One-step, rapid fluorescence sensing of fungal viability based on a bioprobe with aggregation-induced emission characteristics†
Xiaoxue
Ge‡
 ab,
Meng
Gao‡
ab,
Meng
Gao‡
 c,
Bo
Situ‡
c,
Bo
Situ‡
 ab,
Weiwei
Feng
ab,
Bairong
He
ab,
Xiaojing
He
ab,
Shiwu
Li
d,
Zihao
Ou
ab,
Weiwei
Feng
ab,
Bairong
He
ab,
Xiaojing
He
ab,
Shiwu
Li
d,
Zihao
Ou
 ab,
Yiqi
Zhong
ab,
Yahui
Lin
ab,
Xinyi
Ye
ab,
Xiumei
Hu
ab,
Ben Zhong
Tang
ab,
Yiqi
Zhong
ab,
Yahui
Lin
ab,
Xinyi
Ye
ab,
Xiumei
Hu
ab,
Ben Zhong
Tang
 *def and
Lei
Zheng
*def and
Lei
Zheng
 *ab
*ab
aDepartment of Laboratory Medicine, Nanfang Hospital, Southern Medical University, Guangzhou, 510515, China. E-mail: nfyyzhenglei@smu.edu.cn
bGuangdong Engineering and Technology Research Center for Rapid Diagnostic Biosensors, Nanfang Hospital, Southern Medical University, Guangzhou, 510515, China
cNational Engineering Research Center for Tissue Restoration and Reconstruction, South China University of Technology, Guangzhou, 510006, China
dCenter for Aggregation-Induced Emission, State Key Laboratory of Luminescent Materials and Devices, South China University of Technology, Guangzhou, 510640, China
eDepartment of Chemistry, Hong Kong Branch of Chinese National Engineering Research Center for Tissue Restoration and Reconstruction, The Hong Kong University of Science & Technology, Clear Water Bay, Kowloon, Hong Kong, China. E-mail: tangbenz@ust.hk
fHKUST Shenzhen Research Institute, No. 9 Yuexing 1st RD, South Area, Hi-tech Park, Nanshan, Shenzhen, 518057, China
First published on 20th January 2020
Abstract
Fungi play key roles in various fields, including fermentation-based industries, medicine, and public health. The rapid and precise evaluation of fungal viability is critical for fungi-related fields. However, the methods currently applied have a series of limitations, such as time-consuming, laborious, and cumbersome. In this study, we designed and synthesized a water-dispersible fluorescent probe DPASI with typical aggregation-induced emission (AIE) characteristics for selectively lighting up dead Candida cells within 5 minutes. The rapid staining of the dead Candida cells could be ascribed to the efficient binding of DPASI with mitochondria, which was revealed by high-resolution fluorescence images taken by a structured illumination microscope (SIM) and colocalization images taken by a confocal laser scanning microscope (CLSM). Compared with traditional probes for the identification of dead Candida cells, the DPASI probe does not require tedious centrifugation and washing steps and has significant advantages in time-and-labor-saving. It thus provides a facile and promising tool for the rapid evaluation of antifungal susceptibility and screening of new antifungal drugs, which would greatly contribute to the fungal research.
Introduction
Fungi, a type of eukaryotes, are closely related to human beings, animals, and plants.1–3 There are about 1 to 1.5 million kinds of fungi widely existing in nature, including air, water, and soil.4–7 The evaluation of fungal viability is critical in fungi-related fields. The antifungal susceptibility testing in a clinic based on the evaluation of the fungi viability is the basis for the use of suitable antifungal drugs and the treatment of nosocomial infections.8–10 Moreover, the determination of Saccharomyces cerevisiae viability in fermentation-based industries will help to evaluate their growth state and is critical for the quality control of various products, such as beer,11,12 beverage, and fermented foods.13 Therefore, it is highly desirable to develop a rapid method for the evaluation of fungal viability.Two traditional methods are widely used to assess the fungal viability: (1) the culture-based assay by counting the number of colony-forming units (CFUs) after fungi growth over 24 to 48 hours;14 (2) the methylene blue method, which includes the counting of the dead fungal cells with blue color and calculation of the stained ratio.15,16 However, the culture-based assay is laborious and time-consuming, which cannot satisfy the requirements in the clinic. Moreover, underestimated CFUs would be obtained when more than one live cell grow together. Although the methylene blue method can shorten time to some extent, it is arduous for manually counting under an optical microscope. Moreover, it highly depends on the subjective judgment of observers and easily leads to large deviations for the counting results of the same sample. Therefore, these two traditional methods are broadly restricted in practice. During the past decades, a series of fluorescent probes have been developed for the evaluation of cell viability.17 In contrary to color staining, fluorescence imaging has significant advantages in terms of high sensitivity, non-invasiveness, easy operation, low interference, and rapid response.18
Luminogens with aggregation-induced emission (AIE)19–23 characteristics have shown great potential in biomedical studies, such as cell imaging, tumor in vivo visualization, and phototherapy.24–33 This could be ascribed to their significant advantages of large Stokes, high emission intensity, excellent photostability, good biocompatibility, and easy operation. Fluorogens that can differentiate bacterial34 or mammalian cell35 viability have been reported. To tackle the challenge for the rapid identification of fungal viability, we herein developed an AIE-active and water-dispersible probe DPASI for the rapid identification of dead fungal cells through turn-on fluorescence imaging within just 5 minutes (Scheme 1). The probe can be used in a wash-free manner, which greatly simplifies the staining procedure and avoids the loss of fungal cells during the washing steps. Moreover, DPASI will not hinder the cell viability under the working condition. Owing to its brilliant properties, DPASI is a potential bioprobe that can be used as a powerful tool for fungal research through the rapid identification of the fungal viability.36
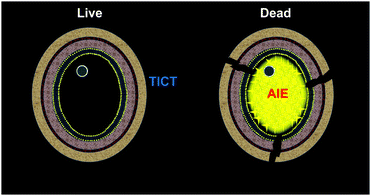 | ||
| Scheme 1 The proposed schematic representation for the identification of fungal viability with DPASI via the turn-on fluorescence imaging of the dead fungal cells. | ||
Experimental section
Materials, chemicals and strains
The RPMI 1640 medium (powder, with L-glutamine, without bicarbonate) was purchased from Sigma. Morpholinepropanesulfonic acid (MOPS) was purchased from MPbio. Both ATB Fungus 3 and Sabouraud Gentamicin Chloramph Agar were purchased from BIOMERIEUX. Toluene, ethyl acetate, tetrahydrofuran (THF), ethanol, methanol, acetonitrile, dimethyl sulfoxide (DMSO), methyl iodide and piperidine were purchased from Sigma-Aldrich or Aladdin. Other commonly used chemical reagents were purchased from Guangzhou Tianjun Biotechnology Co Ltd. Common cell culture consumables such as EP tubes, centrifuge tubes, 96-well plates, and pipette tips were purchased from Axygen and NEST. Milli-Q water was taken from the Milli-Q water purification system (Millipore Corporation, USA) (18.2 MΩ cm−1). Candida albicans (C. albicans, ATCC 66027), Candida glabrata (C. glabrata, ATCC 66032), and Candida krusei (C. krusei, ATCC 6258) were purchased from American Type Culture Collection (ATCC).Synthesis and characterization of DPASI
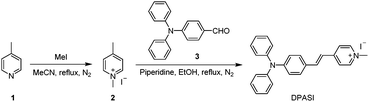
Compound 1 (930 mg, 10 mmol) was first dissolved in MeCN (15 mL), followed by the addition of methyl iodide (2.13 g, 15 mmol). The mixture was heated under reflux for 12 h under nitrogen. After the reaction was completed, the reaction mixture was cooled to room temperature and then dried under a reduced pressure. The residue was added with MeCN (2 mL) under sonication, and a pale yellow precipitate was obtained, which was filtered and dried under vacuum to yield compound 2 (1.59 g, yield 64%).Compound 2 (235 mg, 1.0 mmol) and compound 3 (410 mg, 1.5 mmol) were first dissolved in ethanol (15 mL), and piperidine (400 μL) was then added. The mixture was heated under reflux for 4 h under nitrogen, and a brown-red precipitate was generated in the reaction mixture. After filtration, washing with ethanol and drying under vacuum, pure DPASI was obtained (465 mg, yield 95%).
The 1H and 13C NMR spectra were recorded on a Bruker Avance 500 or 600 MHz NMR spectrometer. The high-resolution mass spectra (HRMS) of compound 2 were recorded on a ThermoFisher Scientific Orbitrap Fusion Tribrid mass spectrometer. The HRMS of DPASI was conducted on a Thermo Fisher ITQ1100 mass spectrometer. UV-vis absorption and photoluminescence (PL) spectra were recorded on a PerkinElmer Lambda 25 UV/vis Spectrometer and PerkinElmer LS 55 Fluorescence Spectrometer, respectively. PL quantum yield (PLQY) was measured on a Hamamatsu absolute PLQY spectrometer C11347 Quantaurus-QY. Particle size was obtained on a Malvern Nanosight NS300 nanoparticle tracking analysis (NTA) system.
Preparation of the RPMI 1640 medium and Candida cell suspension
Powdered RPMI 1640 medium (with L-glutamine, without bicarbonate) 5.2 g was dissolved in 450 mL of pure water. Then, 17.25 g of MOPS (the final concentration was 0.165 mol L−1) was added, and the mixture was stirred until it dissolved. The pH was adjusted to 7.0 ± 0.1 at 25 °C using 1 mol L−1 sodium hydroxide. Additional water was added to make the final volume reach 500 mL. Finally, the medium was sterilized with a 0.22 μm filter and stored at 4 °C.An appropriate amount of Candida colony was picked from the Sabouraud Gentamicin Chloramph Agar stored at 4 °C using an inoculating loop, and was inoculated into a new Sabouraud Gentamicin Chloramph Agar via a partition streak method, then cultured at 35 °C. After 24 hours, several single colonies were picked into normal saline, vortexed and mixed, then counted using an improved Neubauer counting chamber under an optical microscope, and the suspension was adjusted to the concentration required for each experiment, using an RPMI 1640 medium.
Cytotoxicity assay
The C. albicans, C. glabrata, and C. krusei suspensions (final concentration 106 cell per mL) were prepared from the RPMI 1640 medium and added DPASI at concentrations of 0, 2.5, 5, 10, 20, 40, and 80 μM. After incubation at 35 °C for 2 hours or 1 hour, the amount of each concentration was counted using an improved Neubauer counting chamber under an optical microscope. Each concentration was repeated three times. Viability was calculated according to the equation: Viability = C. albicans treated with 2.5 (or 5 or 10 or 20 or 40 or 80) μM DPASI/C. albicans treated with 0 μM DPASI.Fluorescence imaging
The Candida suspensions (the concentration was determined by manual counting with an improved Neubauer counting chamber under an optical microscope) were prepared and divided into two parts, out of which one part was incubated with 10 μM DPASI at room temperature for 5 minutes directly; the other one was heat-treated at 100 °C for 15 minutes, followed by incubating with 10 μM DPASI at room temperature for 5 minutes after cooling down.The fluorescence photograph of live and dead bulk solutions was obtained under 365 nm UV light. To take confocal laser scanning microscope (CLSM) images, about 10 μL of stained Candida solution was transferred onto a glass slide and then covered by a coverslip. The images were collected using Objective Plan-Apochromat 63×/1.40 oil immersion lens. The Candida was imaged under an upright confocal laser scanning microscope LSM 880 (Carl Zeiss, German) using 488 nm excitation lasers. To take structured illumination microscope (SIM) images, about 100 μL of the stained Candida solution was transferred to a 35 mm glass-bottomed culture dish. The images were collected using 100 × SR apochromatic TIRF objective. The Candida was imaged under a structured illumination microscope N-SIM (Nikon, Japan), using 488 nm excitation laser.
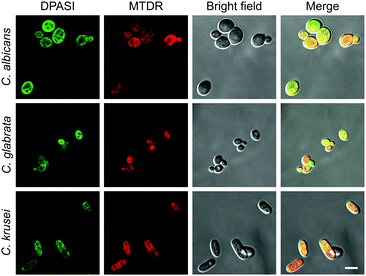
To take colocalization images, Candida suspensions were prepared and were incubated with 200 nM MitoTracker Deep Red (MTDR) at 35 °C for 30 minutes. Then, these suspensions were fixed with 4% paraformaldehyde (PFA) for 15 minutes. After fixing, they were stained with 10 μM DPASI for 5 minutes, respectively. CLSM images were collected using an Objective Plan-Apochromat 100×/1.40 oil immersion lens. C. albicans, C. glabrata, and C. krusei were imaged under an inverted confocal laser scanning microscope LSM 880 with Airyscan (Carl Zeiss, German) using 488 nm and 647 nm excitation laser.
Results and discussion
Synthesis and characterization of DPASI
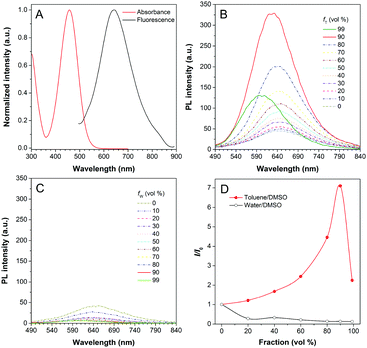
DPASI, comprising of a pyridinium moiety, a carbon–carbon double bond, and a triphenylamine moiety, was synthesized following a simple two-step procedure (Fig. 1). The structures of compound 1, compound 2, and DPASI were well verified by 1H NMR, 13C NMR, and HRMS spectra (Fig. S1–S7, ESI†). In the DMSO solution, DPASI has a maximum absorption peak at 467 nm and a maximum emission peak at 645 nm (Fig. 2A). The PL quantum yield (PLQY) of 10 μM DPASI in the DMSO solution was 1% and was 3% when measured in the thin-film mode. The pyridinium moiety acts as an electron-acceptor, while the triphenylamine moiety acts as an electron-donor. The UV-vis and PL spectra of DPASI were measured in solvents with different polarities along with the increased solvent polarity the red-shift and decreased emission spectra were observed, while no obvious change was observed for the absorption spectra (Fig. S8, ESI†). The solvent-dependent PL spectra change can be ascribed to the typical electron donor–acceptor (D–A) structure of DPASI and the twisted intramolecular charge transfer (TICT) effect.37Since toluene is a poor solvent for DPASI, we next measured the PL spectra of DPASI in DMSO/toluene mixtures with different toluene fractions (fT) (Fig. 2). As shown in Fig. 2B and D, the PL intensity at 645 nm increases by 7 times, upon gradually increasing fT from 0% to 90%. However, the PL intensity decreases sharply, when fT increases to 99%, which can be ascribed to the formation of precipitates of DPASI, in its poor solvent. These results indicate that DPASI exhibits a typical AIE feature. The blue shift of the spectrum in Fig. 2B, when fT increases to 99%, might be ascribed to the TICT effect resulting from the D–A structure of DPASI, with the blue-shifting of emission as the solvent polarity decreases (toluene is a non-polar solvent).38,39 The PL spectra of DPASI in the DMSO/water mixtures with different water fractions (fW) were also measured. As shown in Fig. 2C and D, the PL intensity gradually decreased as the fW increased, thereby indicating that DPASI exhibits good dispersity in water whose polarity was higher than DMSO. All these suggest that DPASI shows TICT property. The NTA result verified the formation of well-dispersed nanoaggregates, with an average diameter of 65.9 nm in the phosphate buffer (Fig. S9, ESI†), further indicating DPASI exhibits good dispersity. The low fluorescence background in the aqueous solution indicates the potential of DPASI for the biological applications.
Rapid differentiation of Candida viability and evaluation of DPASI toxicity
The brilliant properties of DPASI prompt us to explore its biological applications in fungal imaging. C. albicans, the most common pathogenic fungus,40,41 was used as a test model. After incubation of living C. albicans with DPASI, there was almost no enhancement in fluorescence. In sharp contrast, the dead C. albicans stained with DPASI exhibited strong emission with distinct fluorescence enhancement than live ones. This marked fluorescence difference in the fungal cell makes DPASI an excellent candidate as a fluorescent probe for the evaluation of fungal viability. To prove this capability of DPASI, we mixed live and dead C. albicans cells together in equal proportions and incubated with DPASI for 5 minutes. As expected, we could clearly observe that only half of the fungal cells emit strong fluorescence (Fig. 3). Another two kinds of common Candida, C. glabrata and C. krusei, were also investigated. Similarly, dead and viable fungal cells could be well-distinguished by their fluorescence intensity (Fig. S10, ESI†). These results suggest that DPASI was capable of rapid identification of fungal cell viability. A similar assay was conducted on the most common pathogenic bacteria, E. coli.42 Interestingly, we found that DPASI can also indicate E. coli viability (Fig. S11, ESI†), which means that DPASI has the potential in differentiating bacterial viability also, too.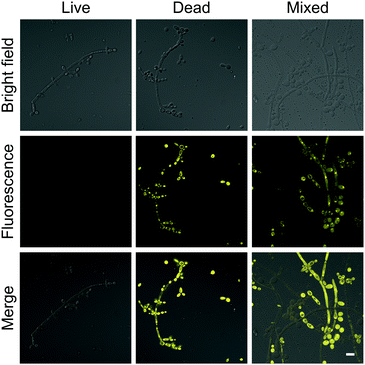 | ||
| Fig. 3 The confocal laser scanning microscope (CLSM) images of C. albicans stained with 10 μM DPASI. λex = 488 nm; scale bar = 10 μm. | ||
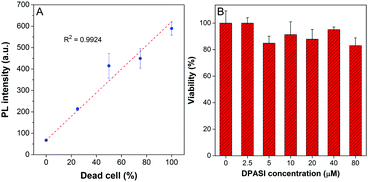
Since dead and live fungal cells can be clearly distinguished by DPASI under a fluorescence microscope, we next investigated whether DPASI can differentiate fungal viability in bulk solutions. As shown in the inset of Fig. S12 (ESI†), the solution containing live C. albicans was almost non-emissive, but it exhibited a strong orange fluorescence in the presence of dead Candida cells, which can be clearly observed by naked eyes. Notably, the whole staining process took only 5 minutes (Fig. S12, ESI†), and no centrifugation and washing steps were required due to its fluorescent turn-on effect on the dead fungal cells. This can greatly simplify the experimental procedure and avoid the loss of fungal cells and some potential contamination during the experimental process. To quantify the fluorescence intensity change, we measured the PL spectra of the solutions containing Candida cells without treatment or heating for 15 minutes by the fluorescence spectrometer. In accordance with the CLSM results, the PL intensity of the solutions containing heat-treatment Candida cells was 3–9 times higher than that of solutions with live cells (Fig. S13, ESI†). We further used another method to interfere with Candida cell viability by treatment of 75% ethanol. As shown in Fig. S13 (ESI†), the PL intensity of solutions with ethanol-treatment Candida cells was 3–13 times higher than that of solutions under control. These results obtained from both heat and ethanol treatment reveals that DPASI can efficiently indicate the fungal viability. In order to find out whether DPASI can quantitatively detect Candida viability, we also measured the PL intensity of DPASI to a different percentage of dead C. albicans cells. To our delight, a linear relationship was observed (R2 is 0.9924) between the PL intensity of DPASI and dead C. albicans cells fractions (Fig. 4A).
The cytotoxicity of a probe is also a crucial parameter because the toxic probe itself may lead to some cell death, which would bring about an inaccurate evaluation of the cell viability. Therefore, cell counting assay was performed with C. albicans, C. glabrata, and C. krusei to evaluate the toxicity of DPASI to Candida cells. The C. albicans, C. glabrata, and C. krusei were cultured at 35 °C with different concentrations (0, 2.5, 5, 10, 20, 40, and 80 μM) of DPASI for 2 hours. As displayed in Fig. 4B and Fig. S14 (ESI†), under the working concentration of DPASI (10 μM), the cell viability was maintained above 80% of control, which illustrates that there was almost no influence on the growth and proliferation of Candida cells, and DPASI was almost non-toxic to Candida under the working condition.
Investigation of mechanism for the binding of DPASI and fungal cell
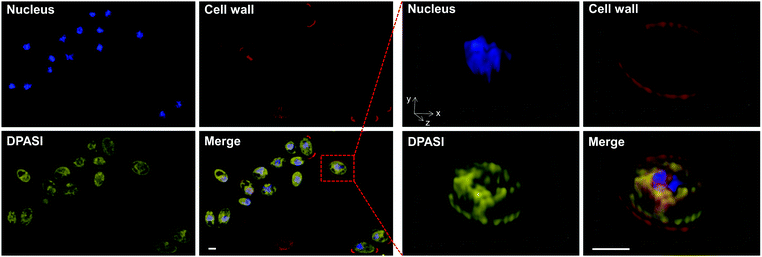
To investigate the potential working mechanism of DPASI, we used structured illumination microscopy (SIM), a kind of imaging pattern of super-resolution microscopy, to observe the subcellular location of DPASI. DAPI, a dye for dead cell nucleus, Calcofluor White Staining, a well-known dye for cell wall, and our probe DPASI were used to observe their relative biodistribution in the dead Candida cell. We found that DPASI mainly located in the cytoplasm of the dead cell, and something with reticular structure was stained, indicating that DPASI was probably binding to the membrane-like organelle inside the cell (Movie S1, ESI†). Moreover, as shown in Fig. 5, we observed that there was no overlap at all between the fluorescence of DPASI and DAPI and Calcofluor White Staining. It was somewhere outside the nucleus but inside the cell wall. Therefore, we speculated that it might selectively accumulate in the cytoplasm. As we all know, an impaired cell wall is one of the characteristics of a dead cell. Given that our probe was AIE-active and positively charged, DPASI may rapidly diffuse into the cytoplasm of a dead cell and aggregate in the intracellular organelles, such as mitochondria, endoplasmic reticulum, and Golgi apparatus. The restriction of the intramolecular motion would then activate the fluorescence emission. Instead, in a viable cell as the cell membrane and cell wall are intact, DPASI cannot diffuse into the cytoplasm and shows almost no fluorescence emission due to the intramolecular motion and TICT effect.To further investigate the DPASI distribution, a colocalization assay was performed. C. albicans, C. glabrata, and C. krusei were stained with 200 nM commercial mitochondria-targeting dye MTDR, followed by fixing with 4% PFA, and then stained with 10 μM DPASI. As shown in Fig. 6, the fluorescence of DPASI was well-colocalized with that of MTDR, which demonstrates that DPASI stains mitochondria of the dead fungal cell. We speculate that DPASI can indicate the fungal viability through binding to the mitochondria of dead fungi as the inner membrane of mitochondria was negatively charged, whereas DPASI was positively charged.
Conclusions
In summary, we developed a rapid and facile method for fungal viability differentiation with the help of a new AIE molecule, namely DPASI. It exhibits typical AIE properties, such as a high signal-to-noise ratio and good water dispersity. Importantly, DPASI can evaluate fungal viability by lighting up fungal cells that have lost viability and emit intense yellow fluorescence, whereas there was almost no fluorescence in the viable ones. This could be ascribed to the positively charged DPASI, which can easily diffuse into the dead cells with an impaired cell wall and cell membrane and bind with mitochondria inside the cytoplasm, while in the viable ones, DPASI is blocked by the intact cell wall and cell membrane and cannot enter into the cells and well-dispersed in the solution with no fluorescence emission. The obvious distinction in the fluorescence intensity can be clearly observed by naked eyes, and the whole staining process takes only 5 minutes. More importantly, since DPASI is water-dispersible, it emits faintly in the aqueous solutions with negligible background, and the centrifugation and washing steps can be avoided, which not only simplifies the whole procedures but also increases the evaluation accuracy. Moreover, DPASI is almost non-toxic to the fungal cell under the working condition. These brilliant properties of DPASI prompt us to further expand the applications of AIE probes in the field of microbiology and to explore their potentials in antifungal susceptibility testing and the screening of new antifungal drugs in our laboratory.Author contributions
B. S. T. guided the entire study, participated in data analysis, manuscript drafting and revision. X. X. G. conducted the research and participated in analyzing data and drafting this manuscript. M. G. and S. W. L. designed, synthesized and characterized DPASI. B. R. H. participated in the characterization of DPASI and manuscript revision. W. W. F., X. J. H., Z. H. O., Y. Q. Z., Y. H. L. and X. Y. Y. participated in the experiments and data analysis. X. M. H. assisted with the data collection and analysis. L. Z. and B. Z. T. designed the experiments, initiated and supervised the project. All authors worked on the manuscript and approved the final manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank Chenglin Xiang, Manna Lin, and Yufang Cheng for their help with the fluorescence imaging. This work was supported in part by the National Natural Science Foundation of China (81672076, 81802116, 81871735, 81601819, 21788102, 51620105009, 21877040, 21602063), the Medical Science and Technology Research Foundation of Guangdong Province (A2017326), the Guangdong Provincial Key Laboratory of Luminescence from Molecular Aggregates (South China University of Technology) (2019B030301003), the Science and Technology Program of Guangzhou (201510010097), the Major Program of Health Care and Innovation of Guangzhou Project (201704020213, 201604020015), the Pearl River S&T Nova Program of Guangzhou (201806010152), the Natural Science Foundation of Guangdong Province (2016A030313852), the Outstanding Youths Development Scheme of Nanfang Hospital, Southern Medical University (2018J002, 2016J013), the Outstanding Youths Development Scheme of Southern Medical University (2019YQPY007), and the College Students' Innovative Entrepreneurial Training Plan Program (201812121045).References
- M. C. Fisher, D. A. Henk, C. J. Briggs, J. S. Brownstein, L. C. Madoff, S. L. McCraw and S. J. Gurr, Emerging fungal threats to animal, plant and ecosystem health, Nature, 2012, 484, 186 CrossRef CAS PubMed.
- M. C. Fisher, N. A. Gow and S. J. Gurr, Tackling emerging fungal threats to animal health, food security and ecosystem resilience, Philos. Trans. R. Soc. London, Ser. B, 2016, 371, 20160332 CrossRef PubMed.
- E. Haneke, Fungal infections of the nail, Semin. Dermatol., 1991, 10, 41 CAS.
- B. Jung, J. Park, N. Kim, T. Li, S. Kim, L. E. Bartley, J. Kim, I. Kim, Y. Kang, K. Yun, Y. Choi, H. H. Lee, S. Ji, K. S. Lee, B. Y. Kim, J. C. Shon, W. C. Kim, K. H. Liu, D. Yoon, S. Kim, Y. S. Seo and J. Lee, Cooperative interactions between seed-borne bacterial and air-borne fungal pathogens on rice, Nat. Commun., 2018, 9, 31 CrossRef PubMed.
- G. Stegen, F. Pasmans, B. R. Schmidt, L. O. Rouffaer, S. Van Praet, M. Schaub, S. Canessa, A. Laudelout, T. Kinet, C. Adriaensen, F. Haesebrouck, W. Bert, F. Bossuyt and A. Martel, Drivers of salamander extirpation mediated by Batrachochytrium salamandrivorans, Nature, 2017, 544, 353 CrossRef CAS PubMed.
- L. Chen, N. G. Swenson, N. Ji, X. Mi, H. Ren, L. Guo and K. Ma, Differential soil fungus accumulation and density dependence of trees in a subtropical forest, Science, 2019, 366, 124 CrossRef CAS PubMed.
- T. J. Thirkell, D. Pastok and K. J. Field, Carbon for nutrient exchange between arbuscular mycorrhizal fungi and wheat varies according to cultivar and changes in atmospheric carbon dioxide concentration, Glob. Chang. Biol., 2019 DOI:10.1111/gcb.14851.
- L. F. Yang, X. Liu, L. L. Lv, Z. M. Ma, X. C. Feng and T. H. Ma, Dracorhodin perchlorate inhibits biofilm formation and virulence factors of Candida albicans, J. Mycol. Med., 2018, 28, 36 CrossRef CAS PubMed.
- S. Dahiya, A. K. Chhillar, N. Sharma, P. Choudhary, A. Punia, M. Balhara, K. Kaushik and V. S. Parmar, Candida auris and nosocomial infection, Curr. Drug Targets, 2020, 21, 365–373 Search PubMed.
- A. Belkin, Z. Gazit, N. Keller, R. Ben-Ami, A. Wieder-Finesod, A. Novikov, G. Rahav and T. Brosh-Nissimov, Candida auris infection leading to nosocomial transmission, Israel, 2017, Emerging Infect. Dis., 2018, 24, 801 CrossRef PubMed.
- B. Gallone, J. Steensels, T. Prahl, L. Soriaga, V. Saels, B. Herrera-Malaver, A. Merlevede, M. Roncoroni, K. Voordeckers, L. Miraglia, C. Teiling, B. Steffy, M. Taylor, A. Schwartz, T. Richardson, C. White, G. Baele, S. Maere and K. J. Verstrepen, Domestication and divergence of Saccharomyces cerevisiae beer yeasts, Cell, 2016, 166, 1397 CrossRef CAS PubMed.
- B. Gallone, S. Mertens, J. L. Gordon, S. Maere, K. J. Verstrepen and J. Steensels, Origins, evolution, domestication and diversity of Saccharomyces beer yeasts, Curr. Opin. Biotechnol., 2018, 49, 148 CrossRef CAS PubMed.
- Z. Li, K. Song, H. Li, R. Ma and M. Cui, Effect of mixed Saccharomyces cerevisiae Y10 and Torulaspora delbrueckii Y22 on dough fermentation for steamed bread making, Int. J. Food Microbiol., 2019, 303, 58 CrossRef CAS PubMed.
- P. G. Sohnle, B. L. Hahn and M. D. Erdmann, Effect of fluconazole on viability of Candida albicans over extended periods of time, Antimicrob. Agents Chemother., 1996, 40, 2622 CrossRef CAS PubMed.
- S. Chermsirivathana, A rapid method of staining for fungus and monilial infection, J. Invest. Dermatol., 1952, 19, 7 CrossRef CAS PubMed.
- R. J. Webb, L. Berger, L. F. Skerratt and A. A. Roberts, A rapid and inexpensive viability assay for zoospores and zoosporangia of Batrachochytrium dendrobatidis, J. Microbiol. Methods, 2019, 165, 105688 CrossRef CAS PubMed.
- M. Tian, Y. Ma and W. Lin, Fluorescent probes for the visualization of cell viability, Acc. Chem. Res., 2019, 52, 2147 CrossRef CAS PubMed.
- B. N. Giepmans, S. R. Adams, M. H. Ellisman and R. Y. Tsien, The fluorescent toolbox for assessing protein location and function, Science, 2006, 312, 217 CrossRef CAS PubMed.
- J. Luo, Z. Xie, J. W. Lam, L. Cheng, H. Chen, C. Qiu, H. S. Kwok, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Aggregation-induced emission of 1-methyl-1,2,3,4,5-pentaphenylsilole, Chem. Commun., 2001, 1740 RSC.
- J. Mei, N. L. Leung, R. T. Kwok, J. W. Lam and B. Z. Tang, Aggregation-induced emission: Together we shine, united we soar!, Chem. Rev., 2015, 115, 11718 CrossRef CAS PubMed.
- J. Yang, Z. G. Chi, W. H. Zhu, B. Z. Tang and Z. Li, Aggregation-induced emission: A coming-of-age ceremony at the age of eighteen, Sci. China: Chem., 2019, 62, 1090 CrossRef CAS.
- Q. Q. Li and Z. Li, The strong light-emission materials in the aggregated state: What happens from a single molecule to the collective group, Adv. Sci., 2017, 4, 1600484 CrossRef PubMed.
- Q. Q. Li and Z. Li, Miracles of molecular uniting, Sci. China Mater., 2020, 63, 177 CrossRef.
- J. Liang, B. Z. Tang and B. Liu, Specific light-up bioprobes based on AIEgen conjugates, Chem. Soc. Rev., 2015, 44, 2798 RSC.
- X. Ni, X. Zhang, X. Duan, H. L. Zheng, X. S. Xue and D. Ding, Near-infrared afterglow luminescent aggregation-induced emission dots with ultrahigh tumor-to-liver signal ratio for promoted image-guided cancer surgery, Nano Lett., 2019, 19, 318 CrossRef CAS PubMed.
- C. Chen, H. Ou, R. Liu and D. Ding, Regulating the photophysical property of organic/polymer optical agents for promoted cancer phototheranostics, Adv. Mater., 2019, e1806331 Search PubMed.
- J. Qi, C. Chen, X. Zhang, X. Hu, S. Ji, R. T. K. Kwok, J. W. Y. Lam, D. Ding and B. Z. Tang, Light-driven transformable optical agent with adaptive functions for boosting cancer surgery outcomes, Nat. Commun., 2018, 9, 1848 CrossRef PubMed.
- C. Y. Yu, H. Xu, S. Ji, R. T. Kwok, J. W. Lam, X. Li, S. Krishnan, D. Ding and B. Z. Tang, Mitochondrion-anchoring photosensitizer with aggregation-induced emission characteristics synergistically boosts the radiosensitivity of cancer cells to ionizing radiation, Adv. Mater., 2017, 29, 1606167 CrossRef PubMed.
- F. Hu, S. Xu and B. Liu,, Photosensitizers with aggregation-induced emission: Materials and biomedical applications, Adv. Mater., 2018, 30, e1801350 CrossRef PubMed.
- R. Hu, F. Zhou, T. Zhou, J. Shen, Z. Wang, Z. Zhao, A. Qin and B. Z. Tang, Specific discrimination of Gram-positive bacteria and direct visualization of its infection towards mammalian cells by a DPAN-based AIEgen, Biomaterials, 2018, 187, 47 CrossRef CAS PubMed.
- G. Feng and B. Liu, Aggregation-induced emission (AIE) dots: Emerging theranostic nanolights, Acc. Chem. Res., 2018, 51, 1404 CrossRef CAS PubMed.
- J. Qi, C. Chen, D. Ding and B. Z. Tang, Aggregation-induced emission luminogens: Union is strength, gathering illuminates healthcare, Adv. Healthcare Mater., 2018, 7, e1800477 CrossRef PubMed.
- H. Q. Gao, X. Y. Zhang, C. Chen, K. Li and D. Ding, Unity makes strength: How Aggregation-induced emission luminogens advance the biomedical field, Adv. Biosyst., 2018, 2, 1800074 CrossRef.
- E. Zhao, Y. Hong, S. Chen, C. W. Leung, C. Y. Chan, R. T. Kwok, J. W. Lam and B. Z. Tang, Highly fluorescent and photostable probe for long-term bacterial viability assay based on aggregation-induced emission, Adv. Healthcare Mater., 2014, 3, 88 CrossRef CAS PubMed.
- R. Hu, T. Zhou, B. Li, R. T. K. Kwok, J. Shen, A. Qin and B. Z. Tang, Selective viable cell discrimination by a conjugated polymer featuring aggregation-induced emission characteristic, Biomaterials, 2020, 230, 119658 CrossRef PubMed.
- A. M. Fuentefria, B. Pippi, D. F. Dalla Lana, K. K. Donato and S. F. de Andrade,, Antifungals discovery: An insight into new strategies to combat antifungal resistance, Lett. Appl. Microbiol., 2018, 66, 2 CrossRef CAS PubMed.
- L. Sun, W. Zhu, W. Wang, F. Yang, C. Zhang, S. Wang, X. Zhang, R. Li, H. Dong and W. Hu,, Intermolecular charge-transfer interactions facilitate two-photon absorption in styrylpyridine-tetracyanobenzene cocrystals, Angew. Chem., Int. Ed., 2017, 56, 7831 CrossRef CAS PubMed.
- R. R. Hu, E. Lager, A. Aguilar-Aguilar, J. Z. Liu, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, Y. C. Zhong, K. S. Wong, E. Pena-Cabrera and B. Z. Tang, Twisted intramolecular charge transfer and aggregation-induced emission of BODIPY derivatives, J. Phys. Chem. C, 2009, 113, 15845 CrossRef CAS.
- S. Gao, G. Wei, S. Zhang, B. Zheng, J. Xu, G. Chen, M. Li, S. Song, W. Fu, Z. Xiao and W. Lu, Albumin tailoring fluorescence and photothermal conversion effect of near-infrared-II fluorophore with aggregation-induced emission characteristics, Nat. Commun., 2019, 10, 2206 CrossRef PubMed.
- A. Tsega and F. Mekonnen, Prevalence, risk factors and antifungal susceptibility pattern of Candida species among pregnant women at Debre Markos Referral Hospital, Northwest Ethiopia, BMC Pregnancy Childbirth, 2019, 19, 527 CrossRef PubMed.
- B. J. Kullberg and M. C. Arendrup, Invasive candidiasis, N. Engl. J. Med., 2015, 373, 1445 CrossRef CAS PubMed.
- J. T. Poolman and M. Wacker, Extraintestinal pathogenic Escherichia coli, a common human pathogen: Challenges for vaccine development and progress in the field, J. Infect. Dis., 2016, 213, 6 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9qm00732f |
| ‡ These authors contributed equally to this work. |
| This journal is © the Partner Organisations 2020 |