Chemically modified nucleic acid biopolymers used in biosensing
Lei
Zhang†
a,
Yanbing
Yang†
 a,
Jie
Tan
a,
Jie
Tan
 *a and
Quan
Yuan
*a and
Quan
Yuan
 *ab
*ab
aInstitute of Chemical Biology and Nanomedicine (ICBN), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, People's Republic of China. E-mail: tanjie0416@hnu.edu.cn
bKey Laboratory of Analytical Chemistry for Biology and Medicine (Ministry of Education), College of Chemistry and Molecular Sciences, Wuhan University, Wuhan 430072, People's Republic of China. E-mail: yuanquan@whu.edu.cn
First published on 18th February 2020
Abstract
Biopolymers are highly designable and are often used in biosensing processes. As a biopolymer, a nucleic acid not only has excellent programmability and biocompatibility, but also has a certain molecular recognition function that can directly realize biological recognition in biosensors. However, the structural variety of natural nucleotides hinders the designability of nucleic acid biopolymers. The introduction of chemical modifications in nucleic acids can enrich the designability of nucleic acid biopolymers, thereby expanding their applications in biosensing. To date, there have been many reviews paying attention to biopolymers used in biosensing, but few reviews have focused on chemically modified nucleic acid biopolymers. Here, we review different kinds of assemblies based on chemically modified nucleic acid biopolymers. We summarize their advances in the field of biosensing. Furthermore, we present challenges and prospects in this field, aiming to provide a promising step for a versatile biosensor platform based on chemically modified nucleic acid biopolymers.
1. Introduction
Biosensing consists of a biorecognition process and a transduction process.1–4 Due to their high designability, biopolymers are often used to connect the biorecognition process with the transduction process.5,6 It is worth noting that biopolymers can form nanostructures through a highly controllable dynamic assembly process, enabling the application of biosensing at the cellular or in vivo level.7–10A nucleic acid, as a biopolymer, has excellent programmability and biocompatibility.11–16 In addition, nucleic acids with specific sequences can show a molecular recognition function and can be directly used in the biological recognition process.17,18
However, the structural variety of natural nucleotide hinders the designability of nucleic acid biopolymers, thereby limiting their applications in biosensing.12,19 The introduction of chemical modifications can enrich the functional groups of nucleic acids and enhance the intermolecular interactions between nucleic acids.20–22 Thus, chemically modified nucleic acids enable the design of nucleic acid biopolymers.23,24 Therefore, chemically modified nucleic acid biopolymers hold great promise in the field of biosensing.25,26
To date, there have been many reviews on biopolymers used in biosensing,27–31 but few reviews have paid attention to chemically modified nucleic acid biopolymers. In this review, we focus on chemically modified nucleic acid biopolymers and their applications in biosensing. Specifically, we introduce the assembly methods of chemically modified nucleic acid biopolymers and the applications in biomolecule detection, controlled drug release, and gene therapy. Finally, the challenges and prospects in this field are also comprehensively discussed, aiming to provide a promising step for a versatile biosensor platform based on chemically modified nucleic acid biopolymers.
2. Assembly of chemically modified nucleic acid biopolymers
Nanoassemblies are often used in biosensing due to their similar size to biomolecules.32,33 Studies have shown that the design of nanostructures in two and three dimensions can greatly increase the sensitivity of biomolecule-targeted biosensors.34,35 Besides complementary base pairing interactions, chemical modifications can introduce new molecular interactions in nucleic acids.36 The new molecular forces enable the design of various geometric structures and bionic structures, creating opportunities for biosensing applications.21,37 In the following section, we mainly introduce assemblies based on chemically modified nucleic acid biopolymers with three assembly methods: complementary base pairing-mediated crosslinking method, hydrophilic–hydrophobic interaction-mediated crosslinking method, and metal ion-associated crosslinking method.2.1 Complementary base pairing-mediated crosslinking method
It has been found that complementary base pairing is one of the major driving forces in DNA self-assembly processes.38–40 Previous research studies have shown that nucleic acids can be assembled into DNA nanostructures with controllable size and shape via complementary base pairing.41–45 The same goes for chemically modified nucleic acids, which could develop several nanostructures via base pairing interactions: nanosheets, hydrogels, tetrahedra, nanotubes, and nanoflowers.DNA origami can be used as a carrier to load various components to expand its application.46 Very recently, Zhao et al. established a delivery platform based on DNA origami nanotechnology, which can transport payloads effectively for tumor treatment.47 They used a long M13mp 18 genomic DNA single strand as a scaffold, mixed with multiple predesigned staple strands and capture strands. These strands were annealed to fold into rectangular DNA origami nanosheets. The size of nanosheets is about 90 nm × 60 nm × 2 nm. The complementary strands on the cytotoxic protein ribonuclease (RNase) A molecules were hybridized with capture strands to generate protein–DNA conjugates (Fig. 1). The nanosheets then transported RNase into the cells to exert the cytotoxicity function. To target cancer cells, the authors used aptamers as a guide. Aptamers are oligonucleotides with high affinity and selectivity for targets, which are obtained from in vitro screening.48 The aptamers targeting mucin 1 (MUC1) on a human breast adenocarcinoma cell line were integrated into the nanosheets, which significantly improved the effect of cancer treatment. Similarly, Chen et al. mixed an M13mp 18 single-strand DNA (ssDNA), tiles, and probes together to self-assemble into rectangular nanosheets.49 Subsequently, they used the nanosheets as a template, and precise assembly of proteases and corresponding coenzymes can be achieved. DNA origami nanostructures can precisely control the horizontal distance between enzymes so as to regulate the catalytic activity of the enzyme cascade. Zhou et al. used a similar method to prepare triangular nanosheets with a complete cavity, and the side length of the cavity was about 50 nm.50 These studies show that the framework design guided by DNA hybridization enables the fabrication of nanosheets of different shapes.
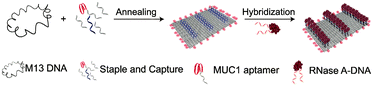 | ||
| Fig. 1 Schematic diagram of two-dimensional nanosheet assembly. Reproduced with permission from ref. 47. Copyright 2019, American Chemical Society. | ||
In addition to 2D origami nanosheets, it has been widely reported that a well-defined DNA hydrogel can be constructed through rational design and precise synthesis of DNA strands.51–53 As a case in point, rapid in situ formation of polypeptide-DNA hydrogels was reported by Li et al.54 First, DNA-grafted polypeptides were prepared by an azide–alkyne click chemistry reaction. Five to six ssDNA motifs were conjugated to the backbone of each peptide, ensuring sufficient crosslinking sites. Additionally, double-stranded DNA served as a DNA linker containing two “sticky ends” that have the same sequence complementary to the DNA motifs. As a consequence of mixing DNA-grafted polypeptides and DNA linkers in a buffer solution, the fluidic solution changed to a transparent supramolecular crosslinking network within a few seconds (Fig. 2a). Tests showed that the mechanical strength of the chemically modified hydrogel was significantly enhanced. In another study, Li et al. also synthesized multifunctional supramolecular hydrogels by DNA-grafted polypeptides.55 The difference is that they used an “X”-shaped DNA as a crosslinker instead of the linear DNA linker. The “X” shaped DNA strands are divided into two parts; one part is used as crosslinking sites, and the other part could be used for further functional modification. Subsequently, the two crosslinking sites are linked to the DNA-grafted polypeptide through precise base pairing to form a hydrogel structure (Fig. 2b). By this method, the resultant hydrogel can integrate various functional molecules to achieve applications in the biomedical field.
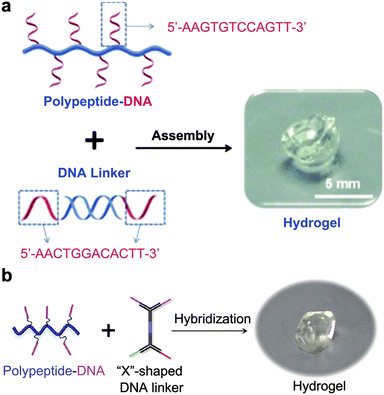 | ||
| Fig. 2 Hydrogel assembly with linear (a) and X-shaped (b) DNA linkers. Figures reproduced with permission from: a, ref. 54. Copyright 2015, Wiley-VCH; b, ref. 55. Copyright 2014, Wiley-VCH. | ||
Three-dimensional DNA tetrahedral nanostructures have been widely studied because of their excellent mechanical stiffness and structural stability.56,57 Pei et al. did an elegant work on the design of DNA tetrahedral nanostructures. They designed four ssDNAs with complementary sequences.58 Among them, three single chains are modified with the thiol group, and the other side chain contains a DNA probe. Four chains are mixed in an equivalent stoichiometric ratio, and the DNA tetrahedron is generated by annealing (heating, then rapid cooling) (Fig. 3a). This assembly process was fast (within two minutes) and the yield was high. This assembly method provides a new idea for constructing DNA tetrahedral probes with ordered orientation, controlled spacing and high stability. Besides, Li et al. assembled a DNA tetrahedron, which was constructed by three carboxyl group modified DNA strands (55 nucleotides) and a DNA strand containing an aptamer sequence (87 nucleotides).59 Four ssDNAs were quickly assembled into a tetrahedron with an edge length of 5.8 nm and each edge was an 18 base pair double helix (Fig. 3b). Polyacrylamide gel electrophoresis (PAGE) results showed that the DNA strands effectively self-assembled in the designed direction (Fig. 3c). The tetrahedral structure keeps the distance distribution of each chain the same, thereby reducing the spatial effect and improving the stability and recognition ability of biomolecules.
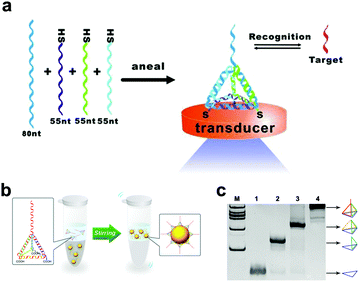 | ||
| Fig. 3 DNA tetrahedra assembled by thiolated DNA strands (a) or carboxyl group modified DNA strands (b). (c) Analysis of DNA tetrahedra by PAGE. Figures reproduced with permission from: a, ref. 58. Copyright 2010, Wiley-VCH; b and c, ref. 59. Copyright 2015, American Chemical Society. | ||
Apart from these structures, chemically modified nucleic acids can be assembled into tubular structures by complementary base pairing. According to the previous ssDNA tile assembly method,60 Kocabey et al. designed 24 functional oligonucleotides; after over 17 hours of annealing, the oligonucleotides were finally assembled into small tubular nanostructures with excellent stability (Fig. 4).61
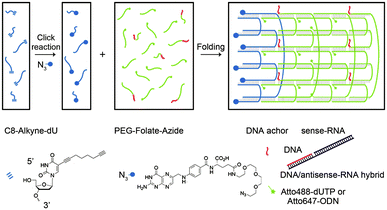 | ||
| Fig. 4 Assembly of DNA nanotubes. Reproduced with permission from ref. 61. Copyright 2014, Molecular Diversity Preservation International. | ||
In addition to the aforementioned DNA nanoassemblies, Zhang et al. synthesized submicron-scale DNA nanoflowers.62 A closed circular DNA containing the complementary sequences of aptamer Sgc8 was used as a template, and Sgc8 tandem repeat DNA sequences were generated by base pairing. The final long DNA sequence can spontaneously form nanoflower structures through crystallization, nucleation and growth processes. Finally, the authors introduced the DNA strands incorporating ferrocene (Fc) bases into the nanoflower (Fig. 5). The research indicated that the size of nanoflowers can be effectively adjusted by the introduction of Fc bases.
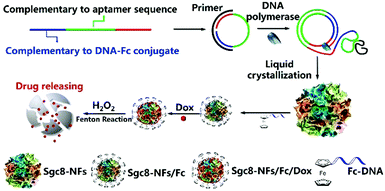 | ||
| Fig. 5 Schematic diagram of nanoflower preparation. Reproduced with permission from ref. 62. Copyright 2019, American Chemical Society. | ||
2.2 Hydrophilic–hydrophobic interaction-mediated crosslinking method
Similar to the assembly of amphiphilic polymers, the hydrophilic–hydrophobic interaction also plays a relevant role in the assembly processes of nucleic acid biopolymers.63 By modifying with hydrophobic groups, hydrophilic nucleic acids can turn into amphiphilic biopolymers, forming different structures through hydrophilic–hydrophobic interactions.64–67 Additionally, assemblies of specific size and shape can be obtained by changing the ratio of hydrophilic to hydrophobic chain length.65,68,69 In this section, we will introduce chemically modified nucleic acid assemblies formed via hydrophilic–hydrophobic interactions.Linking hydrophobic units to hydrophilic units through covalent interactions is the main way to construct amphiphilic motifs.70–72 Liu et al. connected hydrophilic DNA strands to hydrophobic diacyl lipids to form DNA–lipid molecules (Fig. 6a).73 They analysed the effect of DNA strand length on micelles. Atomic force microscopy (AFM) and dynamic light scattering (DLS) results showed that fine-tuning the length of the DNA sequence can achieve the purpose of adjusting the size of the micelles (Fig. 6b and c). In a similar study, Wu et al. linked diacyl lipids to nucleic acid aptamers via polyethylene glycol to assemble a highly ordered micelle-like structure.74 In this structure, the nucleic acid strand not only serves as the basis of the nanostructure but also performs a recognition function for a specific target. These aptamer-lipid assemblies enhance the binding ability to the target in a complex biological environment.
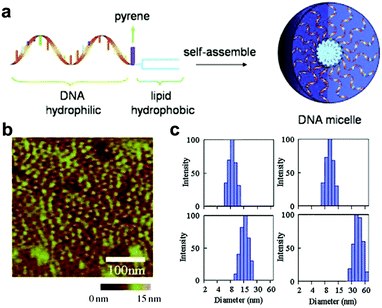 | ||
| Fig. 6 (a) Illustration of assembly of diaycl-DNA micelles. AFM (b) and DLS (c) characterization of DNA micelles. Reproduced with permission from ref. 73. Copyright 2010, Wiley-VCH. | ||
Inspired by the higher hydrophobicity, lower solubility, and stronger aggregation tendency of fluorocarbon groups, Zou et al. replaced the traditional diacyl chain with a fluorocarbon chain to construct micelles (Fig. 7).75 Micelles formed by chain amphiphilic polymers have higher stability, and the critical micelle concentration values are significantly reduced. Thus, creating new hydrophobic building units enables the formation of amphiphilic biomolecules with different properties and expands the versatility of nanomicelles.
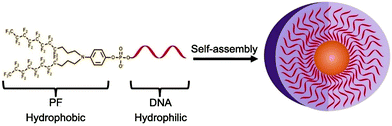 | ||
| Fig. 7 Illustration of diperfluorodecyl–DNA Micelles. Reproduced with permission from ref. 75. Copyright 2018, American Chemical Society. | ||
Moreover, adding chemically modified nucleotides is another way to introduce hydrophobic groups into hydrophilic nucleic acid chains.76,77 Kimoto et al. used the unnatural base 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds) as the hydrophobic moiety in the aptamer selection process.78 In their research, the addition of Ds bases not only improved the performance of aptamers, but also successfully introduced hydrophobicity into the DNA strands. Tan et al. integrated hydrophobic ferrocenes into hydrophilic DNA strands using a similar method and constructed aptamer–ferrocene assemblies (ApFAs) based on hydrophilic–hydrophobic interactions.68 Ferrocene bases are prepared by the reaction of (S)-3amino-1,2-propanediol and ferrocene carboxylic acid. Then using standard phosphoramidite chemistry, the chemically modified nucleic acids are synthesized on the DNA synthesizer by linking ferrocenes to natural bases. By designing synthetic procedures, the authors can synthesize oligonucleotides with specific bases at specific positions. The purified and desalted products can be assembled into micelles after being placed in the DPBS buffer for a certain time. Among them, the hydrophilic nucleic acid fragment is located on the surface of the assemblies in the form of a corona, and the hydrophobic ferrocene fragment forms the core (Fig. 8a). By regulating the number of hydrophobic ferrocene nucleotides, micelles of different sizes can be synthesized. Transmission electron microscopy (TEM) and DLS results showed that as the number of ferrocene nucleotide groups increased, the size of ApFAs increased gradually (Fig. 8b).
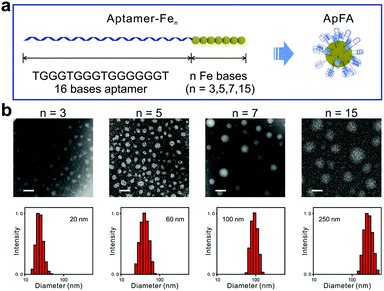 | ||
| Fig. 8 (a) Schematic of ApFAs assembled by aptamers and different numbers of Fe nucleotides. (b) TEM and DLS characterization of the size of ApFAs. Reproduced with permission from ref. 68. Copyright 2019, Elsevier. | ||
2.3 Metal ion-associated crosslinking method
Metal ions have aroused wide interest in biosensing since they play critical roles in biological organisms.79–82 Recently, nucleic acids have also been developed to form nanostructures with the assistance of metal ions.83,84Lyu et al. designed a method for DNA micelle crosslinking in situ with the assistance of metal ions.85 The monomer that makes up the micelle is formed by the covalent attachment of a single stranded DNA and a hydrophobic diacyl lipid. Specifically, the monomer consists of three parts, the most important of which is the part that acts as a template chain (Fig. 9a). The template chain with specific sequences can adsorb different metal ions. The authors selected Cu2+, Ag+, and Au3+ to prepare different DNA micelles. Subsequently, the metal ions were reduced to zero-valent metals, which can be crosslinked into hollow or solid metal nuclei, thereby promoting the crosslinking of DNA micelles (Fig. 9b). In addition to the single-stranded micelles based on diacyl lipids, two cholesterol-modified double-stranded DNAs (dsDNAs) can also act as template domains. The authors have verified this. Compared with the single cholesterol-modified chain, the strong hydrophobic force produced by the two cholesterol-modified dsDNA can stabilize the micelle structure. Using CuSO4 as a copper source and L-ascorbic acid as a reducing agent, it was found that cholesterol-modified dsDNA promoted micelle crosslinking, which can be demonstrated by fluorescence intensity and TEM characterization.
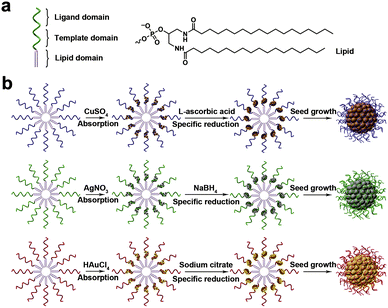 | ||
| Fig. 9 (a) Monomer structure consisting of three domains. (b) Metal ion-assisted crosslinking process of DNA micelles. Reproduced with permission from ref. 85. Copyright 2019, Elsevier. | ||
Inspired by the unique structures of the DNA G-quadruplexes,86 Lu et al. proposed that in the presence of K+ ions, the crosslinking effect of G-quadruplexes was used to promote the formation of hydrogels.87 Acrydite-modified oligonucleotides are polymerized with acrylamide monomers to form copolymer chains under the catalysis of initiators. After adding K+ ions, G-rich nucleic acid sequences self-assemble into G-quadruplex structures, so that polyacrylamide chains are cross-linked to form a DNA hydrogel (Fig. 10). To verify the role of K+ in gel formation, the authors added crown ether that can capture K+ ions, then the crosslinked G-quadruplex broke down, and the DNA hydrogel became a solution. Based on the fact that K+ can stabilize the structure of G-quadruplex, Jin et al. designed a DNA sequence rich in G bases and developed a technique to stabilize the DNA micelle structure.88 With the addition of K+, adjacent guanine bases in the micellar canopy formed G-quadruplexes via Hoogsteen hydrogen bonding, and the hydrophobic lipid tails are confined within the micellar nucleus, thereby stabilizing the structure of micelles. These results showed that K+ can promote the assembly of the gel and achieve reversible switching.
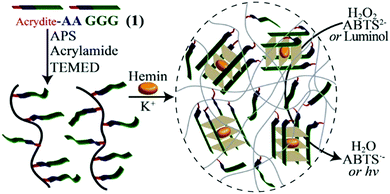 | ||
| Fig. 10 K+-assisted crosslinking process of DNA hydrogels. Reproduced with permission from ref. 87. Copyright 2013, American Chemical Society. | ||
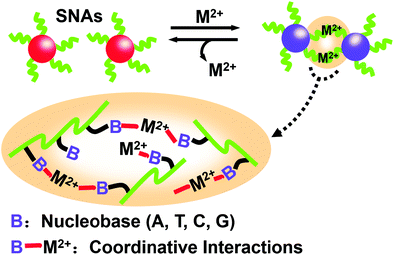
Unlike the works described above, the work by Joo et al. was based on the basic interaction between natural nucleobases and metal ions (M2+s) without any predesigned DNA sequence.89 In other words, even if the nucleic acid sequence of SNAs doesn’t meet the requirements of complementary base pairing, it can induce the assembly of single-type spherical nucleic acids (SNAs). Taking Cu2+ as an example, a spherical nucleic acid solution with gold nanoparticles as the core was reacted with different concentrations of Cu2+ for 12 h. As the concentration of Cu2+ increases, SNAs form larger aggregates. Furthermore, as the crosslinking reaction occurs, changes in the color of the solution can be observed with the naked eye, indicating the formation of assemblies. This research has shown that M2+s can coordinate with multiple bases of SNAs through coordinative interactions, thereby crosslinking into large aggregates (Fig. 11). Once M2+s are chelated by a stronger ligand, the assembled SNAs can be dissociated immediately. These studies showed that metal ions can mediate the formation of assemblies and can be reversibly reduced by removing metal ions.
3. Application in biosensing
Recently, much work has integrated biological functions into biosensing to achieve biomedical functions with high sensitivity and specificity.90–92 This requires that the materials used in biosensing can interact with and respond to target signals uniquely. Nucleic acid biopolymers show recognition ability to target molecules with good selectivity.93 The addition of chemical modifications can impart new chemical properties and biological functions to nucleic acid biopolymers.94,95 Therefore, the chemically modified nucleic acid biopolymers hold great potential to realize multifunctional biosensing. In this section, we will introduce the applications of chemically modified nucleic acid biopolymers in three parts: biomolecule detection, controlled drug release, and gene therapy.3.1 Detection of biomolecules
The detection of biomolecules can provide us with a lot of information, which will help us make more accurate judgments on various life processes.96–98 Nucleic acids with specific sequences have the potential to recognize targets.99 However, the biological environment is so complicated that it limits the use of nucleic acid biopolymers in target recognition.100 Chemically modified nucleic acid biopolymers enable the specific and sensitive detection of targets. Here, we highlight the application of chemically modified nucleic acid biopolymers in biomolecule detection.As exosomes are one of the biomolecules for early diagnosis of cancer, it is of great significance to detect exosomes with high sensitivity.101–103 Wang et al. fixed a nanotetrahedron (NTH) on the gold electrode and designed an aptamer-based biosensor (aptasensor) for detecting hepatocyte exosomes (Fig. 12a).104 When the sensor is incubated with the exosome suspension, the aptamer on the NTH binds to the exosomes specifically. The binding of the aptamer and exosome reduces the electrode surface area, which causes changes in electrical signals. Accordingly, changes in the electrical signal reflect the concentration of exosomes. Compared with the single-chain aptamer functionalized aptasensor, the detection sensitivity of the NTH-assisted aptasensor is increased by about 100 times (Fig. 12b).
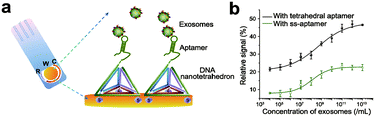 | ||
| Fig. 12 (a) Illustration of the NTH-assisted aptasensor. (b) Comparison of signals between single-chain aptamer-assisted and NTH-assisted sensors. Reproduced with permission from ref. 104. Copyright 2017, American Chemical Society. | ||
In another study, Wu et al. constructed switchable aptamer micelle flares (SAMFs) for the detection of ATP molecules.105 The SAMF hydrophobic diacyl lipid tail is linked to a hydrophilic switchable aptamer head (Fig. 13a). When there are no targets, the aptamer probe retains the loop-stem structure, causing the fluorescence of the fluorophore to be quenched by an adjacent quencher. After binding to the target, the conformation of the aptamer changes and the fluorescence signal is restored. Based on this principle, a series of different concentrations of ATP are used to measure the response of SAMFs. It is observed that as the ATP concentration increases, the fluorescence signal intensity increases firstly and then reaches saturation (Fig. 13b). The lack of the target ATP prevents the stem-loop structure from opening, resulting in a lower fluorescence signal intensity. When the ATP molecules are added at 75 seconds, the fluorescence signal shows a straight upward trend and the response speed is fast (Fig. 13c). This study demonstrated that SAMFs can respond to ATP expression sensitively and detect ATP molecules through changes in fluorescent signals.
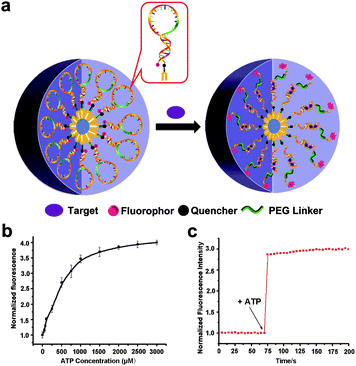 | ||
| Fig. 13 (a) Structure of SAMFs and their working principle. (b) Relationship between the ATP concentration and fluorescent signal. (c) Effect of adding target molecules on fluorescence intensity. Reproduced with permission from ref. 105. Copyright 2013, American Chemical Society. | ||
The detection of mRNA is of great significance for early tumor screening.106 Existing methods for detecting mRNA in cells have many limitations; for example, the probe is susceptible to interference by complex cellular environments, or the stability of the probe is poor.103 He et al. constructed a DNA tetrahedron nanotweezer (DTNT) to solve the above problems.107 To be specific, the DTNT completed the mRNA detection with high sensitivity and high accuracy via the principle of fluorescence resonance energy transfer. They customized four DNA strands, two of which modified the Cy3 or Cy5 fluorophore respectively. In the absence of targets, the two fluorophores in the DTNT are far away, and the FRET is in the “off” state. When the target mRNA exists in the cell, the two fluorophores are located close to each other, which induces the generation of FRET and the signal changes from “off” to “on”.
3.2 Controlled drug release
In recent years, the applications of smart drug delivery systems in the field of biosensing have been widely studied.108–111 Chemically modified nucleic acid biopolymers can also be used to construct smart drug delivery systems.112–114A DNA origami structure is often used as a drug carrier system due to its excellent biocompatibility and spatially addressable functions.41,115 Jiang et al. inserted anticancer drug doxorubicin (DOX) into DNA origami nanostructures through non-covalent bonding to reach a high level of drug loading efficiency.116 The DNA origami structure was effectively internalized into cells and remained in the tumor area for a long time. When the drug-loaded origami is exposed to low environmental pH or DNase, DNA nanostructures are slowly degraded to achieve controlled drug release (Fig. 14). Therefore, using DNA origami to carry drugs greatly improves the internalization ability of DOX, which will help the drug to exert its effect and kill tumor cells. In addition, the amphiphilic block copolymer can form a hydrophilic corona and a lipophilic core, and the core can be effectively loaded with hydrophobic anticancer drugs. Based on this, Alemdaroglu et al. assembled DNA-polypropylene oxide (PPO) into a spherical micelle structure.117 Dox is a hydrophobic drug, and due to the hydrophobic interaction of Dox and PPO, drugs accumulate inside the micelles to form a drug delivery system.
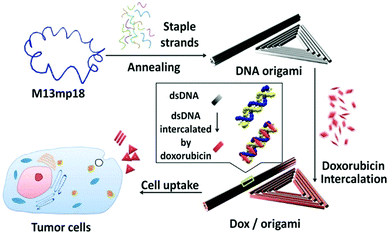 | ||
| Fig. 14 DNA origami-based controlled drug release system. Reproduced with permission from ref. 116. Copyright 2012, American Chemical Society. | ||
Mitochondria play an important regulatory role in tumor growth. Jiang et al. developed a mitochondria-specific DNAtrain (MitoDNAtrs) drug delivery system using DNA as the basic unit (Fig. 15).118 The indocyanine dye Cy5.5 is used to provide the driving force, while the linear DNA strand is used as a carrier for transporting a good deal of antitumor drugs. Guided by Cy5.5, drugs are targeted to the mitochondrial region. The release of the drugs can be detected because Cy5.5 has a visualizable fluorescent signal. Applying MitoDNAtrs to breast cancer cells, and comparing the intracellular fluorescence signals at different times, it is clear that the intracellular fluorescence signals gradually increase. The results mean that drugs are gradually released from MitoDNAtrs, and the imaging method can be used to visualize the drug release process in time. Additionally, Li et al. also designed a controlled drug release system.119 They coupled a nucleolin-targeting aptamer (NucA) with paclitaxel (PTX) through a linker to form a nucleolin aptamer–paclitaxel conjugate (NucA-PTX). NucA directs PTX to ovarian tumor cells for action with little effect on normal cells. Once NucA-PTX enters the tumor cells through endocytosis, the lysosomal protease in the cell will cleave the linker in NucA-PTX, thereby achieving the controlled release of PTX.
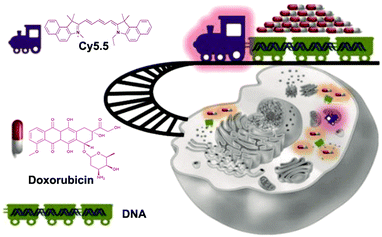 | ||
| Fig. 15 Schematic of the MitoDNAtrs delivery system. Reproduced with permission from ref. 118. Copyright 2019, American Chemical Society. | ||
Interestingly, a high payload delivery system means a powerful lethality to tumor cells.120 As a typical example, Deng et al. proposed a general strategy to construct Aptamer–PolyproDrug Conjugates (ApPDCs); this method involved the assembly of aptamers targeting cancer cells, prodrugs and a biocompatible brush skeleton.121 Compared to traditional drug loading systems, ApPDCs have a high drug-to-aptamer ratio (DApR ≥ 40). After the ApPDCs enter the cell, the reducing environment within the cell can trigger the efficient release of the prodrugs. By using prodrugs with different mechanisms, multi-drug synergy treatment can be achieved, and problems such as multi-drug resistance may be resolved.
Different from the above work, based on molecular engineering technology, the ratio of drugs can be adjusted accurately and multiple drugs can be precisely delivered to tumor cells. In this regard, Zhou et al. coupled circular bivalent aptamers with different drugs to prepare circular bivalent aptamer–drug conjugates (cb-ApDCs) with different drug dosing ratios.122 Drug release of cb-ApDCs can be monitored by confocal laser scanning microscopy; the ester bonds in cb-ApDCs can be cleaved by the esterase present in the late endosomes and lysosomes to realize the controlled release of drugs (Fig. 16b).
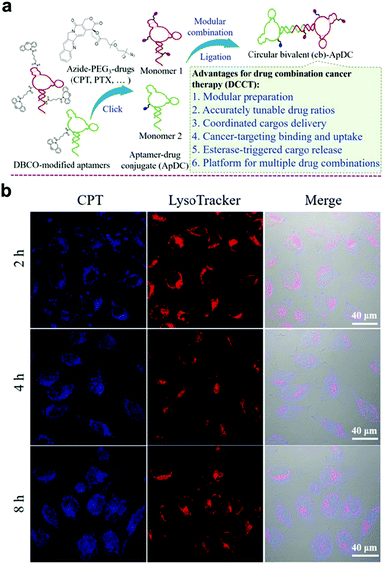 | ||
| Fig. 16 (a) Preparation of cb-ApDCs. (b) Intracellular CPT release was monitored from intracellular cb-ApDCs. Reproduced with permission from ref. 122. Copyright 2019, Wiley-VCH. | ||
3.3 Gene therapy
Chemically modified nucleic acid biopolymers can be combined with gene therapy technology to achieve highly efficient tumor treatment.123Gene therapy has the ability to combat many gene-related diseases and it is a promising cancer treatment strategy.124 Li et al. designed a stimulus-responsive hydrogel for gene therapy, which was cross-linked by a building unit, a blocking and targeting unit and a linking unit (Fig. 17).125 Different functional elements were integrated into the three building units, such as therapeutic genes that can induce apoptosis, DNA ribozymes that can inhibit cell migration, and aptamers that can target cancer cells. Glutathione in cells can effectively cleave the disulfide bonds in the building blocks to achieve dissociation of nanohydrogels and release of multiple therapeutic genes. Moreover, Chen et al. synthesized molecular beacon micelle flares (MBMFs), which were assembled by diacyl lipids with circular DNA probes.126 The premise of using MBMFs for gene therapy is to hybridize with mRNA first, and there are two main therapeutic mechanisms: one is to prevent translation from target mRNAs by providing translational blocking, and the other is to form a DNA/RNA hybrid with the target mRNA, which is subsequently degraded by the RNase H enzyme. Using these two mechanisms can inhibit the growth of cancer cells and realize gene therapy. Liu et al. used gene therapy to enhance antitumor effects.127 Specifically, the authors constructed triangular DNA origami to load Dox, and then hybridized with the tumor therapeutic gene p53 to form a nano-kite assembly (Fig. 18). The assembly can effectively deliver p53 and DOX to tumor areas simultaneously, significantly enhancing the anti-tumor effect.
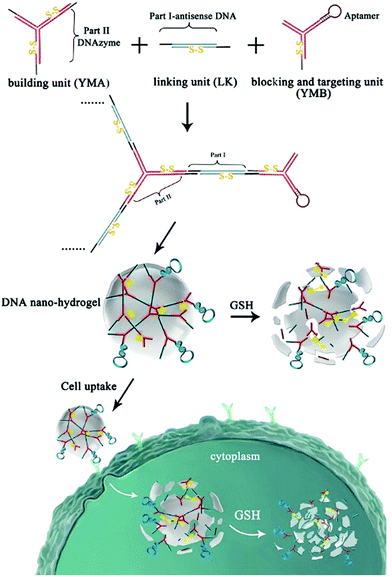 | ||
| Fig. 17 Schematic of the hydrogel formation and gene therapy. Reproduced with permission from ref. 125. Copyright 2015, American Chemical Society. | ||
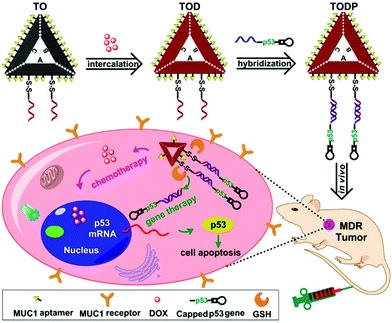 | ||
| Fig. 18 Schematic of gene therapy based on a DNA origami structure. Reproduced with permission from ref. 127. Copyright 2018, American Chemical Society. | ||
4. Conclusion and outlook
The addition of chemical modifications can impart rich structures and functions to nucleic acid biopolymers, attracting widespread attention from researchers. In this review, we introduced different cross-linking forms of chemically modified nucleic acid biopolymer nanostructures. Furthermore, we also summarized the applications of chemically modified nucleic acid biopolymers in biomolecule detection, controlled drug release, and gene therapy. These studies show the importance of chemically modified nucleic acid biopolymers in biosensing.Although great achievements have been made, the application of chemically modified nucleic acid biopolymers in biosensing is still emerging and needs further improvement. For example, the efficiency of chemical modifications of nucleic acids needs to be further improved. Secondly, there is still an urgent requirement to develop nucleic acid biopolymers modified with different functional groups to achieve multifunctional biosensing. Finally, there are few chemically modified nucleic acid biopolymers available for clinical biosensing, so developing practical and safe chemically modified nucleic acid biopolymers is another key point. With the interdisciplinary development, chemically modified nucleic acid biopolymers have the potential to achieve greater promotions and breakthroughs in biosensing.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21925401 and 21904037).Notes and references
- B. A. Cornell, V. Braach-Maksvytis, L. King, P. Osman, B. Raguse, L. Wieczorek and R. J. N. Pace, A biosensor that uses ion-channel switches, Nature, 1997, 387, 580–583 CrossRef CAS PubMed.
- A. J. Haes and R. P. Van Duyne, A Nanoscale Optical Biosensor:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sensitivity and Selectivity of an Approach Based on the Localized Surface Plasmon Resonance Spectroscopy of Triangular Silver Nanoparticles, J. Am. Chem. Soc., 2002, 124, 10596–10604 CrossRef CAS PubMed.
Sensitivity and Selectivity of an Approach Based on the Localized Surface Plasmon Resonance Spectroscopy of Triangular Silver Nanoparticles, J. Am. Chem. Soc., 2002, 124, 10596–10604 CrossRef CAS PubMed. - M. A. Morales and J. M. Halpern, Guide to Selecting a Biorecognition Element for Biosensors, Bioconjugate Chem., 2018, 29, 3231–3239 CrossRef CAS PubMed.
- L. Soleymani and F. Li, Mechanistic Challenges and Advantages of Biosensor Miniaturization into the Nanoscale, ACS Sens., 2017, 2, 458–467 CrossRef CAS.
- J. Sun and R. N. Zuckermann, Peptoid Polymers: A Highly Designable Bioinspired Material, ACS Nano, 2013, 7, 4715–4732 CrossRef CAS PubMed.
- C. Rossner, Q. Tang, O. Glatter, M. Muller and P. Vana, Uniform Distance Scaling Behavior of Planet–Satellite Nanostructures Made by Star Polymers, Langmuir, 2017, 33, 2017–2026 CrossRef CAS.
- K. Doré, S. Dubus, H.-A. Ho, I. Lévesque, M. Brunette, G. Corbeil, M. Boissinot, G. Boivin, M. G. Bergeron, D. Boudreau and M. Leclerc, Fluorescent Polymeric Transducer for the Rapid, Simple, and Specific Detection of Nucleic Acids at the Zeptomole Level, J. Am. Chem. Soc., 2004, 126, 4240–4244 CrossRef PubMed.
- Y. Zhao, S. Hu, H. Wang, K. Yu, Y. Guan, X. Liu, N. Li and F. Liu, DNA Dendrimer–Streptavidin Nanocomplex: an Efficient Signal Amplifier for Construction of Biosensing Platforms, Anal. Chem., 2017, 89, 6907–6914 CrossRef CAS PubMed.
- Y. Wang, L. P. Jiang, S. Zhou, S. Bi and J. J. Zhu, DNA Polymerase-Directed Hairpin Assembly for Targeted Drug Delivery and Amplified Biosensing, ACS Appl. Mater. Interfaces, 2016, 8, 26532–26540 CrossRef CAS PubMed.
- X. Fan, J. Y. Chung, Y. X. Lim, Z. Li and X. J. Loh, Review of Adaptive Programmable Materials and Their Bioapplications, ACS Appl. Mater. Interfaces, 2016, 8, 33351–33370 CrossRef CAS PubMed.
- M. Chandler and K. A. Afonin, Smart-Responsive Nucleic Acid Nanoparticles (NANPs) with the Potential to Modulate Immune Behavior, Nanomaterials, 2019, 9, 611 CrossRef CAS PubMed.
- S. Wan, L. Zhang, S. Wang, Y. Liu, C. Wu, C. Cui, H. Sun, M. Shi, Y. Jiang, L. Li, L. Qiu and W. Tan, Molecular Recognition-Based DNA Nanoassemblies on the Surfaces of Nanosized Exosomes, J. Am. Chem. Soc., 2017, 139, 5289–5292 CrossRef CAS PubMed.
- S. Cai, J. Yan, H. Xiong, Y. Liu, D. Peng and Z. Liu, Investigations on the interface of nucleic acid aptamers and binding targets, Analyst, 2018, 143, 5317–5338 RSC.
- C. Qi, S. Musetti, L. H. Fu, Y. J. Zhu and L. Huang, Biomolecule-assisted green synthesis of nanostructured calcium phosphates and their biomedical applications, Chem. Soc. Rev., 2019, 48, 2698–2737 RSC.
- Y. J. Chen, B. Groves, R. A. Muscat and G. Seelig, DNA nanotechnology from the test tube to the cell, Nat. Nanotechnol., 2015, 10, 748–760 CrossRef CAS.
- C. S. Swenson, A. Velusamy, H. S. Argueta-Gonzalez and J. M. Heemstra, Bilingual Peptide Nucleic Acids: Encoding the Languages of Nucleic Acids and Proteins in a Single Self-Assembling Biopolymer, J. Am. Chem. Soc., 2019, 141, 19038–19047 CrossRef CAS.
- H. Wang, R. Yang, L. Yang and W. Tan, Nucleic Acid Conjugated Nanomaterials for Enhanced Molecular Recognition, ACS Nano, 2009, 3, 2451–2460 CrossRef CAS.
- L. Mo, J. Li, Q. Liu, L. Qiu and W. Tan, Nucleic acid-functionalized transition metal nanosheets for biosensing applications, Biosens. Bioelectron., 2017, 89, 201–211 CrossRef CAS PubMed.
- N. Sugimoto, Designable DNA Functions toward New Nanobiotechnology, Bull. Chem. Soc. Jpn., 2009, 82, 1–10 CrossRef CAS.
- M. Xiao, W. Lai, T. Man, B. Chang, L. Li, A. R. Chandrasekaran and H. Pei, Rationally Engineered Nucleic Acid Architectures for Biosensing Applications, Chem. Rev., 2019, 119, 11631–11717 CrossRef CAS PubMed.
- D. Al Sulaiman, P. Cadinu, A. P. Ivanov, J. B. Edel and S. Ladame, Chemically Modified Hydrogel-Filled Nanopores: A Tunable Platform for Single-Molecule Sensing, Nano Lett., 2018, 18, 6084–6093 CrossRef CAS PubMed.
- M. Kuwahara and N. Sugimoto, Molecular Evolution of Functional Nucleic Acids with Chemical Modifications, Molecules, 2010, 15, 5423–5444 CrossRef CAS PubMed.
- K. A. Lennox and M. A. Behlke, Chemical modification and design of anti-miRNA oligonucleotides, Gene Ther., 2011, 18, 1111–1120 CrossRef CAS PubMed.
- P. A. Lichtor, Z. Chen, N. H. Elowe, J. C. Chen and D. R. Liu, Side chain determinants of biopolymer function during selection and replication, Nat. Chem. Biol., 2019, 15, 419–426 CrossRef CAS PubMed.
- I. K. Astakhova and J. Wengel, Scaffolding along Nucleic Acid Duplexes Using 2′-Amino-Locked Nucleic Acids, Acc. Chem. Res., 2014, 47, 1768–1777 CrossRef CAS PubMed.
- K. Holz, E. Schaudy, J. Lietard and M. M. Somoza, Multi-level patterning nucleic acid photolithography, Nat. Commun., 2019, 10, 3805 CrossRef PubMed.
- L. Uzun and A. P. Turner, Molecularly-imprinted polymer sensors: realising their potential, Biosens. Bioelectron., 2016, 76, 131–144 CrossRef CAS PubMed.
- C. Wu and D. T. Chiu, Highly Fluorescent Semiconducting Polymer Dots for Biology and Medicine, Angew. Chem., Int. Ed., 2013, 52, 3086–3109 CrossRef CAS PubMed.
- Y. Wu, W. Wei and S. Liu, Target-Triggered Polymerization for Biosensing, Acc. Chem. Res., 2012, 45, 1441–1450 CrossRef CAS.
- M. Liu, Z. Li, Y. Li, J. Chen and Q. Yuan, Self-assembled nanozyme complexes with enhanced cascade activity and high stability for colorimetric detection of glucose, Chin. Chem. Lett., 2019, 30, 1009–1012 CrossRef CAS.
- S. Cosnier and M. Holzinger, Electrosynthesized polymers for biosensing, Chem. Soc. Rev., 2011, 40, 2146–2156 RSC.
- H. Tran, K. Ronaldson, N. A. Bailey, N. A. Lynd, K. L. Killops, G. Vunjak-Novakovic and L. M. Campos, Hierarchically Ordered Nanopatterns for Spatial Control of Biomolecules, ACS Nano, 2014, 8, 11846–11853 CrossRef CAS PubMed.
- C. Rodriguez-Quijada, M. Sanchez-Purra, H. de Puig and K. Hamad-Schifferli, Physical Properties of Biomolecules at the Nanomaterial Interface, J. Phys. Chem. B, 2018, 122, 2827–2840 CrossRef CAS PubMed.
- T. Ming, Y. Wang, J. Luo, J. Liu, S. Sun, Y. Xing, G. Xiao, H. Jin and X. Cai, Folding Paper-Based Aptasensor Platform Coated with Novel Nanoassemblies for Instant and Highly Sensitive Detection of 17β-Estradiol, ACS Sens., 2019, 4, 3186–3194 CrossRef CAS PubMed.
- N. M. Robertson, M. S. Hizir, M. Balcioglu, M. Rana, H. Yumak, O. Ecevit and M. V. Yigit, Monitoring the Multitask Mechanism of DNase I Activity Using Graphene Nanoassemblies, Bioconjugate Chem., 2015, 26, 735–745 CrossRef CAS PubMed.
- C. Jin, J. He, J. Zou, W. Xuan, T. Fu, R. Wang and W. Tan, Phosphorylated lipid-conjugated oligonucleotide selectively anchors on cell membranes with high alkaline phosphatase expression, Nat. Commun., 2019, 10, 2704 CrossRef PubMed.
- R. J. Banga, B. Meckes, S. P. Narayan, A. J. Sprangers, S. T. Nguyen and C. A. Mirkin, Cross-Linked Micellar Spherical Nucleic Acids from Thermoresponsive Templates, J. Am. Chem. Soc., 2017, 139, 4278–4281 CrossRef CAS PubMed.
- P. Chidchob, T. G. Edwardson, C. J. Serpell and H. F. Sleiman, Synergy of Two Assembly Languages in DNA Nanostructures: Self-Assembly of Sequence-Defined Polymers on DNA Cages, J. Am. Chem. Soc., 2016, 138, 4416–4425 CrossRef CAS PubMed.
- F. A. Aldaye, A. L. Palmer and H. F. Sleiman, Assembling Materials with DNA as the Guide, Science, 2008, 321, 1795–1799 CrossRef CAS PubMed.
- N. C. Seeman, DNA in a material world, Nature, 2003, 421, 427–431 CrossRef PubMed.
- J. Fu, M. Liu, Y. Liu, N. W. Woodbury and H. Yan, Interenzyme Substrate Diffusion for an Enzyme Cascade Organized on Spatially Addressable DNA Nanostructures, J. Am. Chem. Soc., 2012, 134, 5516–5519 CrossRef CAS PubMed.
- S. Pal, Z. Deng, B. Ding, H. Yan and Y. Liu, DNA-Origami-Directed Self-Assembly of Discrete Silver-Nanoparticle Architectures, Angew. Chem., Int. Ed., 2010, 49, 2700–2704 CrossRef CAS PubMed.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication, Science, 2005, 310, 1661–1665 CrossRef CAS PubMed.
- Y. Yang, M. Endo, K. Hidaka and H. Sugiyama, Photo-Controllable DNA Origami Nanostructures Assembling into Predesigned Multiorientational Patterns, J. Am. Chem. Soc., 2012, 134, 20645–20653 CrossRef CAS PubMed.
- M. Lin, J. Wang, G. Zhou, J. Wang, N. Wu, J. Lu, J. Gao, X. Chen, J. Shi, X. Zuo and C. Fan, Programmable Engineering of a Biosensing Interface with Tetrahedral DNA Nanostructures for Ultrasensitive DNA Detection, Angew. Chem., Int. Ed., 2015, 54, 2151–2155 CrossRef CAS PubMed.
- P. W. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- S. Zhao, F. Duan, S. Liu, T. Wu, Y. Shang, R. Tian, J. Liu, Z. G. Wang, Q. Jiang and B. Ding, Efficient intracellular delivery of RNase A using DNA origami carriers, ACS Appl. Mater. Interfaces, 2019, 11, 11112–11118 CrossRef CAS PubMed.
- N. N. Quang, A. Miodek, A. Cibiel and F. Ducongé, Selection of Aptamers Against Whole Living Cells: From Cell-SELEX to Identification of Biomarkers, Methods Mol. Biol., 2017, 1575, 253–272 CrossRef CAS PubMed.
- Y. Chen, G. Ke, Y. Ma, Z. Zhu, M. Liu, Y. Liu, H. Yan and C. J. Yang, A Synthetic Light-Driven Substrate Channeling System for Precise Regulation of Enzyme Cascade Activity Based on DNA Origami, J. Am. Chem. Soc., 2018, 140, 8990–8996 CrossRef CAS PubMed.
- C. Zhou, Y. Zhang, Y. Dong, F. Wu, D. Wang, L. Xin and D. Liu, Precisely Controlled 2D Free-Floating Nanosheets of Amphiphilic Molecules through Frame-Guided Assembly, Adv. Mater., 2016, 28, 9819–9823 CrossRef CAS PubMed.
- Y. S. Zhang and A. Khademhosseini, Advances in engineering hydrogels, Science, 2017, 356, 6337 Search PubMed.
- J. Wu, P. Li, C. Dong, H. Jiang, X. Bin, X. Gao, M. Qin, W. Wang, C. Bin and Y. Cao, Rationally designed synthetic protein hydrogels with predictable mechanical properties, Nat. Commun., 2018, 9, 620 CrossRef PubMed.
- N. Kong, Q. Peng and H. Li, Rationally Designed Dynamic Protein Hydrogels with Reversibly Tunable Mechanical Properties, Adv. Funct. Mater., 2014, 24, 7310–7317 CrossRef CAS.
- C. Li, A. Faulkner-Jones, A. R. Dun, J. Jin, P. Chen, Y. Xing, Z. Yang, Z. Li, W. Shu, D. Liu and R. R. Duncan, Rapid Formation of a Supramolecular Polypeptide–DNA Hydrogel for In Situ Three-Dimensional Multilayer Bioprinting, Angew. Chem., Int. Ed., 2015, 54, 3957–3961 CrossRef CAS PubMed.
- C. Li, P. Chen, Y. Shao, X. Zhou, Y. Wu, Z. Yang, Z. Li, T. Weil and D. Liu, A Writable Polypeptide–DNA Hydrogel with Rationally Designed Multi-modification Sites, Small, 2015, 11, 1138–1143 CrossRef CAS PubMed.
- D. Zeng, H. Zhang, D. Zhu, J. Li, L. San, Z. Wang, C. Wang, Y. Wang, L. Wang, X. Zuo and X. Mi, A novel ultrasensitive electrochemical DNA sensor based on double tetrahedral nanostructures, Bioelectronics, 2015, 71, 434–438 CrossRef CAS PubMed.
- R. P. Goodman, M. Heilemann, S. Doose, C. M. Erben, A. N. Kapanidis and A. J. Turberfield, Reconfigurable, braced, three-dimensional DNA nanostructures, Nat. Nanotechnol., 2008, 3, 93–96 CrossRef CAS PubMed.
- H. Pei, N. Lu, Y. Wen, S. Song, Y. Liu, H. Yan and C. Fan, A DNA Nanostructure-based Biomolecular Probe Carrier Platform for Electrochemical Biosensing, Adv. Mater., 2010, 22, 4754–4758 CrossRef CAS PubMed.
- J. Li, C. Y. Hong, S. X. Wu, H. Liang, L. P. Wang, G. Huang, X. Chen, H. H. Yang, D. Shangguan and W. Tan, Facile Phase Transfer and Surface Biofunctionalization of Hydrophobic Nanoparticles Using Janus DNA Tetrahedron Nanostructures, J. Am. Chem. Soc., 2015, 137, 11210–11213 CrossRef CAS PubMed.
- B. Wei, M. Dai and P. Yin, Complex shapes self-assembled from single-stranded DNA tiles, Nature, 2012, 485, 623–626 CrossRef CAS PubMed.
- S. Kocabey, H. Meinl, I. S. MacPherson, V. Cassinelli, A. Manetto, S. Rothenfusser, T. Liedl and F. S. Lichtenegger, Cellular Uptake of Tile-Assembled DNA Nanotubes, Nanomaterials, 2014, 5, 47–60 CrossRef PubMed.
- L. Zhang, R. Abdullah, X. Hu, H. Bai, H. Fan, L. He, H. Liang, J. Zou, Y. Liu, Y. Sun, X. Zhang and W. Tan, Engineering of Bioinspired, Size-Controllable, Self-Degradable Cancer-Targeting DNA Nanoflowers via the Incorporation of an Artificial Sandwich Base, J. Am. Chem. Soc., 2019, 141, 4282–4290 CrossRef CAS PubMed.
- Y. Kopel and N. Giovambattista, Comparative Study of Water-Mediated Interactions between Hydrophilic and Hydrophobic Nanoscale Surfaces, J. Phys. Chem. B, 2019, 123, 10814–10824 CrossRef CAS PubMed.
- S. Gao, X. Zheng, B. Jiao and L. Wang, Post-SELEX optimization of aptamers, Anal. Bioanal. Chem., 2016, 408, 4567–4573 CrossRef CAS PubMed.
- S. K. Albert, X. Hu and S. J. Park, Dynamic Nanostructures from DNA-Coupled Molecules, Polymers, and Nanoparticles, Small, 2019, 15, e1900504, DOI:10.1002/smll.201900504.
- Y. Jia, P. Gao, Y. Zhuang, M. Miao, X. Lou and F. Xia, Facile Probe Design: Fluorescent Amphiphilic Nucleic Acid Probes without Quencher Providing Telomerase Activity Imaging Inside Living Cells, Anal. Chem., 2016, 88, 6621–6626 CrossRef CAS PubMed.
- F. E. Alemdaroglu and A. Herrmann, DNA meets synthetic polymers—highly versatile hybrid materials, Org. Biomol. Chem., 2007, 5, 1311–1320 RSC.
- J. Tan, H. Li, X. Hu, R. Abdullah, S. Xie, L. Zhang, M. Zhao, Q. Luo, Y. Li, Z. Sun, Q. Yuan and W. Tan, Size-tunable assemblies based on ferrocene-containing DNA polymers for spatially uniform penetration, Chem, 2019, 5, 1775–1792 CAS.
- D. Wang, Y. Zhou, X. Li, X. Qu, Y. Deng, Z. Wang, C. He, Y. Zou, Y. Jin and Y. Liu, Mechanisms of pH-Sensitivity and Cellular Internalization of PEOz-b-PLA Micelles with Varied Hydrophilic/Hydrophobic Ratios and Intracellular Trafficking Routes and Fate of the Copolymer, ACS Appl. Mater. Interfaces, 2017, 9, 6916–6930 CrossRef CAS.
- P. Wei, T. R. Cook, X. Yan, F. Huang and P. J. Stang, A discrete amphiphilic organoplatinum(II) metallacycle with tunable lower critical solution temperature behavior, J. Am. Chem. Soc., 2014, 136, 15497–15500 CrossRef CAS.
- C. Bao, Y. Yin and Q. Zhang, Synthesis and assembly of laccase-polymer giant amphiphiles by self-catalyzed CuAAC click chemistry, Biomacromolecules, 2018, 19, 1539–1551 CrossRef CAS.
- C. Yao, A. Yuan, Z. Wang, H. Lei, L. Zhang, L. Guo and X. Dong, Amphiphilic two-dimensional graphitic carbon nitride nanosheets for visible-light-driven phase-boundary photocatalysis, J. Mater. Chem. A, 2019, 7, 13071–13079 RSC.
- H. Liu, Z. Zhu, H. Kang, Y. Wu, K. Sefan and W. Tan, DNA-based micelles: synthesis, micellar properties and size-dependent cell permeability, Chem. – Eur. J., 2010, 16, 3791–3797 CrossRef CAS PubMed.
- Y. Wu, K. Sefah, H. Liu, R. Wang and W. Tan, DNA aptamer–micelle as an efficient detection/delivery vehicle toward cancer cells, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5–10 CrossRef CAS.
- J. Zou, C. Jin, R. Wang, H. Kuai, L. Zhang, X. Zhang, J. Li, L. Qiu and W. Tan, Fluorinated DNA micelles: synthesis and properties, Anal. Chem., 2018, 90, 6843–6850 CrossRef CAS PubMed.
- M. Anaya, M. Kwak, A. J. Musser, K. Mullen and A. Herrmann, Tunable hydrophobicity in DNA micelles: design, synthesis, and characterization of a new family of DNA amphiphiles, Chem. – Eur. J., 2010, 16, 12852–12859 CrossRef CAS.
- Y. X. Wu and Y. J. Kwon, Aptamers: The “evolution” of SELEX, Methods, 2016, 106, 21–28 CrossRef CAS PubMed.
- M. Kimoto, R. Yamashige, K. Matsunaga, S. Yokoyama and I. Hirao, Generation of high-affinity DNA aptamers using an expanded genetic alphabet, Nat. Biotechnol., 2013, 31, 453–457 CrossRef CAS PubMed.
- L. Qiu, T. Zhang, J. Jiang, C. Wu, G. Zhu, M. You, X. Chen, L. Zhang, C. Cui, R. Yu and W. Tan, Cell Membrane-Anchored Biosensors for Real-Time Monitoring of the Cellular Microenvironment, J. Am. Chem. Soc., 2014, 136, 13090–13093 CrossRef CAS PubMed.
- K. P. Carter, A. M. Young and A. E. Palmer, Fluorescent Sensors for Measuring Metal Ions in Living Systems, Chem. Rev., 2014, 114, 4564–4601 CrossRef CAS PubMed.
- X. Qian and Z. Xu, Fluorescence imaging of metal ions implicated in diseases, Chem. Soc. Rev., 2015, 44, 4487–4493 RSC.
- Y. Li, H. Zhong, Y. Huang and R. Zhao, Recent Advances in AIEgens for Metal Ion Biosensing and Bioimaging, Molecules, 2019, 24, 4593 CrossRef CAS PubMed.
- Z.-L. Zhang, Y.-Y. Wu, K. Xi, J.-P. Sang and Z.-J. Tan, Divalent Ion-Mediated DNA-DNA Interactions: A Comparative Study of Triplex and Duplex, Biophys. J., 2017, 113, 517–528 CrossRef CAS PubMed.
- Y. Wu, D. Wang, I. Willner, Y. Tian and L. Jiang, Smart DNA Hydrogel Integrated Nanochannels with High Ion Flux and Adjustable Selective Ionic Transport, Angew. Chem., Int. Ed., 2018, 57, 7790–7794 CrossRef CAS PubMed.
- Y. Lyu, Y. Guo, R. Cai, R. Peng, C. Hong, X. Chen, W. Hou, X. Li, J. Tan, Y. Zou, X. Zhang, Q. Liu and W. Tan, Spherically Directed Synthesis and Enhanced Cellular Internalization of Metal-Crosslinked DNA Micelles, Chem, 2019, 5, 913–928 CAS.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Quantitative visualization of DNA G-quadruplex structures in human cells, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- C. H. Lu, X. J. Qi, R. Orbach, H. H. Yang, I. Mironi-Harpaz, D. Seliktar and I. Willner, Switchable Catalytic Acrylamide Hydrogels Cross-Linked by Hemin/G-Quadruplexes, Nano Lett., 2013, 13, 1298–1302 CrossRef CAS PubMed.
- C. Jin, X. Liu, H. Bai, R. Wang, J. Tan, X. Peng and W. Tan, Engineering Stability-Tunable DNA Micelles Using Photocontrollable Dissociation of an Intermolecular G-Quadruplex, ACS Nano, 2017, 11, 12087–12093 CrossRef CAS PubMed.
- J. H. Joo and J. S. Lee, Divalent metal ion-mediated assembly of spherical nucleic acids: the case study of Cu2+, Phys. Chem. Chem. Phys., 2015, 17, 30292–30299 RSC.
- Z. Tian, L. Ding, K. Li, Y. Song, T. Dou, J. Hou, X. Tian, L. Feng, G. Ge and J. Cui, Rational Design of a Long-Wavelength Fluorescent Probe for Highly Selective Sensing of Carboxylesterase 1 in Living Systems, Anal. Chem., 2019, 91, 5638–5645 CrossRef CAS PubMed.
- Q. Lin, Z. Li and Q. Yuan, Recent advances in autofluorescence-free biosensing and bioimaging based on persistent luminescence nanoparticles, Chin. Chem. Lett., 2019, 30, 1547–1556 CrossRef CAS.
- X. Cao, S. C. Flagg, X. Li, N. Chennamsetty, G. Balakrishnan and T. K. Das, Quadrupole Dalton-Based Controlled Proteolysis Method for Characterization of Higher Order Protein Structure, Anal. Chem., 2019, 91, 5339–5345 CrossRef CAS PubMed.
- J. Zhang, T. Lan and Y. Lu, Molecular Engineering of Functional Nucleic Acid Nanomaterials toward In Vivo Applications, Adv. Healthcare Mater., 2019, 8, e1801158, DOI:10.1002/adhm.201801158.
- Q. Liu, C. Jin, Y. Wang, X. Fang, X. Zhang, Z. Chen and W. Tan, Aptamer-conjugated nanomaterials for specific cancer cell recognition and targeted cancer therapy, NPG Asia Mater., 2014, 6, e95, DOI:10.1038/am.2014.12.
- W. Kotkowiak, J. Wengel, C. J. Scotton and A. Pasternak, Improved RE31 Analogues Containing Modified Nucleic Acid Monomers: Thermodynamic, Structural, and Biological Effects, J. Med. Chem., 2019, 62, 2499–2507 CrossRef CAS PubMed.
- H. Zhang, Y. Liu, K. Zhang, J. Ji, J. Liu and B. Liu, Single Molecule Fluorescent Colocalization of Split Aptamers for Ultrasensitive Detection of Biomolecules, Anal. Chem., 2018, 90, 9315–9321 CrossRef CAS PubMed.
- C. Zong, M. Xu, L. J. Xu, T. Wei, X. Ma, X. S. Zheng, R. Hu and B. Ren, Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges, Chem. Rev., 2018, 118, 4946–4980 CrossRef CAS PubMed.
- F. Ma, Y. Li, B. Tang and C.-Y. Zhang, Fluorescent Biosensors Based on Single-Molecule Counting, Acc. Chem. Res., 2016, 49, 1722–1730 CrossRef CAS PubMed.
- J. Liu and Y. Lu, Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes, Nat. Protoc., 2006, 1, 246–252 CrossRef CAS PubMed.
- A. Borrmann and J. C. M. van Hest, Bioorthogonal chemistry in living organisms, Chem. Sci., 2014, 5, 2123–2134 RSC.
- S. A. Melo, L. B. Luecke, C. Kahlert, A. F. Fernandez, S. T. Gammon, J. Kaye, V. S. LeBleu, E. A. Mittendorf, J. Weitz, N. Rahbari, C. Reissfelder, C. Pilarsky, M. F. Fraga, D. Piwnica-Worms and R. Kalluri, Glypican1 identifies cancer exosomes and facilitates early detection of cancer, Nature, 2015, 523, 177–182 CrossRef CAS PubMed.
- R. Nedaeinia, M. Manian, M. H. Jazayeri, M. Ranjbar, R. Salehi, M. Sharifi, F. Mohaghegh, M. Goli, S. H. Jahednia, A. Avan and M. Ghayour-Mobarhan, Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer, Cancer Gene Ther., 2017, 24, 48–56 CrossRef CAS PubMed.
- Y. H. Soung, S. Ford, V. Zhang and J. Chung, Exosomes in Cancer Diagnostics, Cancers, 2017, 9, 8 CrossRef PubMed.
- S. Wang, L. Zhang, S. Wan, S. Cansiz, C. Cui, Y. Liu, R. Cai, C. Hong, I. T. Teng, M. Shi, Y. Wu, Y. Dong and W. Tan, Aptasensor with Expanded Nucleotide Using DNA Nanotetrahedra for Electrochemical Detection of Cancerous Exosomes, ACS Nano, 2017, 11, 3943–3949 CrossRef CAS PubMed.
- C. Wu, T. Chen, D. Han, M. You, L. Peng, S. Cansiz, G. Zhu, C. Li, X. Xiong, E. Jimenez, C. J. Yang and W. Tan, Engineering of Switchable Aptamer Micelle Flares for Molecular Imaging in Living Cells, ACS Nano, 2013, 7, 5724–5731 CrossRef CAS PubMed.
- A. E. Prigodich, P. S. Randeria, W. E. Briley, N. J. Kim, W. L. Daniel, D. A. Giljohann and C. A. Mirkin, Multiplexed Nanoflares: mRNA Detection in Live Cells, Anal. Chem., 2012, 84, 2062–2066 CrossRef CAS PubMed.
- L. He, D. Q. Lu, H. Liang, S. Xie, C. Luo, M. Hu, L. Xu, X. Zhang and W. Tan, Fluorescence Resonance Energy Transfer-Based DNA Tetrahedron Nanotweezer for Highly Reliable Detection of Tumor-Related mRNA in Living Cells, ACS Nano, 2017, 11, 4060–4066 CrossRef CAS PubMed.
- Z. Wang, Y. Ju, Z. Ali, H. Yin, F. Sheng, J. Lin, B. Wang and Y. Hou, Near-infrared light and tumor microenvironment dual responsive size-switchable nanocapsules for multimodal tumor theranostics, Nat. Commun., 2019, 10, 1–12 CrossRef PubMed.
- C. Argyo, V. Weiss, C. Bräuchle and T. Bein, Multifunctional mesoporous silica nanoparticles as a universal platform for drug delivery, Chem. Mater., 2013, 26, 435–451 CrossRef.
- J. Liu, G. Yang, W. Zhu, Z. Dong, Y. Yang, Y. Chao and Z. Liu, Light-controlled drug release from singlet-oxygen sensitive nanoscale coordination polymers enabling cancer combination therapy, Biomaterials, 2017, 146, 40–48 CrossRef CAS PubMed.
- Y. Dai, H. Bi, X. Deng, C. Li, F. He, P. A. Ma, P. Yang and J. Lin, 808 nm near-infrared light controlled dual-drug release and cancer therapy in vivo by upconversion mesoporous silica nanostructures, J. Mater. Chem. B, 2017, 5, 2086–2095 RSC.
- C. Chen, J. Geng, F. Pu, X. Yang, J. Ren and X. Qu, Polyvalent Nucleic Acid/Mesoporous Silica Nanoparticle Conjugates: Dual Stimuli-Responsive Vehicles for Intracellular Drug Delivery, Angew. Chem., Int. Ed., 2011, 50, 882–886 CrossRef CAS PubMed.
- J. Li, C. Fan, H. Pei, J. Shi and Q. Huang, Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures, Adv. Mater., 2013, 25, 4386–4396 CrossRef CAS PubMed.
- B. Shi, M. Zheng, W. Tao, R. Chung, D. Jin, D. Ghaffari and O. C. Farokhzad, Challenges in DNA Delivery and Recent Advances in Multifunctional Polymeric DNA Delivery Systems, Biomacromolecules, 2017, 18, 2231–2246 CrossRef CAS PubMed.
- F. Zhang, S. Jiang, S. Wu, Y. Li, C. Mao, Y. Liu and H. Yan, Complex wireframe DNA origami nanostructures with multi-arm junction vertices, Nat. Nanotechnol., 2015, 10, 779–784 CrossRef CAS PubMed.
- Q. Jiang, C. Song, J. Nangreave, X. Liu, L. Lin, D. Qiu, Z. G. Wang, G. Zou, X. Liang, H. Yan and B. Ding, DNA Origami as a Carrier for Circumvention of Drug Resistance, J. Am. Chem. Soc., 2012, 134, 13396–13403 CrossRef CAS PubMed.
- F. E. Alemdaroglu, N. C. Alemdaroglu, P. Langguth and A. Herrmann, DNA Block Copolymer Micelles – A Combinatorial Tool for Cancer Nanotechnology, Adv. Mater., 2008, 20, 899–902 CrossRef CAS.
- T. Jiang, L. Zhou, H. Liu, P. Zhang, G. Liu, P. Gong, C. Li, W. Tan, J. Chen and L. Cai, Monitorable Mitochondria-Targeting DNAtrain for Image-Guided Synergistic Cancer Therapy, Anal. Chem., 2019, 91, 6996–7000 CrossRef CAS PubMed.
- F. Li, J. Lu, J. Liu, C. Liang, M. Wang, L. Wang, D. Li, H. Yao, Q. Zhang, J. Wen, Z. K. Zhang, J. Li, Q. Lv, X. He, B. Guo, D. Guan, Y. Yu, L. Dang, X. Wu, Y. Li, G. Chen, F. Jiang, S. Sun, B. T. Zhang, A. Lu and G. Zhang, A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer, Nat. Commun., 2017, 8, 1390 CrossRef PubMed.
- L. Pan, Q. He, J. Liu, Y. Chen, M. Ma, L. Zhang and J. Shi, Nuclear-Targeted Drug Delivery of TAT Peptide-Conjugated Monodisperse Mesoporous Silica Nanoparticles, J. Am. Chem. Soc., 2012, 134, 5722–5725 CrossRef CAS PubMed.
- Z. Deng, Q. Yang, Y. Peng, J. He, S. Xu, D. Wang, T. Peng, R. Wang, X. Q. Wang and W. Tan, Polymeric Engineering of Aptamer–Drug Conjugates for Targeted Cancer Therapy, Bioconjugate Chem., 2020, 31, 37–42 CrossRef CAS PubMed.
- F. Zhou, P. Wang, Y. Peng, P. Zhang, Q. Huang, W. Sun, N. He, T. Fu, Z. Zhao, X. Fang and W. Tan, Molecular Engineering-Based Aptamer–Drug Conjugates with Accurate Tunability of Drug Ratios for Drug Combination Targeted Cancer Therapy, Angew. Chem., Int. Ed., 2019, 58, 11661–11665 CrossRef CAS PubMed.
- O. Madkhali, G. Mekhail and S. D. Wettig, Modified gelatin nanoparticles for gene delivery, Int. J. Pharm., 2019, 554, 224–234 CrossRef CAS PubMed.
- W. L. Chew, Immunity to CRISPR Cas9 and Cas12a therapeutics, Wiley Interdiscip. Rev.: Syst. Biol. Med., 2018, 10, 1–23 Search PubMed.
- J. Li, C. Zheng, S. Cansiz, C. Wu, J. Xu, C. Cui, Y. Liu, W. Hou, Y. Wang, L. Zhang, I. T. Teng, H. H. Yang and W. Tan, Self-assembly of DNA Nanohydrogels with Controllable Size and Stimuli-Responsive Property for Targeted Gene Regulation Therapy, J. Am. Chem. Soc., 2015, 137, 1412–1415 CrossRef CAS PubMed.
- T. Chen, C. S. Wu, E. Jimenez, Z. Zhu, J. G. Dajac, M. You, D. Han, X. Zhang and W. Tan, DNA Micelle Flares for Intracellular mRNA Imaging and Gene Therapy, Angew. Chem., Int. Ed., 2013, 52, 2012–2016 CrossRef CAS PubMed.
- J. Liu, L. Song, S. Liu, Q. Jiang, Q. Liu, N. Li, Z. G. Wang and B. Ding, A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy, Nano Lett., 2018, 18, 3328–3334 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © the Partner Organisations 2020 |