Active targeting co-delivery of therapeutic Sur siRNA and an antineoplastic drug via epidermal growth factor receptor-mediated magnetic nanoparticles for synergistic programmed cell death in glioblastoma stem cells†
Xueqin
Wang
 *a,
Ruifang
Li
a,
Yongxia
Zhu
b,
Zichao
Wang
a,
Huiru
Zhang
a,
Liuqing
Cui
a,
Shaofeng
Duan
*c and
Yuqi
Guo
*b
*a,
Ruifang
Li
a,
Yongxia
Zhu
b,
Zichao
Wang
a,
Huiru
Zhang
a,
Liuqing
Cui
a,
Shaofeng
Duan
*c and
Yuqi
Guo
*b
aCollege of Bioengineering, Henan University of Technology, Zhengzhou, Henan 450001, P. R. China. E-mail: wangxq0708@163.com
bHenan Provincial People's Hospital, Zhengzhou 450003, P. R. China. E-mail: yuqi-guo@163.com; Fax: +86 371 65580059; Tel: +86 371 65580059
cSchool of Pharmacy, Henan University, Kaifeng, Henan 475004, P. R. China. E-mail: sduan@henu.edu.cn; Fax: +86 371 22822134; Tel: +86 371 22822134
First published on 14th December 2019
Abstract
Active targeting co-delivery of therapeutic siRNAs and antineoplastic drugs is considered as a very promising approach to specifically improve the therapeutic effect by simultaneously inhibiting the expression of tumorigenic genes and reducing the systemic toxicity of anticancer drugs. This strategy for effectively targeting cancer stem cells (CSCs) has gained considerable attention in the therapeutic treatment of human cancers. The objective of this study was to fabricate and characterize the therapeutic survivin siRNAs (Sur siRNA) and anti-cancer drug doxorubicin (DOX)-loaded superparamagnetic iron oxide nanoparticles (SPIONPs) that are surface-conjugated to the specific ligand epidermal growth factor receptor (EGFR) which is highly expressed on brain glioblastoma stem cells (GSCs), using carboxymethyl chitosan (CMCS), polyethylenimine (PEI) and heparin-mediated cross linking agents. Our in vitro and in vivo studies demonstrated that the fabricated EGFR-targeted nanoparticles exhibited excellent targeting specificity and enhanced delivery efficiency, and could specifically co-deliver therapeutic siRNAs and DOX into GSCs, thus greatly improving the therapeutic efficacy by effectively silencing survivin gene expression and enhancing the treatment sensitivity of GSCs to anticancer drug DOX. Therefore, these fabricated EGFR-targeted nanoparticles would provide an efficient targeting vector for co-delivery of therapeutic siRNAs and anticancer drugs, and could be used as a promising active targeted nanocarrier for the selective treatment of human brain cancer in the future.
1. Introduction
Glioblastoma multiforme (GBM) is considered as one of the most common malignant cancers within the central nervous system, and accounts for about half of all intracranial cancers with high morbidity and mortality.1,2 Despite the use of conventional treatments such as surgery, chemotherapy and radiation therapy, these treatments seem to only effectively remove differentiated cells, and due to the presence of glioblastoma stem cells (GSCs), the prognosis of patients with malignant glioma remains very different.3,4 Non-GSCs in cancerous tissues may also produce GSCs randomly, and the standard chemotherapy for these GSC treatments is often ineffective because it often leads to GSC enrichment and tumor recurrence.2,3 Although GSCs are a rare subpopulation of glioblastoma cells, there is increasing evidence that GSCs are related to the occurrence, recurrence, persistence and treatment resistance of glioblastoma cells.5,6 Due to the failure to eliminate tumor recurrence in glioma patients, GSCs have received extensive attention in the field of oncology.7,8 Therefore, reasonable targeted GSC therapy and specific killing of GSCs become important strategies to improve the clinical efficacy of GBM.9–11 To achieve this goal, many new GSC treatment strategies have been developed, including inhibiting the development of GSCs by blocking the necessary self-renewal signals, forcing GSCs to differentiate into vulnerable large tumor cells for standard treatment, and identifying specific anti-GSC candidates.12,13SURVIVIN protein is a newly discovered apoptosis-related inhibitor,14,15 which exists in human GBM and is closely related to the differentiation, proliferation, invasion and metastasis of glioblastoma cells in tumor cells.16,17 Thus, effective silencing of SURVIVIN expression by small interfering RNAs (siRNAs) can negatively regulate SURVIVIN translation, and ultimately promote apoptosis of glioblastoma cells.18,19
Polyethylenimine (PEI) conjugated nanomaterials hold great siRNA delivery potential in vitro and in vivo, as the “proton sponge” effect of PEI can fully absorb siRNA and ensure that siRNA escapes from intracellular lysosomal degradation.20,21 However, non-specific delivery of PEI-conjugated nanomaterials exaggerates cytotoxicity in non-malignant tissues, so how to introduce appropriate molecules into PEI conjugated nanomaterials to improve biocompatibility and target specificity has been extensively studied.22,23
Cell-targeted drug delivery via superparamagnetic iron oxide nanoparticles (SPIONPs) coated with cell-specific ligands (antibodies and other small molecules) has emerged as a potential clinical tool for cancer treatment.24–26 Particularly, SPIONPs ranging from 10 to 25 nm exhibit unique magnetic properties that allow them to escape the immune system.27,28 Epidermal growth factor receptor (EGFR) is a tyrosine kinase cell transmembrane receptor associated with abnormal survival and poor prognosis.29,30 Recent evidence suggests that EGFR is overexpressed in most GBM and GSCs, and is a major target for tumor therapy.31–33 Therefore, the preparation of EGFR targeting SPIONPs may be an ideal approach for targeting siRNA and anticancer drugs in clinical cancer therapy.
In this study, we proposed a strategy to develop an active targeting, nano-scale theranostic platform for co-delivery of therapeutic siRNAs and anti-cancer drugs to enhance the antitumor efficacy. In this strategy, the SPIONPs are coated with a CMCS layer and then further modified with PEI to increase siRNA loading and heparin-mediated EGFR affinity to form nanocomplexes (EGF-hep-PEI-CMCS @SPIO, i.e. eMNNS). Finally, the MNNS nanocomposites were loaded with DOX/Sur siRNA and co-delivered to GSCs with high expression of EGFR in vivo and in vitro employing tumor-bearing mice as the animal model.
2. Materials and methods
2.1. Materials
The human glioblastoma U251 cell line was purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell culture medium and fetal bovine serum (FBS) were purchased fromGibco Invitrogen Corporation (CA, USA). Recombinant human epidermal growth factor (rhEGF) and basic fibroblast growth factor (bFGF) were purchased from Peprotech (Rocky Hill, NJ, USA). Leukemia inhibitory factor (LIF) was purchased from Chemicon (Temecula, CA, USA). B27 was received from Invitrogen (Carlsbad, CA, USA). Vincristine was obtained from Haizheng Pharmaceutical Co., Ltd (Zhejiang, China). CMCS, PEI, epidermal growth factor (EGF), heparin, DOX, 3-(4,5-dimethylthiazol-2-diphenyl-tetrazolium) bromide (MTT), fluorescein diacetate (FDA), propidium iodide (PI), Hoechst H33258, acridine orange (AO), dimethyl sulfoxide (DMSO), Triton X-100 solution, and RBITC, were purchased from Sigma-Aldrich (St. Louis, MO, USA). Fluorescent dye 4,6-diamidino-2-phenylindole (DAPI) was purchased from Molecular Probes Inc. (Eugene, OR, USA). The specific primary antibodies against CD133, nestin and EGFR (mouse monoclonal IgG) were obtained from Abcam (Cambridge, UK, USA). The specific primary antibodies against glial fibrillary acidic protein (GFAP) (mouse monoclonal IgG) were from Sigma-Aldrich (St. Louis, MO, USA). The fluorescence-labeled secondary antibodies goat anti-mouse antibody (IgG-RBITC) and goat anti-mouse antibody (IgG-FITC) were obtained from Boster (Wuhan, China). Other reagents and chemicals were purchased from local commercial suppliers and were of analytical reagent grade, unless otherwise stated. Deionized (DI) water (Milli-Q, Millipore, Bedford, MA) was used to prepare aqueous solutions. The duplexed unlabeled Sur siRNA, 5′-carboxyfluorescein (FAM) labeled and scrambled Sur siRNA were synthesized with the following sequences by Shanghai Gene Pharma Co., Ltd (sense, 5′-CAC CGC AUC UCU ACA UUC ATT-3′; antisense, 5′-UGA AUG UAG AGA UGC GGU GTT-3′; negative control, sense, 5′-UUC UCC GAA CGU GUC ACG UTT-3′; antisense, 5′-ACG UGA CAC GUU CGG AGAATT-3′).2.2. Animals
Pathogen-free female BALB/c nude mice aged about 6 weeks were obtained from School of Medicine of Zhengzhou University (Zhengzhou, China) and maintained in the absence of specific pathogens. All animal experiments were conducted in accordance with the guidelines set by the Animal Care and Use Committee of School of Medicine of Zhengzhou University (Zhengzhou, China).2.3. Cell culture, identification and EGFR protein expression assay
The culture of GSCs followed the previous literature.34 Briefly, GSCs were suspended in serum-free Dulbecco's Modified Eagle Medium/F12 (DMEM/F12), and B27 (1×), 20 ng mL−1 hEGF, 20 ng mL−1 bFGF, 10 ng mL−1 leukemia inhibitory factor, and 8 ng mL−1 vincristine were added. GSCs were then collected by gentle centrifugation and then dissociated into single cells. EGFR expression of the GSCs was analyzed by Western blot and immunocytochemical staining.2.4. Synthesis and characterization of PEI modified magnetic nanocarriers
In the present study, SPIONPs, i.e. magnetic γ-Fe2O3NPs with a diameter of 10–15 nm, were synthesized as the carrier core according to the previously reported methods,35,36 and the details can be seen in the ESI.† The biocompatible CMCS-modified SPIONPs (CMCS@SPIO) were fabricated via TPP crosslinking as previously reported.37 In brief, about 50 mL of TPP solution (1 mg mL−1) was gradually added with 10 mg mL−1 of SPIONP suspension, and then stirred vigorously for 30 min at 60 °C. The mixture was kept at room temperature (RT) for 12 h and then washed three times with deionized water (DI) to give TPP@SPIONPs, followed by the addition of 10 mL of CMCS solution in acetic acid (1% w/v). After the reaction proceeded in a phacoemulsifier for 30 min, the CMCS@SPIONPs were rinsed three times with DI water and stored at 4 °C for the next use.To coat the CMCS@SPIONPs with PEI, the PEI aqueous solution was firstly mixed with MNNS solution (v/v, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) in MES buffer, and then magnetically stirred at RT for 24 h. The prepared PEI-MNNS mixture was washed with PBS (pH 7.4) under a magnetic field, and suspended in PBS (pH 7.4). The conjugation of PEI with the MNNS was determined by the ninhydrin chromogenic method as previously described,38 and the details can be seen in the ESI.†
2) in MES buffer, and then magnetically stirred at RT for 24 h. The prepared PEI-MNNS mixture was washed with PBS (pH 7.4) under a magnetic field, and suspended in PBS (pH 7.4). The conjugation of PEI with the MNNS was determined by the ninhydrin chromogenic method as previously described,38 and the details can be seen in the ESI.†
2.5. Preparation of the EGF-targeted MNNS
The EGF-targeted MNNS were prepared according to the electrostatic adsorption principle. After 200 μg Heparin and 20 μg HB-EGF were added to 10 mg MNNS, the reaction was kept for 5 min at RT. The stability and magnetic response of the prepared MNNS and eMNNS in different media were analyzed, and more information is provided in the ESI.†2.6. DOX and siRNA loading onto eMNNS
After 1 mL of DOX solution (1 mg mL−1) was added into 1 mL of eMNNS suspension (1 mg mL−1), the mixture was stirred for 24 h in PBS (pH 7.4) at RT, and then the resulting DOX/eMNNS was washed with PBS buffer solution (pH 7.4) three times under a magnetic field. The loading degree of DOX was determined with the absorbance at 480 nm using a UV-Vis spectrophotometer (UV-1000, Shanghai, China). The drug loading was defined as the following equation:where Wt is the DOX weight in the NPs, Ws is the NP weight, and W0 is the total DOX weight used in the system.
For siRNA loading, eMNNS were diluted to a series of concentrations for desired N/P ratios and added to the 2 μL Sur siRNA solution (20 μmol L−1). The mixture was mixed gently and shaken for 15 min at 37 °C, allowing for sufficient binding of Sur siRNA molecules to the eMNNS. The binding ability of Sur siRNA to eMNNS was determined by ultraviolet-visible spectrophotometry and gel retardation assay. The formed Sur siRNA–eMNNS suspension was centrifuged, followed by magnetic separation. The supernatant was analyzed by UV-Vis spectrophotometer (Nano Drop 2000, thermo) at A260nm to reveal siRNA content. Meanwhile, the Sur siRNA–eMNNS were loaded onto a 2% agarose gel for electrophoresis in TAE buffer at a constant voltage of 45 V for 20 min, and then the agarose gel was stained with a 0.5 μg mL−1 ethidium bromide solution for 15 min to visualize the siRNA bands using a UV gel image system (InGenius LHR, SynGene, UK). The particle size of the Sur siRNA–eMNNS was characterized by a Malvern Zetasizer Nano ZS90.
2.7. Hemolysis assay
The hemocompatibility of the prepared eMNNS was assessed as previously reported.39 Briefly, 5 mL of fresh blood solution containing anticoagulant heparin sodium was centrifuged at 1000 rpm for 10 min to remove the supernatant. After that, the blood sample was washed with PBS (pH 7.4) solution three times to obtain the red blood cells (HRBCs). Then, 100 μL of HRBC suspension was diluted with 900 μL water (positive control group), 900 μL PBS (pH 7.4, negative control group), and 900 μL eMNNS solution at a concentration of 10–200 μg mL−1, respectively. After gently shaking, the mixture was reacted at RT for 3 h and then centrifuged for 1 min. The absorbance of the supernatant at 541 nm and a photograph of the mixture were recorded to measure the hemolysis of eMNNS.2.8. Cell cytotoxicity, cell viability, and cell cycle assay
The cytotoxicity of the prepared eMNNS/MNNS was evaluated by the MTT assay. We performed a standard MTT method to assess the proliferation capacity of the treated GSCs with different formulations. After reaching 200 cells per sphere, the GSCs were harvested to prepare a single cell suspension. The cells were then seeded at a density of 1 × 104 cells per well in a 96-well plate. When 10 cells per sphere were reached, different formulations were added to the wells. After incubation for 24 h, 48 h, 72 h and 96 h, the culture medium was removed, and 200 mL of MTT solution (0.5 mg mL−1; Sigma-Aldrich Co.) was added to each well. The cells were then incubated for 4 h, followed by the addition of 150 μL of DMSO and the incubation of the plates for another 15 min. Finally, the absorbance of the wells was measured at 570 nm on a microplate spectrophotometer (Bio Tek Instrument Inc., USA).The cell viability of the eMNNS-treated GSCs was measured with a fluorescein diacetate (FDA) and PI double-staining protocol.40 Briefly, the GSCs were seeded at a density of 2 × 104 cells per well in a 24-well plate. After reaching 10 cells per sphere, an eMNNS solution at a concentration of 20 μg mL−1 was added to the 24-well plate. Then, the treated GSCs were added to RPMI-1640 with 10% FBS after reaching 200 cells per sphere, and cultured for 1 d, 2 d, 3 d, 4 d, 5 d and 6 d. Subsequently, an FDA solution at a final concentration of 1 μg mL−1 and a PI solution at a final concentration of 20 μg mL−1 were successively introduced to the culture plates. Cell viability was then analyzed after incubation at RT for 5 min. The samples were analyzed with an inverted fluorescence microscope. The living cells were stained green by FDA, whereas the dead cells were stained red by fluorescent dye PI, using untreated GSCs as a control.
The cell cycle of the eMNNS-treated GSCs was analyzed by flow cytometry. The treated GSCs were harvested and dissociated into a single-cell suspension, and 1 × 106 cells were re-suspended in 500 μL of PBS buffer (pH 7.4). The cells were then fixed with 500 μL of 70% ice-cold ethanol and then incubated overnight at 4 °C. The prepared cell suspension was then centrifuged to discard the fixative, and then resuspended in 2 mL of PBS (pH 7.4) solution. The pelleted cells were resuspended in 1 mL of 200 μg mL−1 PI solution containing 20 μg mL−1 RNase for cell staining. The cells were incubated in the dark at 4 °C for 1 h and were analyzed with a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA), and analyzed with CELLQUEST software (BD Biosciences).
2.9. Fluorescent RBITC labeling and cellular uptake analysis
The prepared MNNS/eMNNS were labeled using RBITC with excellent fluorescence properties.41 The isothiocyanato group on RBITC can be directly bound to the amino group on the MNNS/eMNNS to produce fluorescently labeled nanocarriers. 10 mg MNNS/eMNNS was suspended in 1 mL boric acid buffer solution (pH 8.5), and then 100 μL RBITC solution (1 mg mL−1, dissolved in DMSO) was added into the suspension, and then kept on the shaker at RT in the dark for 2 h to prepare RBITC-labeled MNNS/eMNNS. The residual substance was washed three times using PBS under a magnetic field, and then the obtained RBITC-labeled eMNNS/MNNS were stored at 4 °C for future use.After GSCs were cultured for 72 h, 20 μg mL−1 RBITC-labeled eMNNS or MNNS were added to the cells for 4 h. After treatment, the GSCs were stained with Hochest 33258 (1 μg mL−1) for 15 min. The cells were washed three times using PBS (pH 7.4), and then the cells were observed with an inverted fluorescence microscope.
2.10. Assay of anti-proliferation capacity
An MTT assay was employed to assess the antiproliferative capacity of GSCs after treatment with different drug-loaded formulations. Briefly, 20 μg mL−1 of the various drug loading formulations was added to the microwell plates, which were seeded with GSCs, and then incubated for 24 h. 200 μL of the MTT solution (0.5 mg mL−1) was then added and incubated for 4 h, and 150 μL of dimethyl sulfoxide (DMSO) was successively added and incubated for another 15 min. Finally the absorbance of the sample was measured at 570 nm on a microplate spctrophotometer (Bio Tek Instrument Inc., USA).The cell viability of the treated GSCs with different drug formulations was assessed using the FDA/double-staining protocol. Briefly, 20 μg mL−1 of the different formulations was added to the microwell plates that were seeded with GSCs, followed by incubation for 24 h. 1 μg mL−1 of FDA solution and 20 μg mL−1 of PI solution were successively introduced to the culture plates. Cell viability was then analyzed after incubation at RT for 5 min. The samples were analyzed using an inverted fluorescence microscope.
The cell cycle of the treated GSCs was also analyzed using flow cytometry as mentioned above.
2.11. Apoptosis assay of the treated GSCs
Fluorescence Hoechst H33258 staining was performed to visualize apoptotic cells. Briefly, the treated GSCs were fixed with 4% paraformaldehyde for 15 min, washed three times with PBS (pH 7.4), and then stained with bisbenzimide dye Hoechst H33258 solution (2 μg mL−1) at RT for 10 min. The stained GSCs were then rinsed three times with PBS (pH 7.4) and observed using an inverted fluorescence microscope.Fluorescein Annexin V-FITC-PI double labeling was performed using the Annexin V-FITC Apoptosis Detection Kit (Keygen Biotech, Nanjing, China) to detect the apoptosis effect of different formulations on GSCs. The double labeling was conducted according to the instructions. 20 μg mL−1 of the various nanoparticle complex solutions was added to the microwell plates that were seeded with GSCs, followed by incubation for 24 h. The treated GSCs were harvested and washed using PBS. Subsequently, 1 × 106 cells were stained with 5 μL of Annexin V-FITC and 5 μL of PI in a 500 μL binding solution for 10 min at RT in the dark. The apoptotic cells were then detected by flow cytometry.
2.12. Autophagy assay
The volume of acidic vesicular organelles (AVOs), as a marker of autophagy, was detected by staining with lysosomotropic agent acridine orange (AO). The cytoplasm and nucleus of the stained cells emit bright green fluorescence, while AVO emits bright red fluorescence. The treated GSCs were stained with AO (5 μg mL−1) at 37 °C for 1 min and observed by an inverted fluorescence microscope.2.13. Western blotting
The treated GSCs were lysed in a buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM PMSF, and a protease inhibitor cocktail for 15 min on ice, and then heated at 100 °C in a metal bath for 5 min. The total protein extract (10 μg) was separated by 12% SDS-PAGE and transferred onto a PVDF membrane. The membrane was blocked with 8% (w/v) nonfat dry milk in PBS-Tween 20 (PBST; 0.05%) for 1 h and then incubated with primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 in PBST) at 4 °C overnight, and then incubated with the appropriate HRP-conjugated secondary antibodies (1
5000 in PBST) at 4 °C overnight, and then incubated with the appropriate HRP-conjugated secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 000) for 1 h at RT. The immunoreactive bands were developed with the Pierce ECL (Thermo Fisher Scientic, Waltham, MA, USA) western blotting system. The relative quantity of the proteins was analyzed using the Quantity one software.
10 000) for 1 h at RT. The immunoreactive bands were developed with the Pierce ECL (Thermo Fisher Scientic, Waltham, MA, USA) western blotting system. The relative quantity of the proteins was analyzed using the Quantity one software.
2.14. Orthotopicxenograft model and tumor suppression study
The xenograft tumor model was generated by subcutaneous injection of glioblastoma U251 (2 × 106 cells) into one of the flank regions of the mouse. When the tumor short diameter of the tumor xenograft reached approximately 5 mm, the nude mice were divided into 3 groups randomly (PBS, Sur siRNA–MNNS, DOX/Sur siRNA–eMNNS), and were subcutaneously injected with 52.3 μL of various nanoparticle complexes once every three days. The treatment duration was about 2 weeks. The doses of DOX and Sur siRNA were fixed at 500 μg kg−1 and 20 nmol kg−1 respectively. The tumor length (a) and minor diameter (b) were measured with a vernier caliper before each injection and detailed data were recorded. The tumor volume (V) was calculated based on the following equation:| V = ab2 × 0.5 |
After 2 weeks, all the nude mice were sacrificed, and their tumors were immediately collected, photographed, formalin fixed, paraffin embedded, and haematoxylin & eosin (H&E) stained for further histological analysis.
2.15. Characterization methods
A transmission electron microscope (TEM, JEM-2100, Japan) was employed to evaluate the morphology and size of the prepared SPIONPs and their composites. The crystal structures of SPIONPs and MNNS were analyzed with an X-ray diffractometer (XRD, Philips D/Max-2500, Holland) using a monochromatic X-ray beam with nickel-filtered CuKα radiation. Fourier transform infrared spectroscopy (FT-IR, Nicolet NEXUS 670, USA) was performed to record the FT-IR spectra of the SPIONPs and eMNNS. Magnetization measurements were carried out on a vibrating sample magnetometer (LAKESHORE-7304, USA) by changing H between +1375 and −1375 Oe. UV-Vis spectra were tested with a UV-1000 spectrophotometer (Shanghai, China). The hydrodynamic size of the samples was estimated by DLS using an instrument (Zeta Sizer Nano ZS90, Malvern, UK). The efficacy of the siRNA linked to eMNNS was evaluated by agarose gel electrophoresis. The cell cycle and cellular uptake were analyzed using a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA).2.16. Image acquisition and analysis
Fluorescence images were taken using an inverted fluorescence microscope (Eclipse TE 2000-U, Nikon, Kyoto, Japan) equipped with a high-resolution CCD camera (CV-S3200, JAI Co., Japan). Software Image-Pro Plus® 6.0 (Media Cyternetics) and SPSS 12.0 (SPSS Inc.) were used to perform image analysis and statistical analysis, respectively. The quantitative data are presented as means ± standard deviation (SD) for each experiment. All experiments were performed in triplicate and the results were derived from representative experiments.3. Results and discussion
3.1. Glioblastoma stem cell identification and EGFR protein expression assay
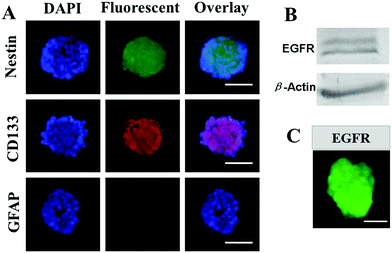
The primary GSC spheres were isolated from glioblastoma U251 cells cultured in serum-free medium supplemented with the anticancer drug vincristine, and the immunocytochemical staining showed that the formed GSCs could express biomarkers of brain cancer stem cells, NESTIN and CD133 (Fig. 1A). Meanwhile, the EGFR expression was also profiled by western blotting and immunocytochemical staining. The results demonstrated that GSCs expressed high levels of EGFR (Fig. 1B and C), indicating that EGFR could be used as a potential target in the therapeutic treatment of GSCs.3.2. Synthesis and characterization of eMNNS
In the present study, the preparation of eMNNS was performed mainly with three processes including: (1) synthesis of the SPIONP core (i.e. γ-Fe2O3) followed by TPP-mediated encapsulation of CMCS; (2) PEI coating via EDC/NHS crosslinking; and (3) conjugation of Heparin and EGF with MNNS to form eMMNS (Scheme 1a). | ||
| Scheme 1 (A) Schematic illustration of the fabrication of the EGFR-targeted theranostic nanoplatform, and (B) targeted co-delivery of therapeutic siRNAs and DOX for GSC treatments. | ||
The morphology and performance of the as-prepared nanoparticles were characterized. TEM analysis demonstrated that the prepared SPIONPs had a suitable particle size distribution, in which the particle size of a single nanoparticle was generally between 10 and 15 nm (Fig. 2A), and the complexes CMCS@SPIONPs also exhibited monodisperse spheres with a uniform particle size of about 40 to 50 nm (Fig. 2B). The XRD results indicated that six marked peaks with the indices (220), (311), (400), (422), (511), and (440) were specifically obtained for the γ-Fe2O3 NPs (Fig. S1, ESI†), which well matched the standard γ-Fe2O3 reflections. In addition, FT-IR spectroscopy was used to determine the composition of the prepared nanoparticles. As illustrated in Fig. 2C, the FT-IR data showed that the C–N stretching vibration of the amide group is at 1626.5 cm−1, the C–O bond vibration of the ether group is at 1087.3 cm−1, and the band of SPIONPs is located at 592.8 cm−1. The peak at 2935.8 cm−1 corresponds to the stretching vibration of methylene residual, and the characteristic band of COOH is reflected at 3420.5 cm−1. Taken together, these results indicate that the SPIONPs were prepared successfully, and could be modified with CMCS, PEI, and EGF respectively.
Fig. 2(D) illustrates that the magnetization curve of the prepared NPs exhibited a symmetrical hysteresis loop. Furthermore, the saturation of eMNNS was estimated at about 46.2 emu g−1, and its coercivity was not detectable. Practically, the data indicated that these prepared NPs become readily magnetized in the presence of a magnetic field, but the removal of the magnetic field would immediately cause a minimum of residual magnetization within the particles.42
The ninhydrin color reaction was used to determine the amount of PEI coupled with nanoparticles, as PEI had a large number of amino groups that react with ninhydrin to produce a substance with the maximum absorption wavelength at 570 nm. The results showed the PEI content as about 31.27% in MNNS, and 10.7% in the CMCS@SPIO (Fig. 3A and B), indicating that the prepared MNNS were successfully coupled with PEI.
 | ||
| Fig. 3 (A) The standard curve of the PEI, and (B) PEI content assay in MNNS. (C) Schematic of siRNA/eMNNS. (D) Particle size distribution of the as-prepared nanoparticles. | ||
As shown in Fig. 3D, the hydrodynamic diameter distribution of the prepared MNNS and eMNNS in an aqueous solution was measured at around 508 nm and 794 nm, and the hydrodynamic diameter of the siRNA/eMNNS complexes was narrow at around 798 nm, which confirmed that the EGF had been successfully coupled with MNNS, and siRNA was loaded onto eMNNS.
3.3. Magnetic responsiveness, stability and drug loading assay
The stability of eMNNS was determined at different time points in three different solutions including DI water, PBS (pH 7.4), and RPMI-1640 culture medium. The results showed that a relatively stable suspension was obtained in DI water and RPMI-1640 medium, but the presence of phosphate and sodium potassium salt probably increased sediments in the PBS buffer after about 15 min of incubation (Fig. S2, ESI†).In addition, aggregation and redispersion of eMNNS in solution was also evaluated under external magnetic fields and gravity conditions applied at different time points. The results showed that eMNNS had excellent responsiveness, and could be easily recycled under an external magnetic field (Fig. S2, ESI†). These results ensure that the prepared eMNNS is suitable for targeted anticancer drug delivery, multi-modal imaging, cancer targeted therapy, etc.
The loading capacity of the drug was measured, according to the optical density of the formed supernatant. The standard curve equation of DOX was Y = 0.00818 + 0.01012X (Fig. S3, ESI†). Finally, the DOX drug loading in DOX–eMNNS was estimated to be 3.8598% approximately.
3.4. Hemolysis assay of the eMNNS
Hemocompatibility is generally considered as an issue in blood-contacting application of nanoparticles.39 Hemolysis was assayed with the eMNNS before they were used as carriers for gene/drug delivery (Fig. 4). The absorbance of the supernatant was measured at 541 nm with the UV-Vis spectra to quantify the hemolysis percentage of the eMNNS at different concentrations (10–200 μg mL−1). The results showed that the eMNNS rarely exhibited hemolytic effects at the measured concentrations, compared with the positive control. The hemolysis percentage of eMNNS at the maximum concentration (200 μg mL−1) was only 1.04%, suggesting that eMNNS had excellent hemocompatibility within the study concentration range and could be used for further applications.3.5. Intracellular internalization
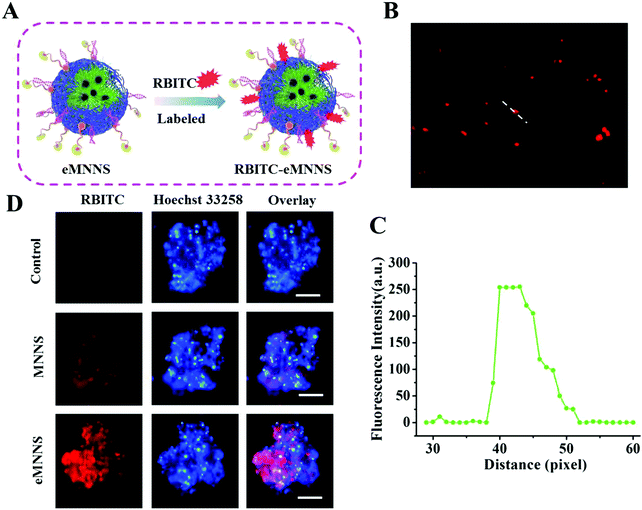
Effective transport into cells is one of the main factors affecting the transfection of siRNA and drugs. Intracellular uptake of eMNNS was performed by incubating the fluorescent RBITC-labeling eMNNS with GSCs for 4 h. The results showed that both MNPs and eMNNS could successfully enter the GSCs after about 4 h co-incubation, and eMNNS could enter the cells more easily and effectively because of the active targeting of HB-EGF, as they demonstrated much stronger red fluorescence (Fig. 5). These results suggested that eMNNS could be a potential vehicle for delivery of siRNA to GSCs.3.6. Cell cytotoxicity, cell viability, and cell cycle assay
The cytotoxicity of eMNNS was evaluated with an MTT assay to assess the cytocompatibility prior to biomedical applications. After 24 h of treatment, the viability of GSCs did not change significantly at all concentrations ranging from 0 to 50 μg mL−1 compared to the cells incubated with PBS (Fig. S4, ESI†). The GSCs were incubated with the eMNNS suspension at 20 μg mL−1 for 24–96 h. The results showed that after 96 h of treatment with eMNNS, the treated GSCs generally exhibited proliferative activity comparable to untreated cells. The MNNS had slight cytotoxicity due to the cationic polymer PEI layer (Fig. 6A). The MTT results implied that the eMNNS were cytocompatible at the studied concentration. Moreover, the cell viability and cell cycle of the GSCs exposed to eMNNS were detected by FDA/PI staining and flow cytometry, revealing similar results to those of the MTT assay (Fig. 6B). As shown in Fig. 6C and D, the flow cytometry analysis showed that with the treatment of MNNS and eMNNS, the number of GSCs and their differentiated progeny increased in the S phase, but the number of cells in the G1 phase decreased. Therefore, these data indicate that the fabricated eMNNS exhibited good cell biocompatibility and high proliferative capacity and could serve as a carrier for further siRNA/drug delivery to GSCs.3.7. Analysis of the ability and transfection efficiency of eMNNS binding to siRNA
In order to evaluate the binding ability of siRNA molecules to eMNNS, we performed quantitative RNA analysis using UV-Vis spectrophotometry and qualitative gel block analysis. As shown in Fig. 7A and B, the amount of bound siRNA molecules increased as more eMNNS was used, and 40 pmol siRNA can bind to 6 μg eMNNS. Furthermore, the negatively charged heparin affected the binding of siRNA molecules to eMNNS. The eMMNS mixed with 1/20 heparin clearly indicates a decrease in the amount of siRNA molecules, but the 1/50 heparin did not affect the binding of siRNA molecules to eMNNS, which was thus used in further analysis.Efficient delivery into cells is one of the major factors affecting the siRNA transfection.43 After 4 h co-incubation with different formulations containing green florescent siRNAFAM. The fluorescence intensity was dramatically enhanced for siRNAFAM nanocomplexes, because the cationic PEI could positively influence intracellular internalization of siRNA with easy access to the cell membrane. Obviously, the fabricated eMNNS was more potent to mediate the cellular uptake of siRNA than MNNS (Fig. 7C).
3.8. In vitro evaluation of anti-proliferation activity
To evaluate the anti-tumor efficacy of the targeted co-delivery of DOX and Sur siRNA in vitro, the proliferative activity of GSCs after DOX/Sur siRNA–eMNNS treatment was investigated by MTT assay after treatment with different formulations for 24 h, 48 h, 72 h, and 96 h. As in Fig. 8A, the MTT results indicated that targeted Sur siRNA–eMNNS could inhibit GSC proliferation more effectively compared with free Sur siRNA. When treated with targeted DOX/Sur siRNA–eMNNS, the cell proliferation of GSCs decreased significantly, indicating that the co-delivery of DOX Sur siRNA by targeted eMNNS had a synergistic effect to arrest cell proliferation.In addition, cell cycle distribution was examined by flow cytometry after treatment with different formulations. The results showed that an increasing number of GSCs were arrested in the S phase after treatment with Sur siRNA–eMNNS, and the co-delivery of DOX/Sur siRNA led to a much higher number of arrested GSCs in the S phase (Fig. 8B), demonstrating that DOX/Sur siRNA–eMNNS could arrest cell cycle progression during DNA synthesis and induce inhibition of cell proliferation.
Cell apoptosis is generally characterized with distinct biological alterations, including blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, chromosomal DNA fragmentation and so on.44,45 Thus, to observe nuclear fragmentation in induced apoptotic cells, GSC cells were stained with Hoechst H33258, a fluorescent dye that binds to the AT-rich region of DNA to detect and relatively quantify DNA from apoptotic cells.46,47 The results showed that the nuclei of GSCs treated with DOX/Sur siRNA–eMNNS were more concentrated and marginalized, with chromosome pyknosis and darker staining (Fig. 8C). AO is a fluorescent dye with cell permeability.48,49 The AO staining results also showed that targeted cells both treated with Sur siRNA–eMNNS and DOX/Sur siRNA–eMNNS became more permeable, thus showing strong red fluorescence, but much more acidic vesicles were detected in cells treated with DOX/Sur siRNA–eMNNS (Fig. 8D), indicating that the addition of DOX onto eMNNS could enhance cell autophagy in treated GSCs.
The cell viability of the treated GSCs was measured by FDA/PI double staining, and the results are shown in Fig. 9A. The red signal in the treated cells indicates that the cell activity was negatively affected. In general, cells treated with DOX/Sur siRNA–eMNNS showed stronger red fluorescence compared to Sur siRNA–eMNNS treatment, indicating that the combined use of DOX and Sur siRNA significantly inhibited cell viability.
Annexin V is a Ca2+ dependent phospholipid binding protein with high affinity to phosphatidylserine (PS) located in the medial membrane of the living cells.50,51 When the cellular apoptosis occurred early, PS valgus could specifically bind to the Annexin V in apoptotic cells. As calculated in Fig. 9B, the apoptotic cell proportion of GSCs treated with Sur siRNA–MNNS reached 13.36%, but the co-delivery of Sur siRNA and DOX produced a much higher apoptosis-inducing effect (apoptosis rate ∼36.95%), which obviously showed the synergistic effects on induction of cell apoptosis.
3.9. Western blotting
Apoptosis is a genetically controlled process, in which Bcl-2 plays a key role. Target genes expressed in cells and tissues were obtained by using antigen–antibody hybridization. Western blot showed that both Bcl-2 and survivin were obviously down-regulated in the cells treated with DOX/Sur siRNA–eMNNS, but Bax was reversely up-regulated after treatment (Fig. 10). Moreover, the expression level of LC3-II increased, while the level of p62 decreased after treatment. These results showed that co-delivery of DOX/Sur siRNA via eMNNS enabled silencing of the survivin gene, making GSCs more sensitive to DOX, and thereby more efficiently inducing cell apoptosis and autophagy in treated GSCs.3.10. Antitumor efficacy of DOX/Sur siRNA–eMNNS in vivo
The above in vitro analyses demonstrated that the growth of GSCs was efficiently inhibited by co-delivery of siRNA and DOX via eMNNS, which led us to in vivo tumor treatments. The xenograft tumor model in BALB/c nude mice was thus established to assess the tumor-suppressing effect of DOX/Sur siRNA–eMNNS in vivo. Nude mice bearing glioblastoma U251 cell-induced tumors were randomly divided into three groups and administrated with different formulations to understand the safety and efficacy of the eMNNS for cancer therapy. The tumor size of the mice treated with Sur siRNA–MNNS, decreased compared to the control group, and the mice treated with DOX/Sur siRNA–eMNNS exhibited the smallest tumor size and the best anti-tumor capacity (Fig. 11A–C). Moreover, the histological analyses (H&E stain) revealed extensive necrosis areas within the tumor mass treated with DOX/Sur siRNA–eMNNS (Fig. 11D). These results demonstrated that the co-delivery of DOX/Sur siRNA via eMNNS could synergically improve anti-tumor capacity in vivo.4. Conclusions
In this study, we have developed an active EGFR-targeted nano-scale eMNNS theranostic platform based on SPIONPs for a co-delivery gene/drug complex. Our experimental results show that EGFR-targeted eMNNS were selectively taken up by GSCs expressing EGFR, and the specificity, sensitivity and efficiency were significantly increased, indicating that this is an ideal vector for gene/drug delivery transport into cells. In addition, the combination of the therapeutic drugs Sur siRNA and DOX with eMNNS had a marked synergistic anti-tumor effect by inducing apoptosis and autophagy, suggesting that the combination therapy was more effective than mono therapy in tumor treatment. In conclusion, the prepared EGFR-conjugated MNNS is highly specific and can be used as a promising gene/drug delivery therapeutic platform for the treatment of human glioblastoma in the future.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31400855), China Postdoctoral Science Foundation (No. 2019M652541), Innovative Research Team from University of Henan Province (No. 19IRTSTHN008), the Scientific and Technological Project of Henan Province (No. 182102210394), the Young Core Instructor Program in Higher Education Institution of Henan province (No. 2018GGJS067), and the Young Core Instructor Program from Henan University of Technology (No. 21420055).References
- P. Y. Wen and D. A. Reardon, Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment, Nat. Rev. Neurol., 2016, 12, 69–70 CrossRef CAS PubMed.
- N. Sanai and M. S. Berger, Surgical oncology for gliomas: the state of the art, Nat. Rev. Clin. Oncol., 2017, 15, 112–125 CrossRef PubMed.
- H. Fan, H. Guo, I. Y. Zhang, B. Liu, L. Luan, S. Xu, X. Hou, W. Liu, R. Zhang, X. Wang and Q. Pang, The different HMGA1 expression of total population of glioblastoma cell line U251 and glioma stem cells isolated from U251, Brain Res., 2011, 1384(15), 9–14 CrossRef CAS PubMed.
- X. Wang, B. C. Prager, Q. Wu, L. J. Y. Kim, R. C. Gimple, Y. Shi, K. Yang, A. R. Morton, W. Zhou and Z. Zhu, et al., Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression, Cell Stem Cell, 2018, 22(4), 514–528 CrossRef CAS PubMed.
- Y. Liu, Q. Guo, H. Zhang, G. H. Li, S. Feng, X. Z. Yu, L. S. Kong, L. Zhao and F. Jin, Effect of siRNA-Livin on drug resistance to chemotherapy in glioma U251 cells and CD133+ stem cells, Exp. Ther. Med., 2015, 10(4), 1317–1323 CrossRef CAS PubMed.
- G. Xi, E. Hayes, R. Lewis, S. Ichi, B. Mania-Farnell, K. Shim, T. Takao, E. Allender, C. S. Mayanil and T. Tomita, CD133 and DNA-PK regulate MDR1 via the PI3K- or Akt-NF-κB pathway in multidrug-resistant glioblastoma cells in vitro, Oncogene, 2016, 35(2), 241–250 CrossRef CAS PubMed.
- K. B. Pointer, P. A. Clark, M. Zorniak, B. M. Alrfaei and J. S. Kuo, Glioblastoma cancer stem cells: Biomarker and therapeutic advances, Neurochem. Int., 2014, 71, 1–7 CrossRef CAS PubMed.
- J. M. Heffernan, J. B. McNamara, S. Borwege, B. L. Vernon, N. Sanai, S. Mehta and R. W. Sirianni, PNIPAAm-co-Jeffamine® (PNJ) scaffolds as in vitro models for niche enrichment of glioblastoma stem-like cells, Biomaterials, 2017, 143, 149–158 CrossRef CAS PubMed.
- I. Nakano, Stem cell signature in glioblastoma: therapeutic development for a moving target, J. Neurosurg., 2015, 122(2), 324–330 Search PubMed.
- D. A. Spencer, B. M. Auffinger, J. P. Murphy, M. E. Muroski, J. Qiao, Y. Gorind and M. S. Lesniak, Hitting a moving target: gliomastem cells demand new approaches in glioblastoma therapy, Curr. Cancer Drug Targets, 2017, 17(3), 236–254 CrossRef CAS PubMed.
- A. Sattiraju, K. K. S. Sai and A. Mintz, Glioblastomastem cells and their microenvironment, Adv. Exp. Med. Biol., 2017, 1041, 119–140 CrossRef CAS PubMed.
- A. L. Marisetty, S. K. Singh, T. N. Nguyen, C. Coarfa, B. Liu and S. Majumder, REST represses miR-124 and miR-203 to regulate distinct oncogenic properties of glioblastoma stem cells, Neuro-Oncology, 2017, 19(4), 514–523 CAS.
- A. Junca, C. Villalva, G. Tachon, P. Rivet, U. Cortes, K. Guilloteau, A. Balbous, J. Godet, M. Wager and L. Karayan-Tapon, Crizotinib targets in glioblastoma stem cells, Cancer Med., 2017, 6(11), 2625–2634 CrossRef CAS PubMed.
- H. Garg, P. Suri, J. C. Gupta, G. P. Talwar and S. Dubey, Survivin: a unique target for tumor therapy, Cancer Cell Int., 2016, 16, 49 CrossRef PubMed.
- D. Li, C. Hu and H. Li, Survivin as a novel target protein for reducing the proliferation of cancer cells, Biomed. Rep., 2018, 8(5), 399–406 CAS.
- P. Wang, H. Zhen, J. Zhang, W. Zhang, R. Zhang, X. Cheng, G. Guo, X. Mao, J. Wang and X. Zhang, Survivin promotes glioma angiogenesis through vascular endothelial growth factor and basic fibroblast growth factor in vitro and in vivo, Mol. Carcinog., 2012, 51(7), 586–595 CrossRef CAS PubMed.
- F. Zhang, J. Chu and F. Wang, Expression and clinical significance of cyclooxygenase 2 and survivin in human gliomas, Oncol. Lett., 2017, 14(2), 1303–1308 CrossRef PubMed.
- S. Hendruschk, R. Wiedemuth, A. Aigner, K. Töpfer, M. Cartellieri, D. Martin, M. Kirsch, C. Ikonomidou, G. Schackert and A. Temme, RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo, Neuro-Oncology, 2011, 13(10), 1074–1089 CrossRef CAS PubMed.
- R. Q. Cruz, C. M. Morais, A. M. Cardoso, S. G. Silva, M. L. Vale, E. F. Marques, M. C. Pedroso de Lima and A. S. Jurado, Enhancing glioblastoma cell sensitivity to chemotherapeutics: A strategy involving survivin gene silencing mediated by gemini surfactant-based complexes, Eur. J. Pharm. Biopharm., 2016, 104, 7–18 CrossRef CAS PubMed.
- X. Wang, L. Zhu, X. Hou, L. Wang and S. Yin, Polyethylenimine mediated magnetic nanoparticles for combined intracellular imaging, siRNA delivery and anti-tumor therapy, RSC Adv., 2015, 5(123), 101569 RSC.
- Y. Jia, D. Niu, Q. Li, H. Huang, X. Li, K. Li, L. Li, C. Zhang, H. Zheng, Z. Zhu, Y. Yao, X. Zhao, P. Li and G. Yang, Effective gene delivery of shBMP-9 using polyethyleneimine-based core-shell nanoparticles in an animal model of insulin resistance, Nanoscale, 2019, 11(4), 2008–2016 RSC.
- Y. Zhao, R. J. Lee, L. Liu, S. Dong, J. Zhang, Y. Zhang, Y. Yao, J. Lu, Q. Meng, J. Xie and L. Teng, Multifunctional drug carrier based on PEI derivatives loaded with small interfering RNA for therapy of liver cancer, Int. J. Pharm., 2019, 564, 214–224 CrossRef CAS PubMed.
- M. Wojnilowicz, A. Glab, A. Bertucci, F. Caruso and F. Cavalieri, Super-resolution imaging of proton sponge-triggered rupture of endosomes and cytosolic release of small interfering RNA, ACS Nano, 2019, 13(1), 187–202 CrossRef CAS PubMed.
- Y. Guo, X. Y. Wang, Y. L. Chen, F. Q. Liu, M. X. Tan, M. Ao, J. H. Yu, H. T. Ran and Z. X. Wang, A light-controllable specific drug delivery nanoplatform for targeted bimodal imaging-guided photothermal/chemo synergistic cancer therapy, Acta Biomater., 2018, 80, 308–326 CrossRef CAS PubMed.
- Q. Liu, L. Song, S. Chen, J. Gao, P. Zhao and J. Du, A superparamagnetic polymersome with extremely high T2 relaxivity for MRI and cancer-targeted drug delivery, Biomaterials, 2017, 114, 23–33 CrossRef CAS PubMed.
- B. Wang, W. Wu, H. Lu, Z. Wang and H. Xin, Enhanced anti-tumor of pep-1 modified superparamagnetic iron oxide/PTX loaded polymer nanoparticles, Front. Pharmacol., 2019, 9, 1556 CrossRef PubMed.
- C. Fan, W. Gao, Z. Chen, H. Fan, M. Li, F. Deng and Z. Chen, Tumor selectivity of stealth multi-functionalized superparamagnetic iron oxide nanoparticles, Int. J. Pharm., 2011, 404(1–2), 180–190 CrossRef CAS PubMed.
- C. Janko, T. Ratschker, K. Nguyen, L. Zschiesche, R. Tietze, S. Lyer and C. Alexiou, Functionalized superparamagneticiron oxide nanoparticles (SPIONs) as platform for the targeted multimodal tumor therapy, Front Oncol., 2019, 9, 59 CrossRef PubMed.
- K. J. Hatanpaa, S. Burma, D. Zhao and A. A. Habib, Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance, Neoplasia, 2010, 12(9), 675–684 CrossRef CAS PubMed.
- S. R. Ho and W. C. Lin, RNF144A sustains EGFR signaling to promote EGF-dependent cell proliferation, J. Biol. Chem., 2018, 293(42), 16307–16323 CrossRef CAS PubMed.
- T. Hide, T. Takezaki, Y. Nakatani, H. Nakamura, J. Kuratsu and T. Kondo, Combination of a ptgs2 inhibitor and an epidermal growth factor receptor-signaling inhibitor prevents tumorigenesis of oligodendrocyte lineage-derived glioma-initiating cells, Stem Cells, 2011, 29(4), 590–599 CrossRef CAS PubMed.
- C. von Achenbach, M. Weller and E. Szabo, Epidermal growth factor receptor and ligand family expression and activity in glioblastoma, J. Neurochem., 2018, 147(1), 99–109 CrossRef CAS PubMed.
- X. Jiang, H. Xing, T. M. Kim, Y. Jung, W. Huang, H. W. Yang, S. Song, P. J. Park, R. S. Carroll and M. D. Johnson, Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity, Stem Cells, 2012, 30(7), 1313–1326 CrossRef CAS PubMed.
- X. Wang, F. Wei, A. Liu, L. Wang, J. Wang, L. Ren, W. Liu, Q. Tu, L. Li and J. Wang, Cancer stem cell labeling using poly(L-lysine)-modified iron oxide nanoparticles, Biomaterials, 2012, 33(14), 3719–3732 CrossRef CAS PubMed.
- S. Qu, H. Yang, D. Ren, S. Kan, G. Zou, D. Li and M. Li, Magnetite nanoparticles prepared by precipitation from partially reduced ferric chloride aqueous solutions, J. Colloid Interface Sci., 1999, 215, 190–192 CrossRef CAS PubMed.
- Y. K. Sun, M. Ma, Y. Zhang and N. Gu, Synthesis of nanometer-size maghemite particles from magnetite, Colloids Surf., A, 2004, 245, 15–19 CrossRef CAS.
- X. Wang, B. Li, R. Li, Y. Yang, H. Zhang, B. Tian, L. Cui, H. Weng and F. Wei, Anti-CD133 monoclonal antibody conjugated immunomagnetic nanosensor for molecular imaging of targeted cancer stem cells, Sens. Actuators, B, 2018, 255, 3447–3457 CrossRef CAS.
- M. Friedman, Applications of the ninhydrin reaction for analysis of amino acids, peptides, and proteins to agricultural and biomedical sciences, J. Agric. Food Chem., 2004, 52, 385–406 CrossRef CAS PubMed.
- W. Hou, F. Xia, G. Alfranca, H. Yan, X. Zhi, Y. Liu, C. Peng, C. Zhang, J. M. de la Fuente and D. Cui, Nanoparticles for multi-modality cancer diagnosis: Simple protocol for self-assembly of gold nanoclusters mediated by gadolinium ions, Biomaterials, 2017, 120, 103–114 CrossRef CAS PubMed.
- B. W. Kristensen, H. Noer, J. B. Gramsbergen, J. Zimmer and J. Noraberg, Colchicine induces apoptosis in organotypic hippocampal slice cultures, Brain Res., 2003, 964, 264–278 CrossRef CAS PubMed.
- X. Wang, Y. Chang, D. Zhang, B. Tian, Y. Yang and F. Wei, Transferrin-conjugated drug/dye-co-encapsulated magnetic nanocarriers for active-targeting fluorescent/magnetic resonance imaging and anti-tumor effects in human brain tumor cells, RSC Adv., 2016, 6, 105661 RSC.
- J. Wang, X. Wang, L. Ren, Q. Wang, L. Lix, W. Liu, Z. Wan, L. Yang, P. Sun, L. Ren, M. Li, H. Wu, J. Wang and L. Zhang, Conjugation of biomolecules with magnetic protein microspheres for the assay of early biomarkers associated with acute myocardial infarction, Anal. Chem., 2009, 81, 6210–6217 CrossRef CAS PubMed.
- H. Z. Jia, W. Zhang, J. Y. Zhu, B. Yang, S. Chen, G. Chen, Y. F. Zhao, J. Feng and X. Z. Zhang, Hyperbranched-hyperbranched polymeric nanoassembly to mediate controllable co-delivery of siRNA and drug for synergistic tumor therapy, J. Controlled Release, 2015, 216, 9–17 CrossRef CAS PubMed.
- C. P. Andreatta, P. Nahreini, A. J. Hanson and K. N. Prasad, Regulated expression of VP16CREB in neuroblastoma cells: analysis of differentiation and apoptosis, J. Neurosci. Res., 2004, 78, 570–579 CrossRef CAS PubMed.
- G. Kiraly, A. S. Simonyi, M. Turani, I. Juhasz, G. Nagy and G. Banfalvi, Micronucleus formation during chromatin condensation and under apoptotic conditions, Apoptosis, 2017, 22(2), 207–219 CrossRef CAS PubMed.
- P. R. Kamath, D. Sunil, M. M. Joseph and A. A. Abdul, Indole-coumarin-thiadiazole hybrids: an appraisal of their MCF-7 cell growth inhibition, apoptotic, antimetastatic and computational Bcl-2 binding potential, Eur. J. Med. Chem., 2017, 136, 442–451 CrossRef CAS PubMed.
- K. Chamaon, J. Stojek, D. Kanakis, S. Braeuninger, E. Kirches, G. Krause, C. Mawrin and K. Dietzmann, Micromolar concentrations of 2-methoxyestradiol kill glioma cells by an apoptotic mechanism, without destroying their microtubule cytoskeleton, J. Neurooncol., 2005, 72(1), 11–16 CrossRef CAS PubMed.
- A. M. Leopoldino, C. H. Squarize, C. B. Garcia, L. O. Almeida, C. R. Pestana, L. M. Sobral, S. A. Uyemura, E. H. Tajara, J. Silvio Gutkind and C. Curti, SET protein accumulates in HNSCC and contributes to cell survival: antioxidant defense, Akt phosphorylation and AVOs acidification, Oral Oncol., 2012, 48(11), 1106–1113 CrossRef CAS PubMed.
- Y. Li, L. Zhang, J. Zhou, S. Luo, R. Huang, C. Zhao and A. Diao, Nedd4 E3 ubiquitin ligase promotes cell proliferation and autophagy, Cell Prolif., 2015, 48(3), 338–347 CrossRef CAS PubMed.
- I. Vermes, C. Haanen, H. Steffens-Nakken and C. Reutelingsperger, A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labeled Annexin V, J. Immunol. Methods, 1995, 184(1), 39–51 CrossRef CAS PubMed.
- S. E. Logue, M. Elgendy and S. J. Martin, Expression, purification and use of recombinant annexin V for the detection of apoptotic cells, Nat. Protoc., 2009, 4(9), 1383–1395 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9qm00666d |
| This journal is © the Partner Organisations 2020 |