Open Access Article
Open Access ArticleEffect of TiO2-nanoparticles on copper toxicity to bacteria: role of bacterial surface
Xiaomin Li a,
Qingquan Maa,
Tong Liua,
Zhaomin Dongab and
Wenhong Fan
a,
Qingquan Maa,
Tong Liua,
Zhaomin Dongab and
Wenhong Fan *ab
*ab
aSchool of Space and Environment, Beihang University, No. 37 Xueyuan Road, Haidian, Beijing 100191, P. R. China. E-mail: fanwh@buaa.edu.cn
bBeijing Advanced Innovation Center for Big Data-Based Precision Medicine, Beihang University, No. 37 Xueyuan Road, Haidian, Beijing 100191, P. R. China
First published on 30th January 2020
Abstract
The impact of titanium dioxide nanoparticles (nano-TiO2) on the aquatic environment is an important issue due to their increasing application. Although nano-TiO2 was reported to show an effect on heavy metal toxicity to aquatic organisms, the underlying mechanism is not well understood. In this study, two bacterial species (Bacillus thuringiensis (B. thuringiensis) and Bacillus megaterium (B. megaterium)) from sediment were selected to study the effects of nano-TiO2 on copper toxicity. Nano-TiO2 was found to inhibit the growth of B. thuringiensis and enhance the oxidative stress damage caused by copper, whereas these effects were not observed for B. megaterium. Transmission electron microscopy and flow cytometry showed that B. thuringiensis has stronger association ability to nano-TiO2 than B. megaterium. The existence of the S-layer on the surface of B. thuringiensis might be the possible reason, leading to the difference in copper toxicity. This indicates that the characteristics of bacterial surfaces might be important to the toxicity responses of nanoparticles.
1. Introduction
Titanium dioxide nanoparticles (nano-TiO2) are widely used in commercial and industrial fields because of their unique physical and chemical properties.1–3 It has been predicted that the annual production of nano-TiO2 will reach 2.5 million tons by 2025.4 Therefore, the release of nano-TiO2 into the aquatic environment will be inevitable, triggering potential risks to the aquatic ecosystem. Although the concentration of nano-TiO2 in the natural aquatic environment is low (3–6 ng L−1),5 studies have demonstrated that nano-TiO2 could be toxic to aquatic organisms.6–11 Furthermore, nano-TiO2 could interact with other contaminants already present in the environment, thus affecting the behavior and toxicity of these contaminants.12–14 Several studies have focused on the effect of nano-TiO2 on heavy metal toxicity to aquatic organisms, such as Daphnia magna, algae and Tetrahymena thermophile.14–18 Only limited studies have reported the toxic influence of nano-TiO2 on heavy metal toxicity to bacteria.The number of bacteria in freshwater can reach 108 ml−1. Bacteria not only play an important role in nutrient cycling (e.g., carbon, nitrogen and phosphorus), biomass decomposition and bioremediation of pollutants in the aquatic environment,11 but also serve as the primary foundation nutrient source for organisms in higher trophic levels.19 The stability of the bacterial community is important for a healthy aquatic ecosystem.20–22 Therefore, it is essential to study the effect of nano-TiO2 on bacteria in this environment.
Several studies have found that nano-TiO2 could inhibit bacterial growth21 and change microbial communities,23 and that the toxicity of nano-TiO2 to bacteria may be impacted by several factors. Firstly, the physical properties of nano-TiO2, such as size and shape, were studied.24,25 Tong et al.26 found that different shapes of nano-TiO2 (nanotubes, nanorods, nanosheets and nanospheres) showed varying degrees of phototoxicity to Escherichia coli and Aeromonas hydrophila due to different alignment of nano-TiO2 at the bacterial surface. In addition, different aqueous media conditions, such as pH, ionic strength and organic matter, could also affect the toxicity of nano-TiO2 to bacteria by changing the agglomeration and sedimentation characteristics of nanoparticles and thus inhibiting the interaction of nanoparticles with bacteria.27–30 These differing results could be due to the different interactions between nano-TiO2 and the bacterial surface. For example, Kumari et al.31 showed that the cytotoxicity of TiO2-NPs to three different bacteria (Bacillus alitudinis, Bacillus subtilis and Pseudomonas aeruginosa) could be due to the adsorption of nano-TiO2 to the bacterial surface, resulting in membrane damage and reactive oxygen species (ROS), which induced cytotoxicity. The interaction between nanoparticles and bacterial surface is, therefore, of great significance for evaluating the toxicity of nanoparticles.26,32
The degree of interaction between nanoparticles and bacteria is related to the surface characteristics of the bacteria. Suresh et al.33 found that Gram-positive bacteria (B. subtilis) were more sensitive than Gram-negative bacteria (E. coli) to nano-Ag due to different cell wall structures. Li et al.34 reported that nano-TiO2 showed higher toxicity to B. subtilis than Pseudomonas putida, because Gram-negative P. putida have a lipopolysaccharide membrane. Therefore, the difference in the microbial cell wall structures may be an important factor in the toxicity of nanoparticles to the bacteria. Moreover, bacterial surfaces are heterogeneous, consisting of complex substances (such as phospholipids, proteins and polysaccharides)35 and are coated with different soluble microbial products (e.g., exopolysaccharides) secreted by the bacteria during metabolic processes.36 There are numerous species of bacteria in the aquatic environment, and their surface characteristics are often diverse.37 Therefore, exploring the interactions between different bacteria and nanoparticles will help to assess the risk of nanoparticles to the aquatic environment.
In this study, two individual bacteria with different surface properties were selected to investigate the effects of nano-TiO2 on the biotoxicity of copper (Cu). Bacterial growth, ROS content, superoxide dismutase (SOD) and malondialdehyde (MDA) were measured to assess the toxic effects. Finally, the interaction of nanoparticles and two bacteria was investigated to elucidate the impacts of different surface characteristics on effect of nanoparticle on copper toxicity to bacteria.
2. Materials and methods
2.1. Nanoparticles preparation and characterization
Aeroxide P25 nano-TiO2 particles were purchased from Acros Organics (Belgium). Stock suspensions of nano-TiO2 (1 g L−1) were prepared by adding 1 g of P25 nano-TiO2 particles to 1000 mL of sterile Milli-Q water. The mixture was placed in an ultrasonic bath (100 W, 40 kHz) for at least 30 min to break up large agglomerates and homogenize the dispersion. The nano-TiO2 stock solution was sonicated for a further 30 min before use. For the prepared suspension, surface areas were calculated using the BET method, with a Nova 2200e BET surface area analyzer (Quantachrome, FL, USA). Size (hydrodynamic diameter) and zeta potential (ζ-potential) of nanoparticles were measured using Zetasizer (Zetasizer Nano Series, Malvern Instruments, UK).2.2. Bacterial cultivation
The two bacterial species, isolated from sediment, used in this study were identified as Bacillus megaterium and Bacillus thuringiensis by the Institute of Microbiology, Chinese Academy of Sciences, according to the morphology, physiological and biochemical characteristics and 16S rRNA gene sequence and ilvD gene sequence data.Bacteria were incubated for 24 h at 37 °C, with shaking (SHP-150, Shanghai Jinghong). The nutrient agar medium composition included 5 g L−1 NaCl, 3 g L−1 beef extract, 10 g L−1 peptone and agar 20 g L−1 (NB solid medium), sterilized for 30 min at 121 °C. After incubation, the culture was washed twice with sterilized physiological saline (0.9%, w/v) following centrifugation at 3500g for 10 min. Finally, we resuspended the cells in sterilized physiological saline for exposure experiment use. The number of bacterial suspensions was found to be about 109 CFU mL−1. Sterilized physiological saline (9 g NaCl with 1 L ultrapure water after sterilization for 30 min at 121 °C) was used as the experimental matrix throughout the study.
2.3. Exposure experiment
The toxicity exposure experiment was carried out in a 50 mL sterilized conical flask. Bacterial suspensions (1 mL; see details of bacterial cultivation) were added and diluted to 20 mL with sterilized physiological saline. The experiments were carried out in two systems (Cu-only and Cu + 1 mg L−1 nano-TiO2) in triplicate, and treatment without Cu (as control). The Cu concentration gradient was: 0, 50, 100, 200, 300 and 400 μg L−1. All flasks were sealed with sterilized film and cultured at 37 °C for 24 h, with shaking.2.4. Bacterial growth
The viability of the bacteria was assessed by the flat colony counting method. To evaluate the toxic effect of nano-TiO2 with Cu on bacterial viability after the exposure experiment, we diluted 1 mL of the experimental suspensions with typically ten-fold dilutions, three replicates of the dilution series, then spread 0.5 mL of different suspension concentrations onto nutrient agar medium plates (NB solid medium). These were cultured in the biochemical incubator at 37 °C. After 24 h, the flat colony counting method was used to calculate the number of CFU. Then, according to the corresponding dilution ratio, we calculated the quantity of bacteria, as CFU mL−1.2.5. Cu accumulation
After 24 h of exposure to nano-TiO2 and Cu, 10 mL bacterial suspensions were collected and washed twice with sterilized physiological saline. The samples were then washed with 10 mM EDTA-Na2 to remove the Cu2+ absorbed onto the cell wall. Subsequently, they were digested with 68% HNO3 (Aristar grade) at 110 °C until the solution became completely transparent. The digested solution was transferred to a volumetric flask and adjusted to a constant volume of 10 mL with 2% HNO3. The Cu concentration in the digested solution was analyzed by inductively coupled plasma mass spectroscopy (ICP MS, VG PQ2 TURBO). The Cu accumulation was expressed based on the CFU of bacteria.2.6. Determination of ROS and enzyme activity
The test kits used to measure ROS, SOD and MDA were purchased from Nanjing Jiancheng Bioengineering Institute, China. ROS generation was tested with the fluorescence probe dichloro-dihydro-fluorescein diacetate (DCFH-DA). Cell suspensions (2 mL) were incubated with DCFH-DA at a concentration of 14 μM at 37 °C for 60 min. The redundant DCFH-DA was removed by centrifugation and then suspended in 2 mL of physiological saline. To estimate the autofluorescence activity of nano-TiO2 that may interfere with the DCFH dye, we tested a negative control of nanomaterials without bacterial cells. ROS generation of microorganisms was assessed by the DCF level using a fluorescence spectrometer.Cells of 10 mL bacterial suspension were collected by centrifugation at 8000 rpm for 10 min, then washed twice with sterilized physiological saline, and transferred into 2 mL centrifugal tubes after centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. The supernatant was removed and then disrupted with an ultrasonic cell crusher (Scientz-II, Ningbo Xinzhi Biological Company) in an ice bath following the addition of 1.5 mL sucrose buffer (0.25 mo1 L−1 sucrose, 0.1 mo1 L−1 Tris–HCl, pH 8.6). The homogenate was then centrifuged (4 °C, 16
000g for 10 min. The supernatant was removed and then disrupted with an ultrasonic cell crusher (Scientz-II, Ningbo Xinzhi Biological Company) in an ice bath following the addition of 1.5 mL sucrose buffer (0.25 mo1 L−1 sucrose, 0.1 mo1 L−1 Tris–HCl, pH 8.6). The homogenate was then centrifuged (4 °C, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g) for 20 min. The supernatant was used to determine SOD activities and MDA content, using the test kits (Nanjing Jiancheng Bioengineering Institute, China) according to the instruction manual.
000 g) for 20 min. The supernatant was used to determine SOD activities and MDA content, using the test kits (Nanjing Jiancheng Bioengineering Institute, China) according to the instruction manual.
2.7. Interaction of nanoparticles and bacteria
Transmission electron microscopy (TEM) and flow cytometry (FCM) were used to investigate the interaction of different bacterial surfaces with nano-TiO2.Bacterial cells were prepared according to bacterial cultivation, bacterial suspension was incubated with 1 mg L−1 TiO2 nanoparticles for 24 h and cultured in dark conditions at 37 °C. TEM samples were prepared by dropping a suspension of bacteria and nano-TiO2 on copper grids (200 mesh, Zhongjingkeyi Technology Co., Ltd.). These were left to stand overnight to ensure adequate fixation, then the morphology of bacterial combined nano-TiO2 was obtained by TEM (JEM-2100) at 100 kV.
The association of nanomaterials and bacterial cells can be evaluated by FCM.38,39 We followed the following procedure: 1 mL bacterial suspension (as described in bacteria cultivation) was diluted to 20 mL with physiological saline, then the appropriate amount of nano-TiO2 was added to obtain a concentration of 1 mg L−1. We removed 300 μL samples from the bacterial suspension at 0, 0.5, 1, 2, 4, and 24 h and placed them into 2 mL centrifugal tubes. For time-dependent experiments, all 2 mL tubes were centrifuged at 1500 g for 15 min, then the bacterial cells were resuspended to 300 μL with phosphate buffer saline to remove the unadsorbed nano-TiO2. In order to eliminate the interference of nano-TiO2, we added 5 μL SYBR Green I nucleic acid dye (PCR) to the bacterial suspension, after mixing under dark conditions for staining on bacterial DNA about 30 min, as a marker of bacteria. Using CytoFLEX flow cytometry (Backman Kurt) we measured all stained samples with light scattering, only the SYBR Green I dye positive signals were used to analyze, using the CytExpert 1.1.10.0 software for processing.
2.8. Statistical analysis
Results were expressed as mean value ± standard deviation (n = 3). Statistical significance was accepted if p value was < 0.05. Statistical analyses (one-way analysis of variance (one-way ANOVA) and two-way analysis of variance (two-way ANOVA)) were conducted using IBM® SPSS® Statistics 20.0.3. Results and discussion
3.1. Material characterization
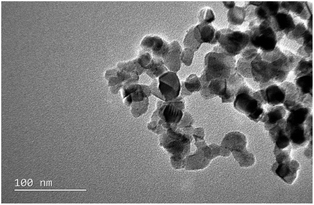
The surface area of Aeroxide P25 (nano-TiO2) was 55.29 m2 g−1. The average hydrodynamic diameter and ζ-potential were 827.1 ± 8.63 nm and 7.20 ± 0.17 mV, respectively. The nano-TiO2 images were obtained by TEM (JEM-2100F, JEOL, Japan) operated at 100 keV. The morphology of nano-TiO2 in aqueous solution was aggregated, as shown in Fig. 1.3.2. Bacterial growth
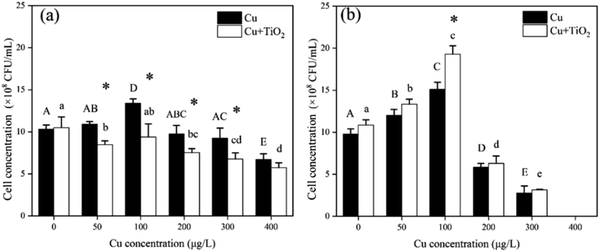
As shown in Fig. 2, Cu exposure can significantly affect the growth of both bacterial species, compared to the control (Cu concentration = 0 μg L−1). In the Cu-only system, low Cu concentration promoted the growth of bacteria. As Cu concentration increased, the growth of the two bacterial species was inhibited. The colony-forming units (CFU) of both bacteria reached a maximum at a Cu concentration of 100 μg L−1. When Cu concentration reached 400 μg L−1, the growth of B. thuringiensis was lower than that of the control (T test p < 0.05), and all B. megaterium died. The exposure to 1 mg L−1 nano-TiO2 (Cu concentration = 0 μg L−1) did not distinctly affect the growth of bacteria compared to the control (T test p > 0.05). For the Cu + nano-TiO2 system, the growth of B. thuringiensis was significantly lower than that in Cu-only system (p < 0.05, two-way ANOVA), indicating that nano-TiO2 inhibited the growth of B. thuringiensis; B. megaterium, however, showed no significant difference compared with the Cu-only system (p > 0.05, two-way ANOVA).3.3. ROS and enzyme activity
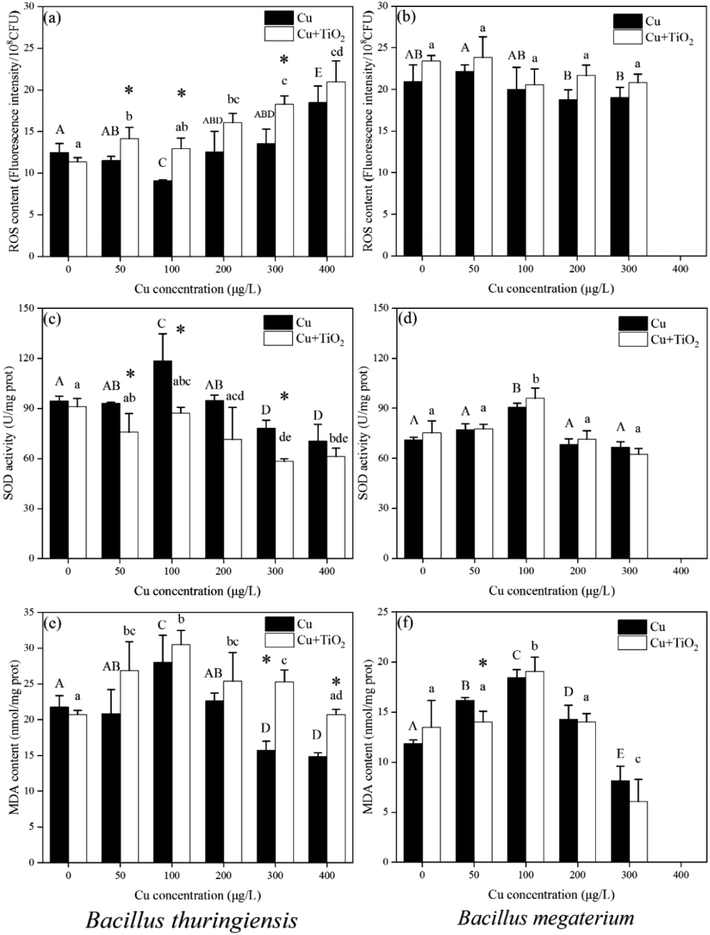
The ROS content of B. thuringiensis and B. megaterium after 24 h exposure is shown in Fig. 3(a) and (b), respectively. For B. thuringiensis in the Cu-only system, the ROS content at a Cu concentration of 100 μg L−1 was significantly lower than that in the control (Cu concentration = 0 μg L−1), while that at a Cu concentration of 400 μg L−1 it was higher than that in the control.The presence of nano-TiO2 could significantly increase the ROS content of B. thuringiensis compared to that in the Cu-only system (p < 0.01, two-way ANOVA). However, for B. megaterium, the addition of nano-TiO2 did not affect the ROS content in two exposure conditions.
As shown in Fig. 3(c) and (d), when the Cu concentration was 100 μg L−1 in the Cu-only system, the SOD activities of both bacteria significantly increased compared to those of the control (Cu concentration = 0 μg L−1). The SOD activity of B. thuringiensis decreased significantly when the Cu concentration was 300 μg L−1 and 400 μg L−1, while no significant changes were observed for B. megaterium. In the Cu + nano-TiO2 system, the SOD activity of B. thuringiensis reduced significantly compared with that in the Cu-only system (p < 0.05, two-way ANOVA), indicating that nano-TiO2 weakened the antioxidant enzyme activity. For B. megaterium, however, there was no significant difference in SOD activity in the absence or presence of nano-TiO2 (p > 0.05, two-way ANOVA).
As the final product of lipid peroxidation, MDA could reflect the extent of intracellular lipid peroxidation.40 As shown in Fig. 3(e) and (f), for B. thuringiensis, the MDA content in the Cu + nano-TiO2 system was significantly higher than that in the Cu-only system when the Cu concentration exceeded 200 μg L−1 (T test, p < 0.05). This result indicated that nano-TiO2 might enhance the oxidative stress of Cu on B. thuringiensis. For B. megaterium, however, no difference was found in either the co-existence or the Cu-only system.
It was reported that Cu could induce oxidative stress in organisms. The ROS content reflects the oxidative stress, while SOD is an important antioxidant enzyme, and MDA is a lipid oxidation product mainly induced by ROS.41,42 In these experiments, it was found that the presence of nano-TiO2 could significantly increase the ROS content, decrease SOD activity, and increase the MDA content in B. thuringiensis; however, the oxidative stress process did not significantly affect B. megaterium whether nano-TiO2 was present or not.
3.4. Interaction between bacteria surface and nano-TiO2
It was found that nano-TiO2 enhanced the toxicity of Cu to B. thuringiensis, but had no significant effect on B. megaterium. This difference in toxicity between the two bacterial species may be due to the differing interactions between nano-TiO2 and their individual bacterial surfaces. It has been reported that the interaction between nanoparticles and bacteria may proceed in two steps: first, the nanoparticles adhere to the bacterial surface, leading to membrane damage; and then the intracellular components of the bacteria leak out, resulting in bacterial death.19 Simondeckers et al.24 reported that the more TiO2 nanoparticles adsorbed onto the bacterial surface, the higher the toxicity of TiO2 nanoparticles to bacteria was found. The association between nanoparticles and bacteria may be a prerequisite in determining the toxicity of nanoparticles to bacteria.To assess our hypothesis on the association of nano-TiO2 with B. thuringiensis and B. megaterium, FCM analysis was carried out. FCM images showed the interaction between the nanoparticles and bacteria during contact. Data were showed as density plots. Gating sections, which were constructed manually using the supplied software of the FCM, were as control to compared with others to show percent cell having increase in side scatter but no increase in forward scatter. As shown in Fig. 4(a), after adding nano-TiO2, the side scatter of B. thuringiensis gradually increased with time (exposure time of 0.5, 1, 2, 4 h corresponding to 5.90%, 6.84%, 6.56%, 11.95%, respectively). It showed that the nano-TiO2 amalgamated with B. thuringiensis gradually. In contrast, the side scatter of B. megaterium increased less after the addition of nano-TiO2 (Fig. 4(b)), indicating that the association between B. megaterium and nano-TiO2 was lower. The results suggest that B. thuringiensis has a stronger association with nano-TiO2 than B. megaterium, which could lead to increased toxicity of Cu to bacteria. Wang et al.43 reported that the association of nanoparticles with bacteria might inhibit intracellular and extracellular substance transfer, cause cellular metabolism disturbance, and thus lead to cell death. The association of nanoparticles with the microbes would also facilitate the production of ROS.44,45 It is certified by the increase in growth inhibition and ROS content in B. thuringiensis in current study.
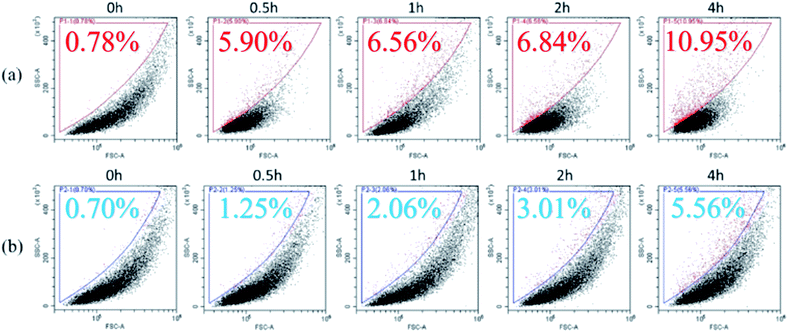 | ||
| Fig. 4 Association of nano-TiO2 with two bacteria (a) Bacillus thuringiensis and (b) Bacillus megaterium over time. | ||
The two bacterial species that we investigated are similar in shape and are both Gram-positive. Thus, it is believed that the difference in interaction behavior is due to the different surface characteristics. B. thuringiensis excretes crystalline proteins during metabolic processes, and its surface is covered by an S-layer, composed of proteins. B. megaterium, however, produces exopolysaccharides, which adhere to the bacterial surface.46 We used TEM to determine the impact of the bacterial surface characteristics on the interactions between nano-TiO2 and the bacteria. The results showed that the surface of B. thuringiensis was indeed enveloped by an S-layer (Fig. 5(a)), and exopolysaccharides aggregated around B. megaterium (Fig. 5(b)). Following exposure to 1 mg L−1 nano-TiO2 for 24 h, more nano-TiO2 attached to the surface of B. thuringiensis (Fig. 5(c)) than B. megaterium, i.e., greater association formed between nano-TiO2 and B. thuringiensis than between nano-TiO2 and B. megaterium. Other researchers also found that proteins have a greater ability to aggregate with nanoparticles than polysaccharides.47
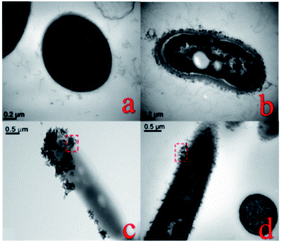 | ||
| Fig. 5 TEM images of bacteria (a) Bacillus thuringiensis and (b) Bacillus megaterium, and (c) Bacillus thuringiensis and (d) Bacillus megaterium after 24 h exposure to 1 mg L−1 nano-TiO2. | ||
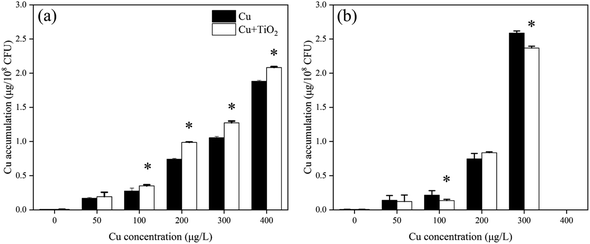
Therefore, nano-TiO2 adsorbed onto the surface of B. thuringiensis may compete with heavy metal ions for binding sites, thus influencing the regulation of heavy metals by the bacteria. When nano-TiO2 and Cu co-existed, more Cu ions were able to enter into the B. thuringiensis cells subsequently, leading to increase of Cu accumulation (Fig. 6), growth inhibition and oxidative stress. For B. megaterium, however, the exopolysaccharides attached to the bacterial surface provided a protective barrier, thus obstructing contact with nano-TiO2, and decreasing the effect of nano-TiO2 on Cu toxicity to B. megaterium.48,49
4. Conclusions
The results suggest that when nanoparticles co-exist with heavy metals, the characteristics of bacterial surface may influence the association of bacteria with nano-TiO2, hence impacting the toxicity of heavy metals to bacteria. The study presented here demonstrates that the interaction between bacterial surface and nanoparticles may be crucial to evaluate the toxicity of the nanoparticles.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant no. 21976010, 51178031).References
- J. Lee, S. Mahendra and P. J. J. Alvarez, ACS Nano, 2010, 4, 3580 CrossRef CAS PubMed.
- M. Auffan, M. Pedeutour, J. Rose, A. Masion, F. Ziarelli, D. Borschneck, C. Chaneac, C. Botta, P. Chaurand, J. Labille and J.-Y. Bottero, Environ. Sci. Technol., 2010, 44, 2689 CrossRef CAS PubMed.
- A. Weir, P. Westerhoff, L. Fabricius, K. Hristovski and N. von Goetz, Environ. Sci. Technol., 2012, 46, 2242 CrossRef CAS PubMed.
- C. O. Robichaud, A. E. Uyar, M. R. Darby, L. G. Zucker and M. R. Wiesner, Environ. Sci. Technol., 2009, 43, 4227 CrossRef CAS PubMed.
- F. Gottschalk, T. Sonderer, R. W. Scholz and B. Nowack, Environ. Sci. Technol., 2009, 43, 9216 CrossRef CAS PubMed.
- J. Chen, X. Dong, Y. Xin and M. Zhao, Aquat. Toxicol., 2011, 101, 493 CrossRef CAS PubMed.
- H. Ma and S. A. Diamond, Environ. Toxicol. Chem., 2013, 32, 2139 CrossRef CAS PubMed.
- I. Amiano, J. Olabarrieta, J. Vitorica and S. Zorita, Environ. Toxicol. Chem., 2012, 31, 2564 CrossRef CAS PubMed.
- F. Li, Z. Liang, X. Zheng, W. Zhao, M. Wu and Z. Wang, Aquat. Toxicol., 2015, 158, 1 CrossRef CAS PubMed.
- S. Jomini, H. Clivot, P. Bauda and C. Pagnout, Environ. Pollut., 2015, 202, 196 CrossRef CAS PubMed.
- C. T. Binh, T. Tong, J. F. Gaillard, K. A. Gray and J. J. Kelly, Environ. Toxicol. Chem., 2014, 33, 317 CrossRef CAS PubMed.
- R. R. Rosenfeldt, F. Seitz, R. Schulz and M. Bundschuh, Environ. Sci. Technol., 2014, 48, 6965 CrossRef CAS PubMed.
- C. D. Torre, F. Buonocore, G. Frenzilli, S. Corsolini, A. Brunelli, P. Guidi, A. Kocan, M. Mariottini, F. Mottola, M. Nigro, K. Pozo, E. Randelli, M. L. Vannuccini, S. Picchietti, M. Santonastaso, V. Scarcelli, S. Focardi, A. Marcomini, L. Rocco, G. Scapigliati and I. Corsi, Environ. Pollut., 2015, 196, 185 CrossRef PubMed.
- W. W. Yang, Y. Wang, B. Huang, N. X. Wang, Z. B. Wei, J. Luo, A. J. Miao and L. Y. Yang, Environ. Sci. Technol., 2014, 48, 7568 CrossRef CAS PubMed.
- W. Fan, M. Cui, H. Liu, C. Wang, Z. Shi, C. Tan and X. Yang, Environ. Pollut., 2011, 159, 729 CrossRef CAS PubMed.
- C. Tan, W. H. Fan and W. X. Wang, Environ. Sci. Technol., 2012, 46, 469 CrossRef CAS PubMed.
- W. W. Yang, Y. Li, A. J. Miao and L. Y. Yang, Ecotoxicol. Environ. Saf., 2012, 85, 44 CrossRef CAS PubMed.
- W. W. Yang, A. J. Miao and L. Y. Yang, PLoS One, 2012, 7(3), e32300 CrossRef CAS PubMed.
- S. J. Klaine, T. F. Fernandes, R. D. Handy, D. Y. Lyon, S. Mahendra, M. J. Mclaughun and J. R. Lead, Environ. Toxicol. Chem., 2010, 31, 1825 Search PubMed.
- Y. Ju-Nam and J. R. Lead, Sci. Total Environ., 2008, 400, 396 CrossRef CAS PubMed.
- L. K. Adams, D. Y. Lyon and P. J. Alvarez, Water Res., 2006, 40, 3527 CrossRef CAS PubMed.
- S. J. Klaine, A. A. Koelmans, N. Horne, S. Carley, R. D. Handy, L. Kapustka, B. Nowack and F. von der Kammer, Environ. Toxicol. Chem., 2012, 31, 3 CrossRef CAS PubMed.
- C. T. Binh, T. Tong, J. F. Gaillard, K. A. Gray and J. J. Kelly, PLoS One, 2014, 9(8), e106280 CrossRef PubMed.
- A. Simondeckers, S. Loo, M. Maynel'Hermite, N. Herlinboime, N. Menguy, C. Reynaud, B. Gouget and M. Carrière, Environ. Sci. Technol., 2009, 43, 8423 CrossRef CAS PubMed.
- X. Lin, J. Li, S. Ma, G. Liu, K. Yang, M. Tong and D. Lin, PLoS One, 2014, 9(10), e110247 CrossRef PubMed.
- T. Tong, A. Shereef, J. Wu, C. T. Binh, J. J. Kelly, J. F. Gaillard and K. A. Gray, Environ. Sci. Technol., 2013, 47, 12486 CrossRef CAS PubMed.
- T. Tong, C. T. Binh, J. J. Kelly, J. F. Gaillard and K. A. Gray, Water Res., 2013, 47, 2352 CrossRef CAS PubMed.
- R. A. French, A. R. Jacobson, B. Kim, S. L. Isley, R. L. Penn and P. C. Baveye, Environ. Sci. Technol., 2009, 43, 1354 CrossRef CAS PubMed.
- I. Chowdhury, D. M. Cwiertny and S. L. Walker, Environ. Sci. Technol., 2012, 46, 6968 CrossRef CAS PubMed.
- A. M. Ng, C. M. Chan, M. Y. Guo, Y. H. Leung, A. B. Djurisic, X. Hu, W. K. Chan, F. C. Leung and S. Y. Tong, Appl. Microbiol. Biotechnol., 2013, 97, 5565 CrossRef CAS PubMed.
- J. Kumari, D. Kumar, A. Mathur, A. Naseer, R. R. Kumar, P. Thanjavur Chandrasekaran, G. Chaudhuri, M. Pulimi, A. M. Raichur, S. Babu, N. Chandrasekaran, R. Nagarajan and A. Mukherjee, Environ. Res., 2014, 135, 333 CrossRef CAS PubMed.
- G. Gogniat, M. Thyssen, M. Denis, C. Pulgarin and S. Dukan, FEMS Microbiol. Lett., 2006, 258, 18 CrossRef CAS PubMed.
- A. K. Suresh, D. A. Pelletier, W. Wang, J. Moon, B. Gu, N. P. Mortensen, D. P. Allison, D. C. Joy, T. J. Phelps and M. J. Doktycz, Environ. Sci. Technol., 2010, 44, 5210 CrossRef CAS PubMed.
- M. Li, M. E. Noriega-Trevino, N. Nino-Martinez, C. Marambio-Jones, J. Wang, R. Damoiseaux, F. Ruiz and E. M. Hoek, Environ. Sci. Technol., 2011, 45, 8989 CrossRef CAS PubMed.
- W. Chen, C. Qian, X. Y. Liu and H. Q. Yu, Environ. Sci. Technol., 2014, 48, 11119 CrossRef CAS PubMed.
- B. J. Ni, B. E. Rittmann and H. Q. Yu, Trends Biotechnol., 2011, 29, 454 CrossRef CAS PubMed.
- A. K. Suresh, D. A. Pelletier and M. J. Doktycz, Nanoscale, 2013, 5, 463 RSC.
- K. S. Butler, B. J. Casey, G. V. M. Garborcauskas, B. J. Dair and R. K. Elespuru, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2014, 768, 14 CrossRef CAS PubMed.
- A. Kumar, A. K. Pandey, S. S. Singh, R. Shanker and A. Dhawan, Chemosphere, 2011, 83, 1124 CrossRef CAS PubMed.
- L. Gaetke, Toxicology, 2003, 189, 147 CrossRef CAS PubMed.
- W. Fan, X. Wang, M. Cui, D. Zhang, Y. Zhang, T. Yu and L. Guo, Environ. Sci. Technol., 2012, 46, 10255 CrossRef CAS PubMed.
- W. Fan, R. Peng, X. Li, J. Ren, T. Liu and X. Wang, Water Res., 2016, 105, 129–137 CrossRef CAS PubMed.
- Z. Wang, Y. H. Lee, B. Wu, A. Horst, Y. Kang, Y. J. Tang and D. R. Chen, Chemosphere, 2010, 80, 525 CrossRef CAS PubMed.
- T. T. B. Chu, C. G. Peterson, T. Tong, K. A. Gray, J. F. Gaillard and J. J. Kelly, PLoS One, 2015, 10(4), e0125613 CrossRef PubMed.
- D. Lin, J. Ji, Z. Long, K. Yang and F. Wu, Water Res., 2012, 46, 4477 CrossRef CAS PubMed.
- S. R. Chowdhury, R. K. Basak, R. Sen and B. Adhikari, Carbohydr. Polym., 2011, 86, 1327 CrossRef CAS.
- G. Huang, T. W. Ng, T. An, G. Li, B. Wang, D. Wu, H. Y. Yip, H. Zhao and P. K. Wong, Water Res., 2018, 129, 522 CrossRef CAS PubMed.
- C. M. Hessler, M. Y. Wu, Z. Xue, H. Choi and Y. Seo, Water Res., 2012, 46, 4687 CrossRef CAS PubMed.
- T. T. More, J. S. Yadav, S. Yan, R. D. Tyagi and R. Y. Surampalli, J. Environ. Manage., 2014, 144, 1 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |