Open Access Article
Open Access Articleπ–π stacked DNA G-wire nanostructures formed by a short G-rich oligonucleotide containing a 3′–3′ inversion of polarity site†
Maria
Marzano
 a,
Andrea P.
Falanga
a,
Principia
Dardano
a,
Andrea P.
Falanga
a,
Principia
Dardano
 b,
Stefano
D'Errico
b,
Stefano
D'Errico
 c,
Ilaria
Rea
c,
Ilaria
Rea
 b,
Monica
Terracciano
b,
Monica
Terracciano
 a,
Luca
De Stefano
a,
Luca
De Stefano
 b,
Gennaro
Piccialli
b,
Gennaro
Piccialli
 a,
Nicola
Borbone
a,
Nicola
Borbone
 *a and
Giorgia
Oliviero
*a and
Giorgia
Oliviero
 d
d
aDepartment of Pharmacy, University of Naples Federico II, Via D. Montesano 49, 80131 – Naples, Italy. E-mail: nicola.borbone@unina.it
bInstitute of Applied Sciences and Intelligent Systems, National Council Research of Italy, Via P. Castellino 111, 80131 – Naples, Italy
cCESTEV, University of Naples Federico II, Via T. De Amicis 95, 80145 – Naples, Italy
dDepartment of Molecular Medicine and Medical Biotechnologies, University of Naples Federico II, Via S. Pansini 5, 80131 – Naples, Italy
First published on 6th July 2020
Abstract
The interest in DNA based nanostructures arises from their potential applications in diagnostics and drug delivery and in the development of new hybrid and conducting materials. Guanine-rich oligonucleotides can multimerize forming long and stable supramolecular structures, known as G-wires, based on the G-quadruplex (G4) motif. Herein, we report a method to easily obtain long DNA G-wires based on a new tetramolecular G4 subunit formed by the d(5′-CGG-3′-3′-GGC-5′) sequence containing a 3′–3′ inversion of polarity site. The formation of the G-wire assembly exploits the multimerization ability of G4s presenting the CGG motif at their 5′ end via π–π stacking interactions between flanking G4 subunits. The structures and the stability of the resulting G-wires were investigated by HPLC, size exclusion chromatography, polyacrylamide gel electrophoresis, circular dichroism, 1H NMR and atomic force microscopy studies.
Introduction
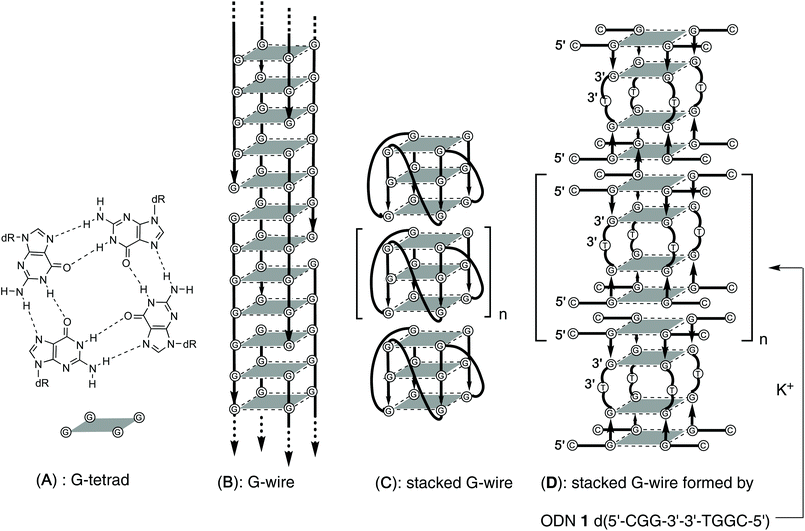
The discovery of new biomaterials based on ordered supramolecular structures is a hot topic in the development of nanotechnologies.1–7 Among the various biomaterials able to build supramolecular structures, the DNA strand is a very attractive building-block having the capability of forming supramolecular complexes whose dimensions and shapes can be predicted ab initio by varying the length and sequence of the DNA strand.8–11 DNA-based supramolecular structures are built by exploiting the self-assembling driving forces of DNA strands which bind to each other following simple base-pairing patterns based on Watson–Crick, Hoogsteen and reverse Hoogsteen hydrogen-bonding rules. Depending on the combination of the hydrogen bonding patterns involved, DNA strands can form predictable and well-defined secondary structures (e.g. duplex, triplex, DNA origami, G4).12–14 G4s are unusual RNA and DNA secondary structures which form when G-rich ON strands are annealed in the presence of suitable cations. The binding motif of G4s is the G-quartet (Fig. 1A), a planar arrangement formed by four guanine bases held together by eight Hoogsteen hydrogen bonds.15–17 π–π interactions generated between the stacked G-quartets and the presence of coordinating cations, such as potassium or sodium, give stability to the whole G4 structure.18–21G4s are involved in many critical biological processes, including the expression of many proto-oncogenes, the maintenance of telomere length and the recognition and binding of several proteins,22–26 and are also emerging as self-assembling scaffolds to be used in supramolecular chemistry applications and nanotechnology.27 Furthermore, G4s possess higher conductivity than DNA duplexes, thus suggesting their use in bioelectronics.28–31
Supramolecular structures based on a G4 scaffold can be obtained directly through the self-assembling of the G-rich strands (Fig. 1B) or by multimerization of G4 building blocks held together by end-to-end π–π stacking (Fig. 1C). The G-wires reported, in Fig. 1B, characterized by the cooperative binding of interlocked slipped strands, have a rod-shaped superstructure that can reach the length of thousands of nanometres along the axis perpendicular to the G-tetrad planes.32–35 The multimerization of a G4 building block is a process that produces stacked G-wires (Fig. 1C), which are supramolecular structures having a shape similar to the G-wires.18,36–38 Other kinds of G-wires containing both these structural motifs are also reported.39–43 The factors that influence this multimerization process are: the molecularity of the G4, the base composition of the loops, the presence of overhanging nucleotides at 5′ or 3′ end of the G-rich strand, the number of G-tetrads in the G4 monomer and the kind of π–π stacking among the monomeric building blocks.44,45
In previous studies, we reported that the 7-mer d(CGGXGGT) (X = T, A, C, G) ONs in the presence of K+ ions form tetramolecular d(CGGXGGT)4 G4s (from here on referred as the Q1 monomeric building blocks) which rapidly evolve to the corresponding dimeric Q2 complexes via π–π end-stacking interaction between the unusual G(:C):G(:C):G(:C):G(:C) planar octad formed at the 5′-CGG end of each Q1 building block (Fig. S1, ESI†).46,47 In a successive paper, this finding was exploited in the formation of π–π stacked G-wires (Fig. 1D) obtained through the multimerization of G4 monomeric building blocks having the 5′-CGG sticky moiety at both ends. To this aim, we synthesized the d(5′-CGGT-3′-3′-GGC-5′) (1, Fig. 1D) ODN sequence incorporating an inversion of polarity site at the 3′ side of the central T.48 In this paper, we continue our study on 5′–5′ stacked G-wires, enlarging their repertoire with those formed by the shorter symmetric d(5′-CGG-3′-3′-GGC-5′) ODN sequence (2, Fig. 2), as demonstrated through size exclusion chromatography (HPLC-SEC), polyacrylamide gel electrophoresis (PAGE), circular dichroism (CD), 1H NMR and atomic force microscopy (AFM) evidence.
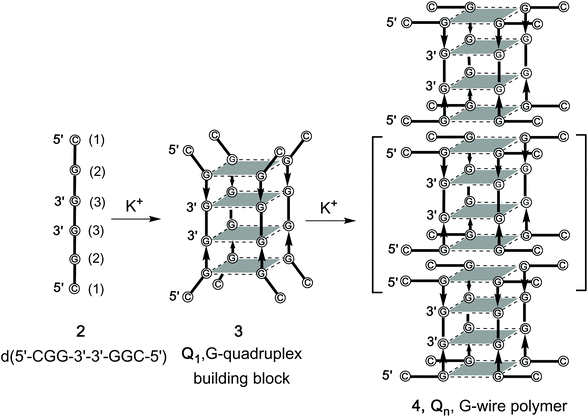 | ||
| Fig. 2 Formation of the G-quadruplex building block 3 (Q1) and its multimerization into G-wire polymers 4 (Qn) starting from 2. | ||
The comparison of stability and length distribution between the G-wires formed by 1 and 2 revealed the crucial role played by the presence of the thymidine at the inversion of polarity site.
Results and discussion
Synthesis of d(5′-CGG-3′-3′-GGC-5′) and formation of π–π stacked G-wires
d(5′-CGG-3′-3′-GGC-5′) (2) was synthesized using a solid phase automated DNA synthesizer as described in the Experimental section. The 3′–3′ phosphodiester bond was achieved by performing the first three coupling cycles using 5′-phosphoramidites, and the remaining three using standard 3′-phosphoramidites. After purification by HPLC in a SAX column, the structure and purity of 2 were confirmed by ESI MS data and 1H NMR spectroscopy. In particular, the enhanced resolution ESI-MS spectrum of 2 showed a single peak corresponding to the single-stranded double-charged pseudomolecular ion [M − 2H]2− (m/z 915.0, Fig. S2†). The 1H NMR spectrum recorded at 50 °C showed the expected three aromatic and three anomeric proton signals because of the symmetric nature of the ODN sequence (Fig. S3†). The G4 building block 3 and the G-wire 4 (Fig. 2), from now on respectively referred as Q1 and Qn, were obtained by dissolving 2 in 1.0 M K+-containing buffer at the single strand (ss) concentration of 0.05, 1.0 and 3.6 mM followed by heating at 90 °C for 10 min, fast cooling at 4 °C and storage for 24 before further investigation. The formation of Q1 and its dimer Q2 was also confirmed by ESI-MS spectrometry which showed multicharged peaks corresponding to the G4 species stabilized respectively by three or six ammonium ions (Fig. S4†).Polyacrylamide gel electrophoresis (PAGE) studies
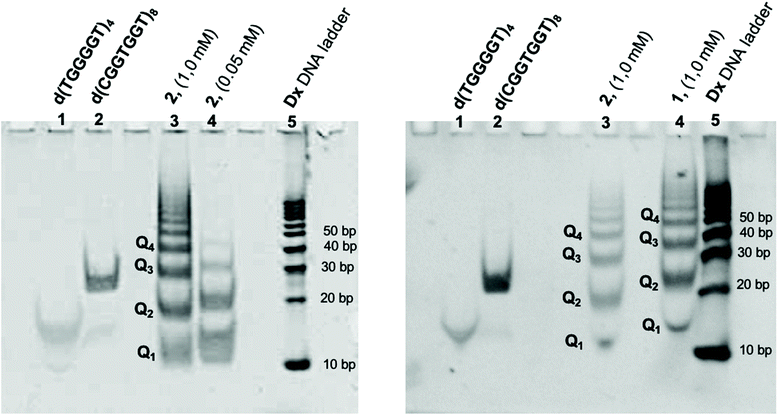
ODN 2 after the annealing procedure was analyzed by PAGE to investigate on the G4 formation and its multimerization to form the Qn multimers. We compared the electrophoretic mobility of 2 (lane 3 and 4, in Fig. 3A) with those of the G4s d(TGGGGT)4 (lane 1) and d(CGGTGGT)8 (lane 2), used as size markers for the G4 building block Q1 and its dimer Q2, respectively. ODN 2 annealed at 1.0 mM ODN single strand concentration in 1.0 M K+ buffer migrated as a well-defined ladder of bands (lane 3) in which the shorter terms of the Qn polymer (Q1–4) could be identified. The fastest band (lane 3) migrated with almost the same mobility as the G4 monomer marker (lane 1) and was attributed to the Q1 species (3, Fig. 2) also considering the DNA ladder reference (lane 5). As expected, Q2 species (48 nucleotides) migrated a little faster than the dimer marker (lane 2, 56 nucleotides). Furthermore, we observed that the efficiency and progression of the polymerization process of 2 depend on its concentration. When 2 was annealed at 0.05 mM in the same buffer, it showed a different distribution of the multimers (lane 4), in which only the first three terms (Q1–3) of the G-wires distribution could be clearly observed.These data strongly suggest that 2 in the presence of K+ ions self-assembles to give a distribution of G-wire species of different length (4, Fig. 2) by the sequential 5′-stacking of the symmetrical tetramolecular G4 building block 3. The PAGE in Fig. 3B reports the comparison between the electrophoretic mobility of the G-wires formed by 1 and 2. As expected, all bands formed by the longer (7-mer) ODN 1 had lower mobility than the corresponding bands formed by the shorter (6-mer) ODN 2.
Size exclusion chromatography (SEC) studies
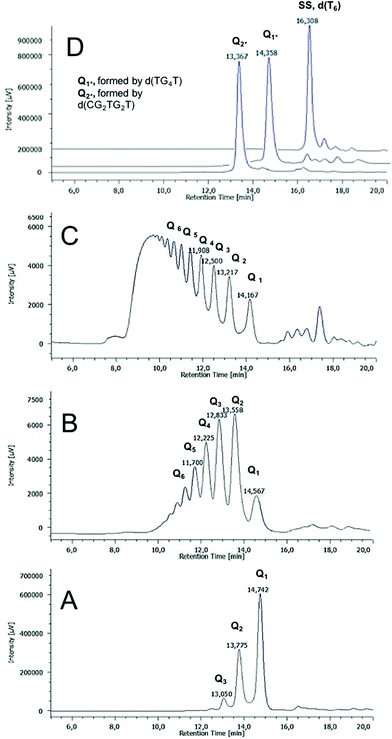
The G-wires distribution formed by 2 was further analyzed by size exclusion chromatography at room temperature on a Yarra 2000 HPLC-SEC column. The HPLC profile of 2 annealed at 1.0 mM ss concentration in 1.0 M K+-containing buffer is reported in Fig. 4B. It showed a distribution of peaks in which lower molecular weight species had greater retention times.As molecular weight markers, we used d(T6), d(TGGGGT)4 and d(CGGTGGT)8 respectively for the single-stranded ODN and for the monomeric (Q1*) and dimeric (Q2*) G4 assemblies (Fig. 4D). The above mentioned HPLC profiles indicated that in the reported conditions 2 forms a distribution of Qn multimers composed by all members of the series with n ≤ 7. Longer G-wires (n > 7) and the ss ODN were present only as traces (Fig. 4B). The HPLC-SEC analysis of the multimers formed annealing ODN 2 at lower concentration (0.05 mM ss concentration, Fig. 4A) agreed well with the PAGE data. Only three peaks (Q1–3) were observed, thus indicating that the polymerization process is slow at lower ODN concentrations. When 2 was annealed at 3.6 mM ss concentration, we saw, as expected, the shift towards higher molecular weight species of the Qn distribution (Fig. 4C). Neither of the HPLC profiles showed in Fig. 4A–C changed prolonging the annealing time of the injected sample up to 48 h and 72 h, thus suggesting that in those conditions the polymerization–depolymerization process reaches an equilibrium within the first 24 h from the annealing (data not shown). We also tested the capability of ODN 2 to form Qn multimers in the presence of NH4+ and Na+ cations. In both cases, we detected a slower but similar polymerization process that was monitored by HPLC-SEC and PAGE experiments (data not shown). To assess the purity and stability at time and temperature of the shorter members of the Qn distribution 4, we collected the HPLC-SEC peaks of Q1–3 and stored them at r.t. in the collected eluent. Each isolated peak was reinjected in the same conditions 30 min or 24 h after the isolation to check the propensity to convert either into higher or lower M.W. assemblies (Fig. S5†). The results revealed that Q1 was obtained as an almost pure species that remained stable for 24 h when only a little amount of the Q2 species (5%) was observed. On the contrary, the reinjection of Q2 and Q3 30 min after their isolation disclosed the formation of shortening fragments, whose amounts increased considerably after 24 h. In any case, we did not observe the re-formation of the single strand 2. The effect of the temperature on the distribution of Qn multimers formed by 2 was studied by injecting the annealed 2 before and after its heating at 45, 65 and 85 °C (Fig. S6B†), and the results were compared with the corresponding data reported for Qn multimers formed by 1 (Fig. S6A†).41 In both cases, the heating induced the melting of longer G-wires with the concomitant increase of the G4 monomer Q1. However, we observed a noteworthy difference in the amount of ss species formed by the two ODNs. In the HPLC-SEC profile of 1, the most retained peak corresponding to the ss ODN was observed at all the studied temperatures and represented the most intense or the only relevant peak respectively at 65 and 85 °C (Fig. S6A†). Conversely, in the case of 2, we did not observe a significant amount of the ss species even at 85 °C, where Q1 and Q2 were the sole observable species in the 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The HPLC-SEC study evidenced the higher thermal stability of the G4 monomer formed by 2, likely due to the absence of the central thymine base that, instead, weakens the G-tetrads stacking in the corresponding G4 building block formed by 1.
1 ratio. The HPLC-SEC study evidenced the higher thermal stability of the G4 monomer formed by 2, likely due to the absence of the central thymine base that, instead, weakens the G-tetrads stacking in the corresponding G4 building block formed by 1.
Circular dichroism (CD)
CD spectroscopy furnishes useful information about the formation and topology of G4s. It is well documented that the CD profiles of parallel G4s display a maximum around 265 nm and a minimum around 240 nm; while those of antiparallel G4s show a maximum around 295 nm and a minimum around 260 nm. These profiles result mainly from the different combination of the glycosidic bond conformation of guanosines participating in the G-tetrad core. In parallel G4s all guanosines engaged in a G-tetrad adopt the anti conformation, whereas in antiparallel G4s they adopt alternating syn–anti glycosidic bond conformation along the stem.18,49–51 In the case of Qn formed by 2, we observed a CD profile that results from the peculiar structural features of these structures, i.e. the presence of a 3′–3′ inversion of polarity site in the G4-forming ODN and the π–π stacking between the 5′-ending octads belonging to the different G4 building blocks participating in the formation of Qn G-wires.The CD profile of the Qn distribution showed positive signals at 250 and 300 nm and negative signals at 230 and 280 nm (Fig. 5), in agreement with those reported for the Qn multimer formed by 148 and the G4 formed by d(5′-TGG3′-3′GGT-5′),52 both containing a 3′–3′ inversion of polarity site in the ODN sequence. We also recorded the CD spectra of the HPLC-SEC peaks corresponding to Q1, Q2 and Q3, as well as that of all other less-retained peaks collected together (Q4−n), to assess the influence of the degree of multimerization on the CD signature of Qn assemblies (Fig. 6).
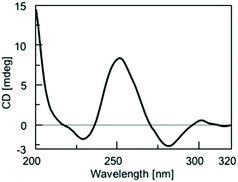 | ||
| Fig. 5 CD spectrum recorded at 25 °C of the Qn distribution obtained by annealing 2 in 100 mM K+ buffer. | ||
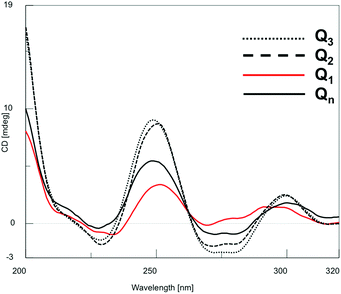 | ||
| Fig. 6 CD spectra of Q1, Q2, Q3 and Q4−n species isolated from the HPLC-SEC fractionation showed in Fig. 4B. | ||
The results confirmed the G4 nature of all the eluted species, as well as the absence of significant perturbations due to polymerization/depolymerization equilibria. To estimate the thermal behavior of the Qn distribution formed by 2, we performed CD melting experiments. The melting curve recorded at 252 nm (Fig. S4†) well evidenced that the G-wires formed by 2 are stable at room temperature and start to disassemble at temperatures higher than 30 °C. The presence of three to four inflection points in the curve disclosed the progressive shortening of the G-wire assembly. Considering that the melting process was not complete even at 90 °C, we attributed the last observed inflection point (at around 75 °C) to the formation of the Q1 building block which, differently from the corresponding Q1 species formed by 1, did not melt in the explored temperature range.
Nuclear magnetic resonance (NMR)
1H NMR spectroscopy is an important technique to investigate the formation, the stability and the topology of G4s. The downfield region of water-suppressed 1H NMR spectra of G4s is characterized by the presence of the diagnostic exchange-protected H1 imino protons of guanine bases engaged in G-tetrads formation. These protons do not exchange with water solvent protons because of the hydrogen bond which they form with either the O6 or N7 atom of the G-tetrad flanking guanosine (Fig. 1A).51,53 Each G-tetrad can originate one, two or four imino proton signals, depending on the magnetic equivalence of the four participating imino protons.54 This magnetic behavior arises from the symmetry of the whole G4 structure and the N-glycosidic torsion angles. Fig. S5† shows the downfield regions of the water suppressed 1H NMR spectra (recorded at 25, 45, 65 and 85 °C) of the Qn distribution obtained by annealing 2 at 1.5 mM ss concentration in 1.0 M K+-containing buffer. The presence of many imino protons signals in the 10.6–11.5 ppm region confirmed the existence of G4 species in all the explored temperature range. The great number of the observed signals was somewhat unexpected, considering the symmetric nature of the Q1 building block (3 in Fig. 2) and the previously reported NMR spectrum of d(5′-TGG3′-3′GGT-5′) which showed only two imino proton signals52 (thus confirming the magnetic equivalence of G-tetrads belonging to the two halves of the G4). However, taking into account the previously reported NMR data regarding the formation of the G4 dimer by the 5′-TGGAGGT-3′ ODN sequence, which evidenced modifications in the resonance frequency of all G4 imino protons before and after G4 dimerization,47 we believe that the exchange-protected signals observed at 25, 45 and 65 °C in Fig. S5† can be attributed to the G-tetrads imino protons of stacked polymeric Qn species. This hypothesis was further corroborated by the general reduction in the number and intensity of imino signals heating the sample from 25 to 85 °C as a consequence of the thermally induced transition from the Qn polymer to the monomeric Q1 building block, in agreement with HPLC-SEC and CD data. Indeed, in the NMR spectrum recorded at 85 °C, we observed only the two expected imino signals belonging to the two couples of symmetric G-tetrads in 3, thus confirming the higher thermal stability of the Q1 formed by ODN 2 with respect to the Q1 obtained from ODN 1, which resulted completely melted at that temperature.48Atomic force microscopy (AFM)
AFM is a powerful technique successfully used to explore the various topologies of G-wire.35,55,56 By using this technique, we investigated the morphology of the Qn G-wire species obtained from 2 annealed in 1.0 M K+ buffer. We used muscovite mica as the AFM support both for the super-hydrophilic property of its surface, that guarantees a lower interaction between suspended molecules in aqueous solution during the evaporation of the solvent, and for its flatness (less than 0.5 nm of root mean-square roughness for a 1000 × 1000 nm2 surface area). AFM images in Fig. 7 shows the typical arrangements of Qn multimers formed by ODN 2; in particular, Non-Contact Mode (NCM) amplitude (left) and phase (right) reveal a kind of elementary units that arranges in formations of linear shape. The analysis of a wider AFM field (Fig. S6†) disclosed the presence of more than 300 rod-shaped structures having several lengths (10–400 nm) but sharing the same height (1.8 ± 0.2 nm) and a mean width of 21 ± 7 nm (the statistical analysis is reported in Fig. S6†). The AFM length measurements are compatible with the hypothesized G-wire polymers formed by Qn multimers of different length. Moreover, the homogeneity of height values of aggregates agrees with the width value of the Q1 building block, suggesting the formation of self-assembled G-wires monolayers only in the x-y plane. Though slightly smaller than expected, the height of the Qn aggregates formed by 2 agrees with the AFM data of the G-wires obtained from 1 (height 2.0 nm),48 and lies in the range of heights reported in the literature for the G-wires (1.5–3.5 nm)35,43,56,57 which is usually smaller than the solution diameter of G4s (about 2.8 nm). Differently from height measurements, the curvature radius of the AFM tip affects the length and width measurements. We believe that the curvature radius of the AFM tip and the evaporation of the buffer solution, which could promote the lateral aggregation of Qn multimers, are both responsible for the larger width of the G-wire filaments observed in the AFM field (21 ± 7 other than the expected 2.0 nm).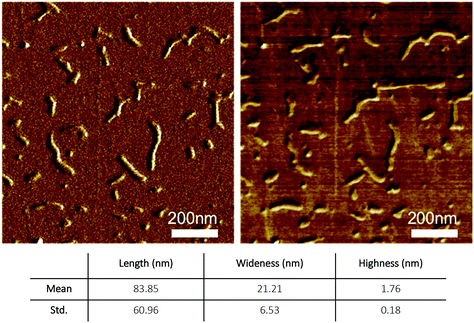 | ||
| Fig. 7 AFM imaging of Qn multimers formed by ODN 2: non-contact mode (NCM) amplitude (left) and phase (right) of the sample. | ||
Conclusions
The here reported results demonstrate that rod-shaped G4 assemblies can also be easily obtained by annealing the symmetric 6-mer ODN sequence d(5′-CGG-3′-3′-GGC-5′) (2) in K+- and NH4+-containing buffers. We selected the ODN 2 because it represented the shortest possible ODN sequence possessing the 5′-CGG moiety at both ends of the ODN strand which in our previous studies allowed the obtainment of G4 multimers via π–π end-stacking interactions. The thermal stability of the G4 building block 3 and the length distribution of the resulting G4 multimers were compared with those of the corresponding G4 structures obtained from the closest related longer analogue d(5′-CGGT-3′-3′-GGC-5′) (1) ODN. The results of this comparative study indicated that the thermal stability and length distribution of the obtained G-wires depend on the thermal stability of the G4 building block, which in turn depends on the sequence of the annealed G-rich ODN. The data indicated that the more stable is the G4 building block, the less stable is the resulting G-wire polymer. This finding suggests that it is possible to build stacked G-wires with customizable stability by varying the length and sequence of the parent ODN. In summary, our results contribute to the understanding of the processes that govern the multimerization of G-quadruplexes and extend the DNA toolbox for the construction of self-assembled supramolecular entities.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was supported in part by MIUR PRIN 2017 project 2017YJMPZN.Notes and references
- L. Yang, X. Tan, Z. Wang and X. Zhang, Supramolecular Polymers: Historical Development, Preparation, Characterization, and Functions, Chem. Rev., 2015, 115, 7196–7239 CrossRef CAS PubMed.
- L. Wang, C. Gong, X. Yuan and G. Wei, Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review, Nanomaterials, 2019, 9, 285 CrossRef CAS PubMed.
- G. A. Ozin, K. Hou, B. V. Lotsch, L. Cademartiri, D. P. Puzzo, F. Scotognella, A. Ghadimi and J. Thomson, Nanofabrication by Self-Assembly, Mater. Today, 2009, 12, 12–23 CrossRef CAS.
- E. Busseron, Y. Ruff, E. Moulin and N. Giuseppone, Supramolecular Self-Assemblies as Functional Nanomaterials, Nanoscale, 2013, 5, 7098–7140 RSC.
- M. Martín-Arroyo, A. Prado, R. Chamorro, N. Bilbao and D. González-Rodríguez, Elucidating Noncovalent Reaction Mechanisms: G-Quartet as an Intermediate in G-Quadruplex Assembly, Angew. Chem., Int. Ed., 2020, 59, 9041–9046 CrossRef PubMed.
- V. Vázquez-González, M. J. Mayoral, R. Chamorro, M. M. R. M. Hendrix, I. K. Voets and D. González-Rodríguez, Noncovalent Synthesis of Self-Assembled Nanotubes through Decoupled Hierarchical Cooperative Processes, J. Am. Chem. Soc., 2019, 141, 16432–16438 CrossRef PubMed.
- P. S. Kwon, S. Ren, S. J. Kwon, M. E. Kizer, L. Kuo, M. Xie, D. Zhu, F. Zhou, F. Zhang and D. Kim, et al., Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition, Nat. Chem., 2020, 12, 26–35 CrossRef CAS PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, DNA Origami: Scaffolds for Creating Higher Order Structures, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- Y. Cao, Y. Kuang, L. Yang, P. Ding and R. Pei, Construction of One- and Two-Dimensional Nanostructures by the Sequential Assembly of Quadruplex DNA Scaffolds, Biomacromolecules, 2019, 20, 2207–2217 CrossRef CAS PubMed.
- N. C. Seeman and H. F. Sleiman, DNA nanotechnology, Nat. Rev. Mater., 2018, 3, 17068 CrossRef CAS.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- S. Kogikoski, W. J. Paschoalino and L. T. Kubota, Supramolecular DNA Origami Nanostructures for Use in Bioanalytical Applications, TrAC, Trends Anal. Chem., 2018, 108, 88–97 CrossRef CAS.
- A. E. Marras, L. Zhou, H.-J. Su and C. E. Castro, Programmable Motion of DNA Origami Mechanisms, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 713–718 CrossRef CAS PubMed.
- V. Linko and H. Dietz, The Enabled State of DNA Nanotechnology, Curr. Opin. Biotechnol., 2013, 24, 555–561 CrossRef CAS PubMed.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Quadruplex DNA: Sequence, Topology and Structure, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed.
- H. L. Lightfoot, T. Hagen, N. J. Tatum and J. Hall, The Diverse Structural Landscape of Quadruplexes, FEBS Lett., 2019, 593, 2083–2102 CrossRef CAS PubMed.
- S. Kolesnikova and E. A. Curtis, Structure and Function of Multimeric G-Quadruplexes, Molecules, 2019, 24, 3074 CrossRef CAS PubMed.
- N. Smargiasso, W. Hsia, P. Colson, E. S. Baker, M. T. Bowers and E. De Pauw, G-Quadruplex DNA Assemblies: Loop Length, Cation Identity, and Multimer Formation, J. Am. Chem. Soc., 2008, 135, 10208–10216 CrossRef PubMed.
- A. Risitano and K. R. Fox, Influence of Loop Size on the Stability of Intramolecular DNA Quadruplexes, Nucleic Acids Res., 2004, 32, 2598–2606 CrossRef CAS PubMed.
- P. L. T. Tran, A. De Cian, J. Gros, R. Moriyama and J.-L. Mergny, Tetramolecular Quadruplex Stability and Assembly, Top. Curr. Chem., 2013, 330, 243–273 CrossRef CAS PubMed.
- D. Pavc, B. Wang, L. Spindler, I. Drevenšek-Olenik, J. Plavec and P. Šket, Ends Control Topology of DNA G-Quadruplexes and Their Cation-Dependent Assembly, Nucleic Acids Res., 2020, 48, 2749–2761 CrossRef PubMed.
- D. Rhodes and H. J. Lipps, Survey and Summary G-Quadruplexes and Their Regulatory Roles in Biology, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Quantitative Visualization of DNA G-Quadruplex Structures in Human Cells, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- C. Platella, C. Riccardi, D. Montesarchio, G. N. Roviello and D. Musumeci, G-Quadruplex-Based Aptamers against Protein Targets in Therapy and Diagnostics, Biochim. Biophys. Acta, Gen. Subj., 2016, 1861, 1429–1447 CrossRef PubMed.
- C. Roxo, W. Kotkowiak and A. Pasternak, G-Quadruplex-Forming Aptamers—Characteristics, Applications, and Perspectives, Molecules, 2019, 24, 3781 CrossRef CAS PubMed.
- A. P. Falanga, V. Cerullo, M. Marzano, S. Feola, G. Oliviero, G. Piccialli and N. Borbone, Peptide Nucleic Acid-Functionalized Adenoviral Vectors Targeting G-Quadruplexes in the P1 Promoter of Bcl-2 Proto-Oncogene: A New Tool for Gene Modulation in Anticancer Therapy, Bioconjugate Chem., 2019, 30, 572–582 CrossRef CAS PubMed.
- J. L. Mergny and D. Sen, DNA Quadruple Helices in Nanotechnology, Chem. Rev., 2019, 119, 6290–6325 CrossRef CAS PubMed.
- G. I. Livshits, A. Stern, D. Rotem, N. Borovok, G. Eidelshtein, A. Migliore, E. Penzo, S. J. Wind, R. Di Felice, S. S. Skourtis, J. C. Cuevas, L. Gurevich, A. B. Kotlyar and D. Porath, Long-Range Charge Transport in Single G-Quadruplex DNA Molecules, Nat. Nanotechnol., 2014, 9, 1040–1046 CrossRef CAS PubMed.
- W. Sun, D. Varsano and R. Di Felice, Effects of G-Quadruplex Topology on Electronic Transfer Integrals, Nanomaterials, 2016, 6, 184 CrossRef PubMed.
- A.-M. Chiorcea-Paquim, R. Eritja and A. M. Oliveira-Brett, Electrochemical and AFM Characterization of G-Quadruplex Electrochemical Biosensors and Applications, J. Nucleic Acids, 2018, 2018, 5307106 Search PubMed.
- S. P. Liu, S. H. Weisbrod, Z. Tang, A. Marx, E. Scheer and A. Erbe, Direct Measurement of Electrical Transport through G-Quadruplex DNA with Mechanically Controllable Break Junction Electrodes, Angew. Chem., Int. Ed., 2010, 49, 3313–3316 CrossRef CAS PubMed.
- T. C. Marsh, J. Vesenka and E. Henderson, A new DNA nanostructure, the G-wire, imaged by scanning probe microscopy, Nucleic Acids Res., 1995, 23, 696–700 CrossRef CAS PubMed.
- A. B. Kotlyar, N. Borovok, T. Molotsky, H. Cohen, E. Shapir and D. Porath, Long, Monomolecular Guanine-Based Nanowires, Adv. Mater., 2005, 17, 1901–1905 CrossRef CAS.
- Y. Krishnan-Ghosh, D. Liu and S. Balasubramanian, Formation of an Interlocked Quadruplex Dimer by d(GGGT), J. Am. Chem. Soc., 2004, 126, 11009–11016 CrossRef CAS PubMed.
- K. Bose, C. J. Lech, B. Heddi and A. T. Phan, High-Resolution AFM Structure of DNA G-wires in Aqueous Solution, Nat. Commun., 2018, 9, 1959 CrossRef PubMed.
- Y. Shi, H. Q. Luo and N. B. Li, A Highly Sensitive Resonance Rayleigh Scattering Method to Discriminate a Parallel-Stranded G-Quadruplex from DNA with Other Topologies and Structures, Chem. Commun., 2013, 49, 6209–6211 RSC.
- C. Saintomé, S. Amrane, J. L. Mergny and P. Alberti, The Exception That Confirms the Rule: A Higher-Order Telomeric G-Quadruplex Structure More Stable in Sodium than in Potassium, Nucleic Acids Res., 2016, 44, 2926–2935 CrossRef PubMed.
- A. Matsugami, K. Ouhashi, M. Kanagawa, H. Liu, S. Kanagawa, S. Uesugi and M. Katahira, An Intramolecular Quadruplex of (GGA)4 Triplet Repeat DNA with a G:G:G:G Tetrad and a G(:A):G(:A):G(:A):G Heptad, and Its Dimeric Interaction, J. Mol. Biol., 2001, 313, 255–269 CrossRef CAS PubMed.
- N. Ma'Ani Hessari, L. Spindler, T. Troha, W. C. Lam, I. Drevenšek-Olenik and M. Webba Da Silva, Programmed Self-Assembly of a Quadruplex DNA Nanowire, Chem. – Eur. J., 2014, 20, 3626–3630 CrossRef PubMed.
- T. Ilc, P. Šket, J. Plavec, M. Webba Da Silva, I. Drevenšek-Olenik and L. Spindler, Formation of G-wires: The Role of G:C-Base Pairing and G-Quartet Stacking, J. Phys. Chem. C, 2013, 117, 23208–23215 CrossRef CAS.
- P. Tóthová, P. Krafčíková and V. Víglaský, Formation of Highly Ordered Multimers in G-Quadruplexes, Biochemistry, 2014, 53, 7013–7027 CrossRef PubMed.
- P. Podbevšek and J. Plavec, KRAS Promoter Oligonucleotide with Decoy Activity Dimerizes into a Unique Topology Consisting of Two G-Quadruplex Units, Nucleic Acids Res., 2016, 44, 917–925 CrossRef PubMed.
- T. Troha, I. Drevenšek-Olenik, M. Webba Da Silva and L. Spindler, Surface-Adsorbed Long G-Quadruplex Nanowires Formed by G:C Linkages, Langmuir, 2016, 32, 7056–7063 CrossRef CAS PubMed.
- S. Kolesnikova, M. Hubálek, L. Bednárová, J. Cvačka and E. A. Curtis, Multimerization Rules for G-Quadruplexes, Nucleic Acids Res., 2017, 45, 8684–8696 CrossRef CAS PubMed.
- A. M. Varizhuk, A. D. Protopopova, V. B. Tsvetkov, N. A. Barinov, V. V. Podgorsky, M. V. Tankevich, M. A. Vlasenok, V. V. Severov, I. P. Smirnov, E. V. Dubrovin, D. V. Klinov and G. E. Pozmogova, Polymorphism of G4 Associates: From Stacks to Wires via Interlocks, Nucleic Acids Res., 2018, 46, 8978–8992 CrossRef CAS PubMed.
- N. Borbone, J. Amato, G. Oliviero, V. D'Atri, V. Gabelica, E. De Pauw, G. Piccialli and L. Mayol, D(CGGTGGT) Forms an Octameric Parallel G-Quadruplex via Stacking of Unusual G(:C):G(:C):G(:C):G(:C) Octads, Nucleic Acids Res., 2011, 39, 7848–7857 CrossRef CAS PubMed.
- V. D'Atri, N. Borbone, J. Amato, V. Gabelica, S. D'Errico, G. Piccialli, L. Mayol, G. Oliviero, V. D'Atri, N. Borbone, J. Amato, V. Gabelica, S. D'Errico, G. Piccialli, L. Mayol and G. Oliviero, DNA-Based Nanostructures: The Effect of the Base Sequence on Octamer Formation from d(XGGYGGT) Tetramolecular G-Quadruplexes, Biochimie, 2014, 99, 119–128 CrossRef PubMed.
- G. Oliviero, S. D'Errico, B. Pinto, F. Nici, P. Dardano, I. Rea, L. De Stefano, L. Mayol, G. Piccialli and N. Borbone, Self-Assembly of G-Rich Oligonucleotides Incorporating a 3′-3′ Inversion of Polarity Site: A New Route Towards G-Wire DNA Nanostructures, ChemistryOpen, 2017, 6, 599–605 CrossRef CAS PubMed.
- M. Vorlíčková, I. Kejnovská, J. Sagi, D. Renčiuk, K. Bednářová, J. Motlová and J. Kypr, Circular Dichroism and Guanine Quadruplexes, Methods, 2012, 57, 64–75 CrossRef PubMed.
- V. Esposito, A. Virgilio, A. Pepe, G. Oliviero, L. Mayol and A. Galeone, Effects of the Introduction of Inversion of Polarity Sites in the Quadruplex Forming Oligonucleotide TGGGT, Bioorg. Med. Chem., 2009, 17, 1997–2001 CrossRef CAS PubMed.
- R. Jin, B. L. Gaffney, C. Wang, R. A. Jones and K. J. Breslauer, Thermodynamics and Structure of a DNA Tetraplex: A Spectroscopic and Calorimetric Study of the Tetramolecular Complexes of d(TG3 T) and d(TG3T2G3 T), Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 8832–8836 CrossRef CAS PubMed.
- V. Esposito, A. Virgilio, A. Randazzo, A. Galeone and L. Mayol, A New Class of DNA Quadruplexes Formed by Oligodeoxyribonucleotides Containing a 3′-3′ or 5′-5′ Inversion of Polarity Site, Chem. Commun., 2005, 43, 3953–3955 RSC.
- C. Dalvit, Efficient Multiple-Solvent Suppression for the Study of the Interactions of Organic Solvents with Biomolecules, J. Biomol. NMR, 1998, 11, 437–444 CrossRef CAS.
- M. Adrian, B. Heddi and A. T. Phan, NMR Spectroscopy of G-Quadruplexes, Methods, 2012, 57, 11–24 CrossRef CAS PubMed.
- J. Vesenka and T. Marsh, The Diameter of Duplex and Quadruplex DNA Measured by Scanning Probe Microscopy, Scanning Microsc., 1998, 12, 329–342 Search PubMed.
- A.-M. Chiorcea-Paquim, P. V. Santos, R. Eritja and A. M. Oliveira-Brett, Self-Assembled G-Quadruplex Nanostructures: AFM and Voltammetric Characterization, Phys. Chem. Chem. Phys., 2013, 15, 9117–9124 RSC.
- A. D. Rodrigues Pontinha, A. M. Chiorcea-Paquim, R. Eritja and A. M. Oliveira-Brett, Quadruplex Nanostructures of D(TGGGGT): Influence of Sodium and Potassium Ions, Anal. Chem., 2014, 86, 5851–5857 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0qo00561d |
| This journal is © the Partner Organisations 2020 |