v/v) Tween 20) for 2 h and then incubated with primary antibodies overnight at 4 °C. After three times washing in TBST, the membranes were incubated with corresponding secondary antibodies at rt for 1 h. Immunoreactive signals were detected using enhanced chemiluminescence reagents with Chemiluminescence imager (Tanon 5200, Shanghai, China). Densitometry analysis was performed using Quantity One Software (Bio-Rad Laboratories, USA). Antibodies used including phospho (Thr172)- and total-AMPKα (Cell Signaling, Danvers, USA), ACC1 (Cell Signaling, Danvers, USA), FAS (Cell Signaling, Danvers, USA), SCD-1 (Cell Signaling, Danvers, USA), α-Tubulin (Sigma, St. Louis, USA), phospho (Ser428)-LKB1 (Cell Signaling, Danvers, USA), SIRT1 (Cell Signaling, Danvers, USA), PCG-1α (Cell Signaling, Danvers, USA), UCP1 (Cell Signaling, Danvers, USA), PPARα (Abcam, UK), GAPDH (Bioworld, China).2.8 Statistical analysis
All data are expressed as the mean ± SEM. Data between two groups were analyzed by Student's t-test using Graphpad Prism (Graphpad Software Inc, California, USA). The significant difference was statistically analyzed using one-way ANOVA followed by Tukey's HSD post hoc test. Statistical significance was set at P < 0.05.3. Results
3.1 Harmane is a lipid-lowering agent
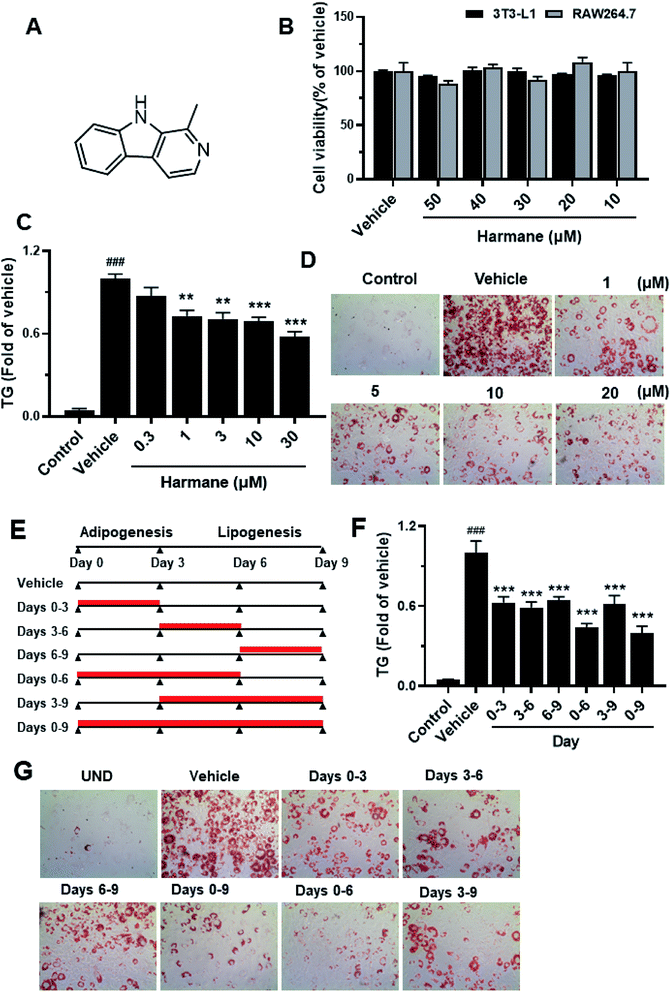
Harmane is not toxic, its effect on cell viability in 3T3-L1 adipocytes and RAW264.7 cells were measured via MTT assay as shown in Fig. 2B. Harmane was evaluated with 3T3-L1 cell model18 for the lipid-lowering activity. As shown in Fig. 2C, harmane decreases lipid accumulation in 3T3-L1 adipocytes in a dose-dependent manner. The lipid inhibitory effects of harmane were also confirmed by ORO staining experiments (Fig. 2D).To determine whether harmane reduced lipid accumulation by suppressing adipogenesis and/or lipogenesis, we treated 3T3-L1 cells with 10 μM harmane in various periods, that is, days 0 to 3, 3 to 6, 6 to 9, 0 to 6, 3 to 9, and 0 to 9, which represent different stages of 3T3-L1 differentiation (Fig. 2E). On day 9, the TG content was measured by TG assay, representative images were captured. Our results demonstrated that harmane reduced TG content at all the stage of adipocyte differentiation and lipid accumulation (Fig. 2F and G).
3.2 Harmane inhibits lipid accumulation through suppressing adipogenic and lipogenic factors in 3T3-L1 cells
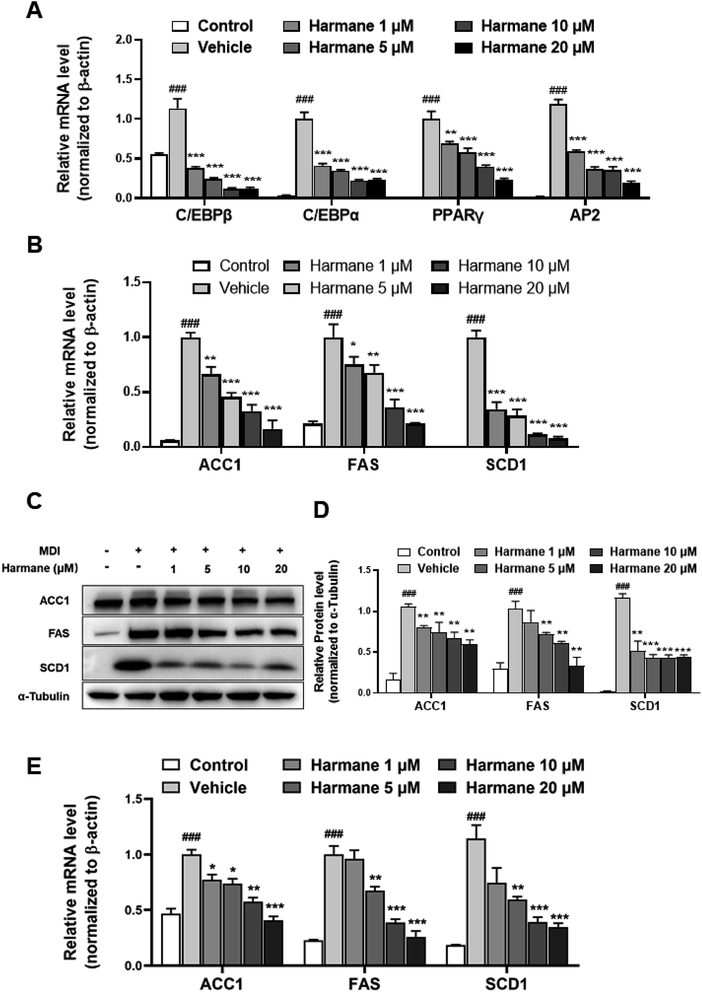
Adipogenesis is regulated by a multiple differentiation factor network, which initiates the process and activates the expression of downstream fatty acid synthesis markers. At the early stage of differentiation (day 0 to day 3), the C/EBPβ is rapidly induced to express and subsequently activate C/EBPα expression. C/EBPα can activate the expression of downstream target genes such as PPARγ and AP2.19,20To prove the regulation effect of harmane in the differentiation stage, we performed qPCR on 3T3-L1 cells with harmane treatment. Our results demonstrate that harmane decreases the mRNA levels of C/EBPβ with the treatment for 1 day. Subsequently, the mRNA levels of PPARγ, C/EBPα and AP2, were attenuated in a dose-dependent manner with harmane treatment compared to the vehicle (Fig. 3A). These results indicate that harmane inhibits lipid accumulation by suppressing adipogenic factor expression.
C/EBPα and PPARγ are the upstream regulators of the lipogenic process, which regulates fatty acid synthesis through activating the expression of genes related with lipid metabolism such as ACC1, FAS and SCD-1.21 Correspondingly, the levels of fatty acid synthesis related mRNAs were decreased dose-dependently by the treatments of harmane for 6 days (Fig. 3B). Furthermore, harmane strongly downregulated lipogenesis-related markers protein expression with 6 days treatment, supporting the qPCR data (Fig. 3C and D). These results indicate that harmane inhibits lipid accumulation by suppressing lipogenic factor expression.
3.3 Harmane decreases lipid accumulation through inducing adipocyte browning in 3T3-L1 cells
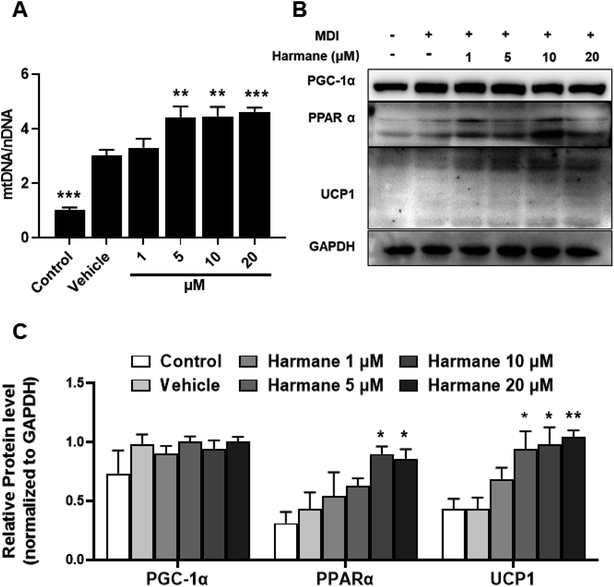
BAT is a key site of heat production in mammals that has been considered an attractive target to promote weight loss.22 UCP1 is responsible for the unique function of brown adipose tissue. PPARα may coordinate expression of fatty acid oxidation enzymes with expression of UCP1. Besides, PGC-1α causes an increase expression for UCP1.23,24 To prove the browning effect of harmane in the differentiation stage, we performed WB on 3T3-L1 cells with harmane treatment. Consistently, treatment with harmane increased the copy numbers of mtDNA (Fig. 4A), mitochondrial transcription factor PGC-1α and PPARα. Furthermore, there was a robust increase in the expression levels of UCP1 at translational levels (Fig. 4B and C). These results suggest that harmane decrease lipid accumulation and induce browning in 3T3-L1 cells, involved in the anti-obesity effect.3.4 Harmane decreases lipid accumulation through activating SIRT1-LKB1-AMPK pathway
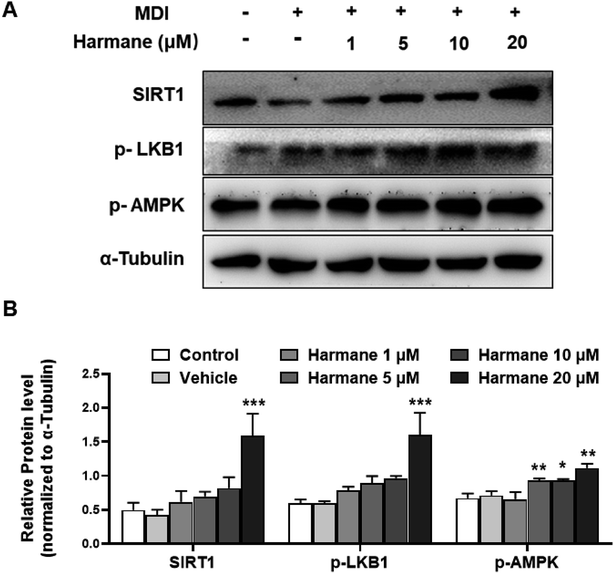
AMP-activated protein kinase (AMPK) is a significant player in regulating energy metabolism. Activated AMPK was reported to be upstream mechanism of the inhibition of adipogenesis and lipogenesis.9,25 AMPK also enhances SIRT1 activity, resulting in the modulation of the activity of downstream SIRT1 targets that include the PGC-1α.16 LKB1 is a key kinase controlling AMPK activity and phosphorylation of AMPK.26 As shown in Fig. 5A and B, activation of AMPK and LKB1 was observed with harmane treatment in a dose-dependent manner. Besides, the activity of SIRT1 is significantly improved. These results demonstrate that harmane decrease TG accumulation with the activation of the SIRT1-LKB1-AMPK pathway.4. Conclusion
Harmane, a natural product derived from Peganum harmala L., is a lipid accumulation inhibitor. This study proves that harmane inhibits TG accumulation though down-regulating the expression of adipogenic and lipogenic factors, up-regulating adipocyte browning markers and activating SIRT1-LKB1-AMPK pathway. Hence, harmane can be a lead to be developed into a therapeutic agent against obesity and related metabolic diseases.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been funded by the National Natural Science Foundation of China (No. 81870608), the Science and technology Planning Project of Guangdong Province (No. 2016A020217002), the National Key R&D Program of China (2017YFB0203403) and Science and Technology of Jiangmen (No. 2016350100170008351, 2018110100330005446).References
- D. W. Haslam and W. P. James, Lancet, 2005, 366, 1197–1209 CrossRef.
- M. Ng, T. Fleming, M. Robinson, B. Thomson, N. Graetz, C. Margono, E. C. Mullany, S. Biryukov, C. Abbafati and S. F. Abera, et al., Lancet, 2014, 384, 766–781 CrossRef.
- M. Shimabukuro, Y.-T. Zhou, M. Levi and R. H. Unger, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 2498–2502 CrossRef CAS PubMed.
- S. J. Olshansky, D. J. Passaro, R. C. Hershow, J. Layden, B. A. Carnes, J. Brody, L. Hayflick, R. N. Butler, D. B. Allison and D. S. Ludwig, N. Engl. J. Med., 2005, 352, 1138–1145 CrossRef CAS PubMed.
- E. D. Rosen and O. A. MacDougald, Nat. Rev. Mol. Cell Biol., 2006, 7, 885–896 CrossRef CAS PubMed.
- F. M. Gregoire, C. A. Smas and H. S. Sul, Physiol. Rev., 1998, 78, 783–809 CrossRef CAS.
- M. Ahmadian, J. M. Suh, N. Hah, C. Liddle, A. R. Atkins, M. Downes and R. M. Evans, Nat. Med., 2013, 19, 557–566 CrossRef CAS PubMed.
- E. D. Rosen, C. J. Walkey, P. Puigserver and B. M. Spiegelman, Genes Dev., 2000, 14, 1293–1307 CAS.
- B. B. Zhang, G. Zhou and C. Li, Cell Metab., 2009, 9, 407–416 CrossRef CAS PubMed.
- Y.-H. Tseng, A. M. Cypess and C. R. Kahn, Nat. Rev. Drug Discovery, 2010, 9, 465–481 CrossRef CAS PubMed.
- M. Rosenwald, A. Perdikari, T. Rulicke and C. Wolfrum, Nat. Cell Biol., 2013, 15, 659–667 CrossRef CAS PubMed.
- M. Harms and P. Seale, Nat. Med., 2013, 19, 1252–1263 CrossRef CAS PubMed.
- R. S. Padwal and S. R. Majumdar, Lancet, 2007, 369, 71–77 CrossRef CAS.
- H. Khan, S. Patel and M. A. Kamal, Curr. Drug Metab., 2017, 18, 853–857 CAS.
- E. J. Cooper, A. L. Hudson, C. A. Parker and N. G. Morgan, Eur. J. Pharmacol., 2003, 482, 189–196 CrossRef CAS PubMed.
- C. Canto, Z. Gerhart-Hines, J. N. Feige, M. Lagouge, L. Noriega, J. C. Milne, P. J. Elliott, P. Puigserver and J. Auwerx, Nature, 2009, 458, 1056–1060 CrossRef CAS PubMed.
- C. Li, B. Cheng, S. Fang, H. Zhou, Q. Gu and J. Xu, Eur. J. Med. Chem., 2018, 143, 114–122 CrossRef CAS PubMed.
- X. Y. Zeng, X. Zhou, J. Xu, S. M. Chan, C. L. Xue, J. C. Molero and J. M. Ye, Biochem. Pharmacol., 2012, 84, 830–837 CrossRef CAS PubMed.
- T. Hishida, M. Nishizuka, S. Osada and M. Imagawa, Biochimie, 2009, 91, 654–657 CrossRef CAS PubMed.
- E. Chang and C. Y. Kim, Molecules, 2019, 24(6), 1157 CrossRef PubMed.
- M. M. Mihaylova and R. J. Shaw, Nat. Cell Biol., 2011, 13, 1016–1023 CrossRef CAS PubMed.
- B. Cannon and J. Nedergaard, Physiol. Rev., 2004, 84, 277–359 CrossRef CAS PubMed.
- M. J. Barbera, A. Schluter, N. Pedraza, P. Iglesias, F. Villarroya and M. Giralt, J. Biol. Chem., 2001, 276, 1486–1493 CrossRef CAS.
- Z. Wu, P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell and R. C. Scarpulla, et al., Cell, 1999, 98, 115–124 CrossRef CAS PubMed.
- B. Viollet, B. Guigas, J. Leclerc, S. Hébrard, L. Lantier, R. Mounier, F. Andreelli and M. Foretz, Crit. Rev. Biochem. Mol. Biol., 2010, 196, 81–98 Search PubMed.
- S. A. Hawley, J. Boudeau, J. L. Reid, K. J. Mustard, L. Udd, T. P. Makela, D. R. Alessi and D. G. Hardie, J. Biol., 2003, 2, 28 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |