 Open Access Article
Open Access ArticleHigh-order structures from nucleic acids for biomedical applications
Alyssa C.
Hill
 * and
Jonathan
Hall
* and
Jonathan
Hall
 *
*
Institute of Pharmaceutical Sciences, Department of Chemistry and Applied Biosciences, ETH Zürich, Switzerland. E-mail: alyssa.hill@pharma.ethz.ch; jonathan.hall@pharma.ethz.ch
First published on 6th February 2020
Abstract
Over the past 40 years, research in the fields of DNA nanotechnology and RNA nanotechnology has taken nucleic acid molecules out of their biological contexts and harnessed their unique base-pairing and self-assembly properties to generate well-defined, organized, and functional supramolecular architectures. Capitalizing on an intrinsic biocompatibility and the ability to tailor size, shape, and functionality from the bottom up, recent work has positioned high-order nucleic acid structures as powerful biomedical tools. This review summarizes advances in nanotechnology that have enabled the fabrication of synthetic nucleic acid structures. Nucleic acid-based platforms for biosensing and therapeutic drug delivery are highlighted. Finally, an outlook that considers the limitations and future challenges for this field is presented.
Advances in DNA nanotechnology
DNA junctions
Deoxyribonucleic acid (DNA) is well known as the macromolecule that encodes genetic information. Taken out of its biological context, DNA is also an attractive material for bottom-up fabrication. First, the composition of DNA is known. DNA sequences are made up of four nucleotides: adenine, cytosine, guanine, and thymine (A, C, G, and T, respectively). Second, DNA participates in some of the most predictable interactions of any natural or synthetic molecule.1 Indeed, DNA sequences form hydrogen bonds according to Watson–Crick base pairing rules (i.e., C pairs with G and A pairs with T), and these interactions confer on DNA the capacity for precise molecular recognition and programmable self-assembly.1,2 Finally, the structure of DNA is defined at the nanometer (nm) scale: DNA helices adopt B-form geometry, with 10.5 base pairs (bp) per turn, a diameter of 2 nm, a helical pitch of 3.4 nm, and a persistence length of 50 nm.3 However, in nature, DNA exists predominantly as a duplex with a linear helical axis, which is poorly suited for fabrication in three dimensions (3D).In 1982, Nadrian Seeman conceived of using branched DNA molecules, or junctions, to assemble DNA structures in 2- and 3D.4 Seeman's inspiration was the Holliday structure, a mobile intermediate of genetic recombination that consists of a single strand exchange, or crossover, between two DNA duplexes. In a seminal publication, Seeman proposed rules for constructing immobile DNA junctions from multiple strands and suggested fitting the junctions with ‘sticky ends’.4 Sticky ends are single-stranded overhangs, and cohesion between sticky ends in DNA generates a helix with standard B-form local geometry.5 By programming DNA junctions to self-assemble via sticky-ended cohesion (Fig. 1A), Seeman imagined the creation of extended DNA arrays, including 2D lattices and 3D crystals.4 Seeman's vision was that DNA crystals with embedded recognition motifs could be used as hosts to organize proteins and other macromolecule guests for structure determination by X-ray crystallography (Fig. 1B).4 In the years following Seeman's innovative proposal, a variety of DNA structures were created, including multi-way junctions,6,7 geometric shapes,8 knots9,10 (Fig. 1C), Borromean rings11 (Fig. 1D), and polyhedra12,13 (Fig. 1E and F), and thus the field of structural DNA nanotechnology was established.
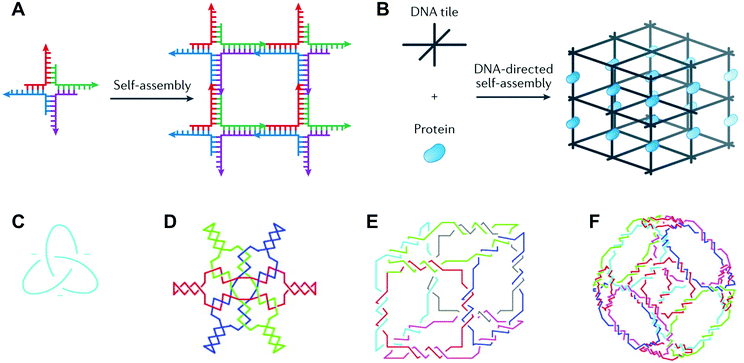 | ||
| Fig. 1 DNA junctions as units of assembly in extended DNA arrays and discrete DNA topologies. (A) A DNA four-way junction with sticky ends self-assembles into a 2D DNA lattice. (B) A 3D DNA crystal (black) scaffolds proteins (blue) for structure determination by X-ray crystallography. (C) A DNA trefoil knot. (D) Borromean rings made of DNA. (E) A DNA cube. (F) A DNA octahedron. Panels (A) and (B) reprinted from ref. 1 by permission from Springer Nature: Nature Reviews Materials. Copyright 2017. Panels (C–F) reprinted from ref. 14 by permission from Elsevier: Trends in Biotechnology. Copyright 1999. | ||
DNA tiles
While early work in the field of DNA nanotechnology demonstrated that target topologies could be generated by manipulating flexible junctions, the creation of specific 2- and 3D geometries hinged on the development of more rigid motifs.15 To this end, a variety of DNA ‘tiles’ with high structural integrity have been developed.16 One notable example is the double-crossover (DX) tile.17 In contrast to the Holliday junction, which features one crossover between two DNA duplexes, the DX tile features two crossovers (Fig. 2A). Accordingly, the DX tile has a stiffness that is twice that of linear, double-stranded DNA.18 DX tiles with sticky ends have been shown to self-assemble into periodic19 and aperiodic20 2D lattices (Fig. 2A). Further research has produced triple crossover (TX)21 and paranemic crossover (PX)22 tiles, multi-point stars23–25 (Fig. 2B–D), double-decker tiles26 (Fig. 2E), T-junctions27 (Fig. 2F), Wang tiles,28 and other tiles for assembling DNA lattices of varying patterns and periodicities, 3D DNA objects with controlled sizes29 (Fig. 2G), and even DNA-based nano-mechanical devices.30,31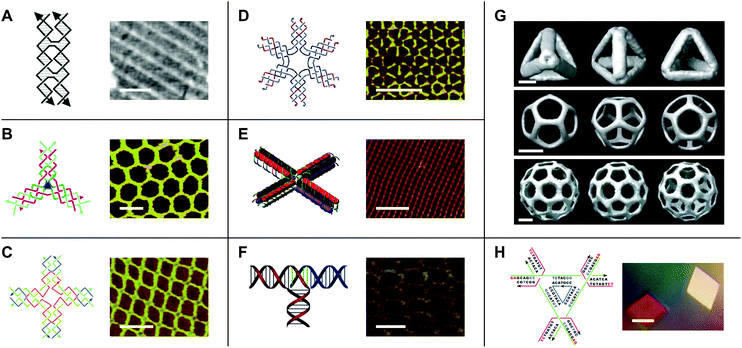 | ||
| Fig. 2 DNA tiles and tile-based structures. (A) Double-crossover (DX) tile (left) and a periodic DNA lattice self-assembled from the DX tile (right). Scale bar: 150 nm. (B) Three-point star motif (left) and a periodic DNA lattice self-assembled from the three-point star motif (right). Scale bar: 50 nm. (C) Four-point star motif (left) and a periodic DNA lattice self-assembled from the four-point star motif (right). Scale bar: 50 nm. (D) Six-point star motif (left) and a periodic DNA lattice self-assembled from the six-point star motif (right). Scale bar: 50 nm. (E) Double-decker tile (left) and a periodic DNA lattice self-assembled from the double-decker tile (right). Scale bar: 200 nm. (F) T-junction (left) and a periodic DNA lattice self-assembled from the T-junction (right). Scale bar: 25 nm. (G) Cryo-electron microscopy (cryo-EM) reconstructed models of DNA polyhedra self-assembled from the three-point star motif. Top panel: DNA tetrahedron, middle panel: DNA dodecahedron, bottom panel: DNA buckyball. Scale bars: 5 nm, 20 nm, and 20 nm, respectively. (H) Tensegrity triangle (left) and DNA crystals self-assembled from the tensegrity triangle (right). Scale bar: 500 µm. Panel (A) tile reprinted from ref. 41 by permission from Springer Nature: Methods in Molecular Biology. Copyright 2005. Panel (A) lattice reprinted from ref. 19 by permission from Springer Nature: Nature. Copyright 1998. Panel (B) reprinted with permission from ref. 24. Copyright 2005 American Chemical Society. Panel (C) from ref. 23. Reprinted with permission from AAAS. Panel (D) reprinted with permission from ref. 25. Copyright 2006 American Chemical Society. Panel (E) reprinted with permission from ref. 26. Copyright 2011 American Chemical Society. Panel (F) reprinted with permission from ref. 27. Copyright 2009 John Wiley and Sons. Panel (G) reprinted from ref. 29 by permission from Springer Nature: Nature. Copyright 2008. Panel (H) reprinted from ref. 33 by permission from Springer Nature: Nature. Copyright 2009. | ||
Additionally, in 2009, Seeman's group used a tile known as the tensegrity triangle to produce the first rationally designed, self-assembled DNA crystal.32,33 In the tensegrity triangle, seven oligonucleotide strands come together to form a structure comprising three struts and three four-way junction vertices, while two-nucleotide sticky ends mediate self-assembly in 3D (Fig. 2H).32,33 The development of self-assembling DNA crystals constituted an important step toward realizing Seeman's aforementioned vision for the field of DNA nanotechnology. However, the crystals diffracted to only 4 Å resolution.33 Similarly, self-assembling DNA crystals designed to contain two tensegrity triangles per asymmetric unit diffracted to only 5 Å resolution.34 In recent years, improvements to resolution and crystal stability have been made by incorporating 5′ terminal phosphates into certain component DNA strands,35 by changing the length36 and composition37 of the sticky ends, and by introducing additional stabilizing DNA strands,38 crosslinks,39 and covalent bonds.40 However, further improvements to resolution will be necessary in order to maximize the utility of DNA crystals as tools for macromolecular structure determination by X-ray crystallography.
DNA origami
In a 2003 publication, Yan et al. described the creation of DNA lattices from DX tiles self-assembled around long (∼300 nucleotide; nt) strands of DNA.20 The following year, Shih et al. reported a DNA octahedron self-assembled from a 1.7 kilobase (kb) strand of DNA and five 40-nt strands.42 Inspired by these advances, in 2006 Paul Rothemund generalized the approach into a method called DNA origami, which consists of folding a large ‘scaffold’ strand of DNA with many short ‘staple’ strands in order to generate defined shapes of arbitrary complexity.43 Specifically, designs are created by raster-filling a shape with the scaffold strand and using staple strands that base pair to the scaffold to hold it in place (Fig. 3A).43 In a now-famous publication, Rothemund folded the ∼7 kb M13 bacteriophage genome with over 200 32-nt DNA strands into rectangles, stars, and smiley faces (Fig. 3B).43 Rothemund's technique revolutionized the field by achieving high yields of the designed structures while simultaneously avoiding requirements for the purification of component strands, multiple assembly steps, and exact stoichiometries.43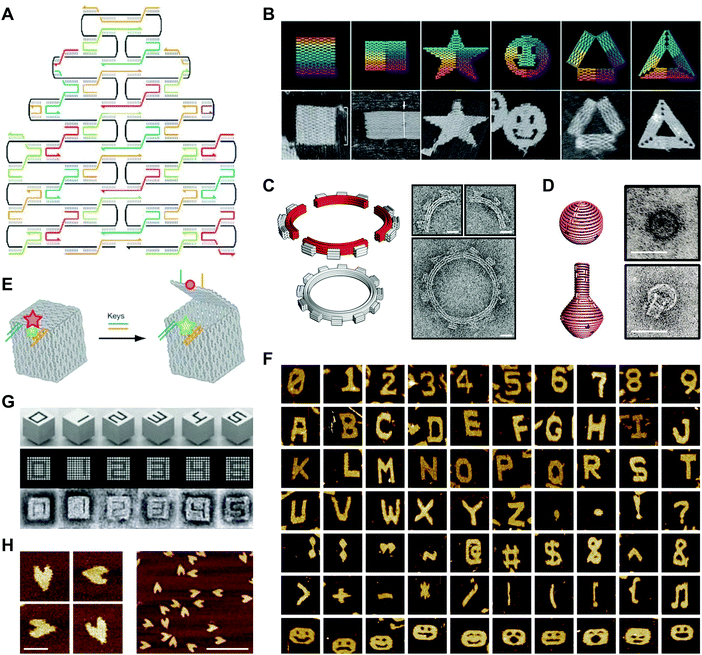 | ||
| Fig. 3 2- and 3D DNA origami shapes. (A) Scaffolded DNA origami: the folding of a long scaffold strand of DNA (black) into a defined 2D shape is accomplished with the help of short staple strands (colors). (B) Complex 2D shapes folded using scaffolded DNA origami. Images are 165 nm × 165 nm. (C) Schematic representation (left) and transmission electron microscopy (TEM) images (right) of a DNA square-tooth gear. Scale bars: 20 nm. (D) Schematic representations of a DNA sphere (top left) and flask (bottom left) and TEM images of the sphere (top right) and flask (bottom right). Scale bars: 50 nm. (E) Illustration of a DNA origami box with a controllable lid. (F) Complex 2D shapes self-assembled from single-stranded DNA tiles. Images are 150 nm × 150 nm. (G) 3D DNA shapes self-assembled from DNA bricks. Each shape is 25 × 25 × 27 nm. (H) Atomic force microscopy (AFM) images of single-stranded DNA origami hearts. Scale bars: left panel, 50 nm; right panel, 200 nm. Panels (A) and (B) reprinted from ref. 43 by permission from Springer Nature: Nature. Copyright 2006. Panel (C) from ref. 46. Reprinted with permission from AAAS. Panel (D) from ref. 47. Reprinted with permission from AAAS. Panel (E) reprinted from ref. 48 by permission from Springer Nature: Nature. Copyright 2009. Panel (F) reprinted from ref. 49 by permission from Springer Nature: Nature. Copyright 2012. Panel (G) from ref. 50. Reprinted with permission from AAAS. Panel (H) from ref. 53. Reprinted with permission from AAAS. | ||
Follow-on research extended Rothemund's design principles into 3D with impressive results. Using DNA origami, Ke et al. created DNA cages,44 and Douglas et al. created DNA monoliths, bridges, and crosses.45 Other studies generated 3D structures that twist and curve at the nano-scale. For example, Dietz et al. produced DNA beach balls and square-tooth gears46 (Fig. 3C), and Han et al. produced DNA spheres, shells, and flasks (Fig. 3D).47 A particularly notable example of 3D DNA origami was reported in a publication by Andersen et al., who used the M13 bacteriophage genome as a scaffold to create fully addressable, self-assembling DNA boxes.48 Each box measured 42 × 36 × 36 nm and was folded from six interconnected sheets of DNA with 220 staple strands bridging the edges.48 Furthermore, by programming the lids of these boxes with DNA ‘locks’, the authors showed that they could control their opening with externally supplied DNA ‘keys’ (Fig. 3E).48 In this case, the locks comprised DNA duplexes with sticky ends that facilitated toehold-mediated strand displacement by auxiliary DNA strands.
Variations on DNA origami
More recent work has produced variations on the DNA origami technique. For example, Wei et al. developed a 42-nt single-stranded DNA tile that self-assembles into complex 2D patterns, including alphanumeric characters, punctuation marks, and smiley faces, without the need for a scaffold strand (Fig. 3F).49 A recent publication from the same group showed that 32-nt single-stranded DNA tiles, or ‘bricks’, self-assemble into prescribed 3D shapes with up to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 unique components (Fig. 3G).50,51 Another recent study using DNA bricks generated polyhedral assemblies with atomic masses up to 1.2 gigadaltons and long, thick tubes similar in size to some bacilli.52 Because they do not require a scaffold strand, these approaches contrast with Rothemund's ‘scaffolded’ DNA origami. A separate approach is single-stranded origami, which folds multi-kilobase nucleic acid strands into complex 2D patterns without using staple strands. In one example, Han et al. folded hearts from a single strand of DNA approximately 3000 nt in length (Fig. 3H).53 Another recent example described the combination of single-stranded origami with DNA tiles to fold highly knotted 2- and 3D topologies.54 Since the advent of DNA origami, semi-automated55,56 and fully-automated57,58 approaches for producing target 2- and 3D geometries have emerged. Today, the ease of the origami technique, combined with the commercial availability of chemically synthesized DNA sequences,59 makes the design and fabrication of complex DNA structures accessible even to non-specialists.1
000 unique components (Fig. 3G).50,51 Another recent study using DNA bricks generated polyhedral assemblies with atomic masses up to 1.2 gigadaltons and long, thick tubes similar in size to some bacilli.52 Because they do not require a scaffold strand, these approaches contrast with Rothemund's ‘scaffolded’ DNA origami. A separate approach is single-stranded origami, which folds multi-kilobase nucleic acid strands into complex 2D patterns without using staple strands. In one example, Han et al. folded hearts from a single strand of DNA approximately 3000 nt in length (Fig. 3H).53 Another recent example described the combination of single-stranded origami with DNA tiles to fold highly knotted 2- and 3D topologies.54 Since the advent of DNA origami, semi-automated55,56 and fully-automated57,58 approaches for producing target 2- and 3D geometries have emerged. Today, the ease of the origami technique, combined with the commercial availability of chemically synthesized DNA sequences,59 makes the design and fabrication of complex DNA structures accessible even to non-specialists.1
Emergence of RNA nanotechnology
In parallel with developments in DNA nanotechnology, the last two decades have witnessed the emergence of the field of ribonucleic acid (RNA) nanotechnology.60,61 Owing to its 2′-hydroxyl group, RNA is more chemically labile than DNA,62 but it nevertheless has several desirable features for nano-scale fabrication. Like DNA, RNA comprises four nucleotides: adenine, cytosine, guanine, and uracil (A, C, G, and U, respectively). It is highly programmable, with molecular recognition and self-assembly properties governed by canonical Watson–Crick interactions (i.e., C:G and A:U). However, unlike DNA, RNA also engages in many non-canonical interactions (e.g., G:U wobble pairs, sheared G:A pairs, reverse Hoogsteen pairs, and G:A imino pairs),63,64 which permit the formation of a breathtaking range of complex 3D structures and the execution of catalytic and recognition functions that rival the activities of proteins.65,66 Indeed, in nature, messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA) have active roles in protein synthesis.67–70 Additionally, ribozymes, riboswitches, small RNAs, and long, noncoding RNAs are central players in genome replication, intron splicing, regulation of gene expression, epigenetic modification and scaffolding, and more.66Moreover, RNA structure is organized on the primary, secondary, and tertiary levels. Primary structure is simply the nucleotide sequence of an RNA molecule. Secondary structure comprises recurrent motifs such as helices, hairpins, bulges, internal loops, and multi-way junctions, and tertiary structure comprises noncovalent interactions that connect these motifs together in 3D.71 Hierarchical folding confers modularity on all levels of RNA structure.72 Therefore, RNA designers can shop the natural repertoire and mix and match different structural and functional elements (‘modules’) in order to create composite RNA structures with tailored functionalities.73 These features, combined with the groundwork laid by research in the field of DNA nanotechnology and advances in chemical RNA synthesis,74 have enabled the construction of a wide variety of synthetic RNA structures. Important advances in the field of RNA nanotechnology are summarized below.
RNA tectonics
In 1996, Westhof et al. proposed ‘RNA tectonics’ to describe the idea that RNA can be resolved into and reassembled from component modules, like a 3D mosaic.75 A few years later, Jaeger and Leontis put this idea into practice by generating synthetic ‘tectoRNA’ units using a hairpin tetraloop and a tetraloop receptor extracted from the Tetrahymena thermophila group I intron.76 Their pioneering work showed that the rational placement of interacting loops and loop-receptors could direct the self-assembly of RNA dimers and 1D arrays.76 Further studies revealed that the self-assembly behavior of tectoRNAs could be fine-tuned by changing the length, helical twist, and flexibility of the linker between interacting motifs.77 Additionally, by modifying the loop:loop-receptor system with a four-way junction derived from the hairpin ribozyme, Nasalean et al. later demonstrated the self-assembly of long, micrometer-scale RNA structures that resemble actin filaments from the protein world (Fig. 4A).78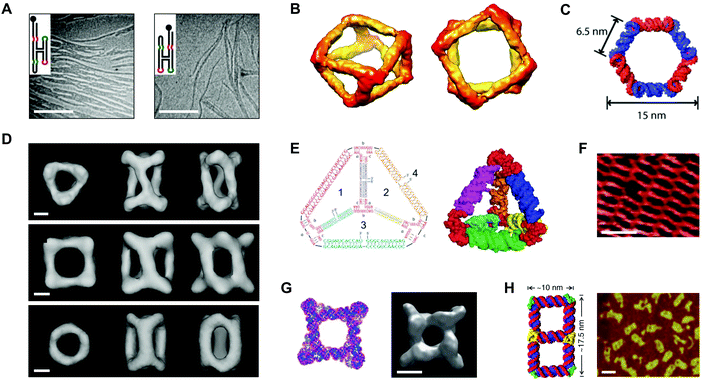 | ||
| Fig. 4 Self-assembled RNA structures. (A) Transmission electron microscopy (TEM) images of RNA filaments self-assembled from tectoRNAs. Scale bars: 400 nm. (B) Views of a cryo-electron microscopy (cryo-EM) reconstructed model of a tRNA polyhedron. (C) 3D model of a hexagonal ring based on the RNAI/IIi kissing complex from E. coli. (D) Cryo-EM reconstructed models of three different RNA cages self-assembled from engineered pRNA. Scale bars: 3 nm. (E) Sequences (left) and a 3D computational model (right) of an RNA tetrahedron with pRNA 3WJ vertices (red). (F) Atomic force microscopy (AFM) image of a periodic RNA lattice self-assembled from the RNA DX tile. Scale bar: 25 nm. (G) Computational model (left) and a cryo-EM reconstructed model (right) of an octameric prism self-assembled from an RNA T-junction. Scale bar: 5 nm. (H) 3D model of an RNA double square (left) and an AFM image of double squares in cell lysates from E. coli (right). Scale bar: 20 nm. Panel (A) reprinted from ref. 78 by permission of Oxford University Press. Panel (B) reprinted from ref. 81 by permission from Springer Nature: Nature Chemistry. Copyright 2010. Panel (C) reprinted with permission from ref. 84. Copyright 2011 American Chemical Society. Panel (D) reprinted from ref. 96 by permission from Springer Nature: Nature Communications. Copyright 2014. Panel (E) reprinted with permission from ref. 106. Copyright 2016 John Wiley and Sons. Panel (F) reprinted from ref. 115 by permission of Oxford University Press. Panel (G) reprinted from ref. 116 by permission from Springer Nature: Nature Communications. Copyright 2015. Panel (H) reprinted from ref. 122. | ||
Using a different approach, Horiya et al. showed that kissing loops taken from the genome of human immunodeficiency virus (HIV) could mediate the formation of large RNA assemblies.79 Kissing loops are short hairpin loops that base pair, and their interactions have been shown to be 102–104 times more stable than loop:loop-receptor interactions.79 Chworos et al. combined HIV kissing loops with a right angle motif conserved in rRNA to generate RNA squares, square patterns, and finite grids of defined size and shape.80 In a separate study, a five-way junction derived from class II tRNA was engineered with HIV kissing loops to generate self-assembling, thermostable RNA polyhedra (Fig. 4B).81 Using 5′-biotinylated tectoRNAs, the authors of this study further illustrated a remarkable degree of spatial control by directing the precise positioning and encapsulation of streptavidins within the RNA structures.81
Yet other studies have adapted a kissing complex from Escherichia coli (E. coli) with a 120° bend to generate multimeric RNA rings with potential drug delivery applications (Fig. 4C).82–84 Notably, these circularized RNA assemblies show increased resistance to ribonucleases relative to their linear RNA counterparts.84 Recent work by Geary et al. has generalized the RNA tectonics approach by composing a ‘syntax’ of structural modules, including kissing loops, tail–tail interactions, triple helices, bulges, and three- and five-way junctions.85 Using this syntax, the authors demonstrated the formation of various RNA shapes, including polygons, ladders, grids, and even hearts85 that together provide a glimpse into the versatility of RNA as a medium for generating complex nanostructures.
Engineering of viral pRNA
Alongside tectoRNAs, another molecule that has significantly shaped the current RNA nanotechnology landscape is prohead or packaging RNA (pRNA).60,61 pRNA is a naturally-occurring RNA molecule that derives from the phi29 bacteriophage and related bacteriophages in the phi29-like family.86 Full-length pRNA is approximately 170 nt long,87 and it has a conserved secondary structure that features six helical regions, a three-way junction (3WJ), and two kissing loops that mediate pRNA self-assembly in the context of a DNA packaging motor, where pRNA performs an essential but yet unknown function.87–89 In 1998, Guo et al. demonstrated that the prototype phi29 pRNA sequence self-assembles in vitro.90 More recent studies on phylogenetically related pRNAs have shown that in vitro self-assembly behavior varies by sequence and depends on the nucleotide composition at the 3WJ.91–93 Engineering of phi29 pRNA has yielded self-assembling RNA dimers, trimers, and arrays in vitro.94,95 The ability to control pRNA self-assembly also has enabled the creation of RNA cages96 (Fig. 4D) and possible vectors for drug delivery.97,98 Recent work has narrowed engineering efforts to the unusually stable 3WJ motif found within the pRNA structure.99–101 The phi29 pRNA 3WJ has been used to construct RNA polygons,102–105 tetrahedra106 (Fig. 4E), and prisms.107 Other studies have developed the 3WJ into a promising platform for the delivery of therapeutic cargoes.108–113 To date, only the phi29 pRNA sequence has been developed for RNA nanotechnology. Related or engineered pRNA sequences with different thermodynamic,100 conformational,93 and self-assembly91,92 properties may prove useful for tailoring the features of pRNA-based structures for specific applications.RNA tiles and RNA origami
As described above, a host of synthetic RNA structures have been engineered by exploiting RNA motifs found in nature. A different approach relies on the de novo design of RNA motifs analogous to the tiles used in DNA nanotechnology. For example, Afonin et al. have demonstrated the self-assembly of RNA PX tiles,114 and Stewart et al. have shown that RNA DX tiles self-assemble into periodic lattices on the micrometer scale (Fig. 4F).115 Moreover, Yu et al. have employed RNA T-junctions in the fabrication of self-assembling octameric prisms (Fig. 4G).116 Another technique that has been developed for RNA nanotechnology is origami. In a recent study, Han et al. demonstrated that single-stranded DNA origami could be adapted for RNA by taking into account the difference in helical periodicity between DNA and RNA molecules.53 Using the adapted technique, the authors were able to produce structures such as rectangles, rhombuses, and hearts from RNA sequences up to 6.3 kb in length.53 In another recent study, Qi et al. combined RNA PX tiles with single-stranded origami to fold intricately knotted 2D RNA structures up to 7.5 kb in length.54Co-transcriptionally folded RNA structures
RNA folding is fast on the timescale of transcription.117 Therefore, one exciting prospect for the field of RNA nanotechnology is encoding RNA structures as DNA and subsequently expressing them in vitro or in vivo. In one example, Afonin et al. showed that rationally designed RNA cubes self-assemble from six or 10 component strands during in vitro transcription.118 In another example, Afonin et al. showed that RNA rings and cubes co-transcriptionally assemble from up to 22 different component strands in vitro.119 An artful study by Geary et al. combined principles of origami with tiles, hairpins, and kissing loops to create hexagonal RNA lattices that co-transcriptionally fold and self-assemble in vitro.120 Other studies have extended these principles into living systems. For example, Delebecque et al. co-transcriptionally assembled RNA scaffolds in E. coli that were capable of organizing proteins in a hydrogen-producing biosynthetic pathway.121 More recently, Li et al. showed that an RNA double square design could be cloned, expressed, and folded in E. coli (Fig. 4H).122 Compared to conventional origami techniques, the gene expression of RNA assemblies avoids costly chemical synthesis as well as lengthy annealing steps120 and moves the field a step closer to one of the holy grails of RNA nanotechnology, i.e., rationally designing RNA objects as large and complex as natural RNA machines.123Biomedical applications
As described above, numerous advances in nucleic acid nanotechnology have contributed to the development of supramolecular DNA and RNA assemblies with precise structural and dynamic control. Here, we highlight examples of high-order nucleic acid structures in two promising biomedical applications: biosensing and therapeutic drug delivery.Biosensing
Biosensors are tools that convert signals from biological analytes (e.g., cells, proteins, nucleic acids, and small metabolites) into recordable signals. They consist of a recognition component (i.e., a probe), a transducer, and a signal amplification device.124 One challenge in biosensing is facilitating maximum interactions between analytes and probes while preventing agglomeration due to an irregular distribution of probes across a sensing surface.124 Rationally designed DNA tetrahedra have proven particularly useful for addressing this problem. DNA tetrahedra are mechanically robust structures that self-assemble rapidly and in nearly quantitative yields from four component strands.125,126 In 2010, Pei et al. developed a sensitive electrochemical biosensor using DNA tetrahedra assembled from three DNA strands bearing a terminal thiol group (–SH) and one DNA strand containing a probe.127 The tetrahedra self-assembled on a gold (Au) electrode by Au–S chemistry and were shown to enforce uniform probe-to-probe spacing.127 Additionally, tetrahedra with pendant DNA probes showed a lower detection limit for target DNA of approximately 1 picomolar (pM), which represented a 250-fold improvement over the same probe without tetrahedron scaffolding.127 Moreover, aptamers are synthetic nucleic acid sequences that are evolved to bind a user-specified target ligand with high affinity using a process known as systematic evolution of ligands by exponential enrichment (SELEX).128,129 Replacement of the pendant DNA probe with a DNA aptamer enabled the detection of thrombin, a potential tumor marker, with a lower detection limit of 100 pM, or three orders of magnitude lower than the detection limit for the aptamer alone.127New work has shown that DNA tetrahedra functionalized with antibodies can detect other disease biomarkers, including tumor necrosis factor alpha (TNF-α),130 a well-known tumor marker, and prostate-specific antigen (PSA),131 a biomarker for prostate cancer. Yet other DNA tetrahedra have been developed for detecting microRNAs (miRNAs).132–136 In one particularly notable example, Wen et al. showed that a DNA tetrahedron-based biosensor could discriminate human let-7 sequences with single nucleotide variations.132 The same system equipped with a different probe could detect the cancer-associated miRNA-21 on the attomolar range.132 Furthermore, using DNA tetrahedra, Zhou et al. were able to detect MCF-7 breast cancer cells.137 Lin et al. since have developed a protocol for the fabrication of DNA tetrahedron-based biosensors for small molecule, nucleic acid, protein, and whole cell detection (Fig. 5A).138 Recent studies have further shown that DNA tetrahedron-based biosensors can detect RNAs,139–142 proteins,143 small molecules,144 and metal ions145 inside living cells.
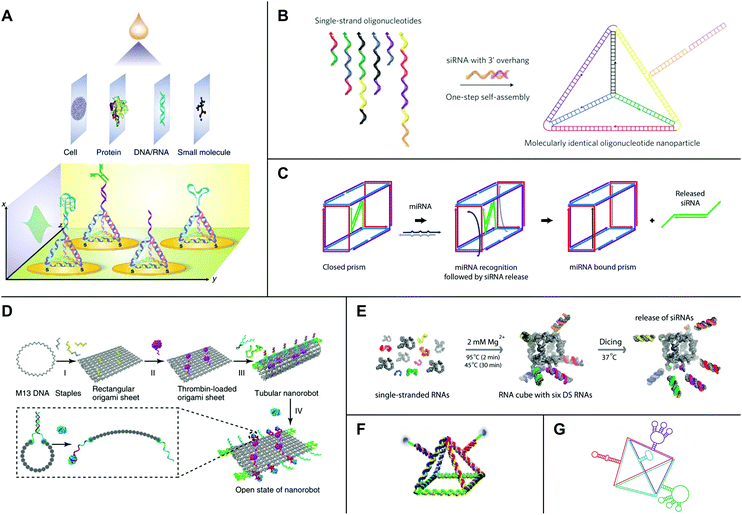 | ||
| Fig. 5 High-order nucleic acid structures for biosensing and drug delivery. (A) A DNA tetrahedron-based biosensing platform for the detection of cells, proteins, nucleic acids, and small molecules. (B) Strands of DNA and siRNA self-assemble into an siRNA-functionalized DNA tetrahedron. (C) A DNA nanosuitcase is triggered to release encapsulated siRNA cargo (green) by a specific miRNA sequence. (D) A DNA origami nanorobot is loaded with thrombin (magenta) and functionalized with nucleolin-binding aptamers (green). (E) Strands of RNA self-assemble into an siRNA-functionalized RNA cube. Processing by Dicer (‘Dicing’) releases siRNAs from the cube. (F) An RNA pyramid with photocleavable drug cargo. (G) An RNA tetrahedron functionalized with aptamers (red, blue, and green) and a ribozyme (purple). Panel (A) reprinted from ref. 138 by permission from Springer Nature: Nature Protocols. Copyright 2016. Panel (B) reprinted from ref. 157 by permission from Springer Nature: Nature Nanotechnology. Copyright 2012. Panel (C) reprinted with permission from ref. 162. Copyright 2016 American Chemical Society. Panel (D) reprinted from ref. 164 by permission from Springer Nature: Nature Biotechnology. Copyright 2018. Panel (E) reprinted with permission from ref. 168, DOI: http://10.1021/nn504508s. Panel (F) reprinted from ref. 169 with permission from Springer Nature: Nano Research. Copyright 2019. Panel (G) reprinted with permission from ref. 106. Copyright 2016 John Wiley and Sons. | ||
Drug delivery
Owing to their intrinsic biocompatibility, nucleic acids also hold promise as platforms for therapeutic drug delivery. In 2006, Erben et al. illustrated the potential of DNA structures to serve as molecular containers for therapeutic cargo by loading cytochrome c into a self-assembled DNA tetrahedron.146 Follow-on research showed that DNA tetrahedra are stable to enzymatic degradation,147 and DNA tetrahedra,148 icosahedra,149 and cages150 may be reconfigured in response to external signals for controlled drug release. Furthermore, many studies have demonstrated that DNA structures are taken up by cultured cells without transfection reagents.151–154 Recently, Wiraja et al. tested a variety of nano-scale DNA structures, including tetrahedra, cylindrical rods, rectangles, and triangles, and showed that highly-ordered structures smaller than 75 nm penetrate the skin.155 Using a mouse model of melanoma, the study further showed that DNA tetrahedra loaded with the chemotherapeutic drug doxorubicin could achieve over 2-fold higher drug accumulation and tumor inhibition relative to topically applied doxorubicin and liposome- or nanoparticle-formulated doxorubicin.155While some DNA structures achieve passive uptake, other platforms have been developed with active targeting mechanisms to promote uptake by specific populations of cells. In one example, Chang et al. showed that DNA icosahedra self-assembled from five- and six-point star motifs could be functionalized with a MUC 1 aptamer for targeted delivery of doxorubicin to MUC 1+ MCF-7 breast cancer cells.156 In a separate, pioneering study, Lee et al. developed a DNA tetrahedron with a small interfering RNA (siRNA) hybridized to each edge (Fig. 5B).157 SiRNAs are short, double-stranded RNA molecules that elicit potent gene silencing by co-opting an endogenous RNA interference (RNAi) pathway.158 Currently, major challenges for the delivery of oligonucleotide drugs such as siRNAs include nuclease protection, systemic delivery, and targeted cellular uptake.159 Notably, siRNA-functionalized tetrahedra administered in a mouse tumor xenograft model displayed longer half-lives in blood circulation relative to siRNAs alone.157 Additionally, tetrahedra with siRNAs bearing folate distributed to several tissues but accumulated in folate receptor-overexpressing KB cells, where they efficiently silenced a luciferase reporter.157 In line with these results, other studies have shown that DNA structures can confer stability on their therapeutic payloads while also maintaining or even improving their therapeutic efficacies.160,161
Yet other DNA platforms have been developed with sophisticated mechanisms for cargo release. In a recent example, Bujold et al. designed DNA ‘nanosuitcases’ that encapsulate siRNA and release it in the presence of specific mRNA or miRNA triggers (Fig. 5C).162 In another example, Douglas et al. developed DNA origami ‘nanorobots’ with AND logic gates.163 These logic gates consisted of two aptamer ‘locks’ that triggered a drastic reconfiguration of the robot and exposed molecular payloads upon binding both antigenic ‘keys’.163 Recent work by Li et al. combined both active targeting and controlled release mechanisms in a DNA nanorobot loaded with thrombin and functionalized with nucleolin-binding aptamers (Fig. 5D).164 Thrombin is a protease that induces coagulation and may be useful for starving tumors of nutrients and oxygen by selective occlusion of tumor blood vessels.164 Nucleolin is a protein that is expressed on tumor-associated endothelial cells.165 Remarkably, the nanorobots were capable of depositing thrombin at tumor cells in mouse models of breast cancer, and they also proved safe and immunologically inert in both mice and miniature pigs.164
Several high-order RNA structures also have been developed for therapeutic drug delivery. Because RNA is more chemically labile than DNA, RNA-based platforms often are chemically modified to improve their stability under physiological conditions. Common modifications include substitutions at the 2′ position of ribose as well as in the internucleotide linkage.159 A series of studies has developed RNA cubes designed in silico118 as a promising platform for drug delivery. The cubes are precisely controlled in terms of size, shape, and composition; they can be chemically modified for downstream applications; and they also are capable of self-assembly in isothermal conditions during in vitro transcription or from several short strands following chemical RNA synthesis.118,119 Computational and experimental analyses have shown that cube designs with 10 bp per edge and single-stranded regions in the corners are not strained and assemble efficiently.166 In 2011, Afonin et al. developed a protocol for the design and self-assembly of siRNA-functionalized RNA cubes using processes that are fully automatable.167 The cubes interact with recombinant human Dicer in vitro to produce siRNAs167 (Fig. 5E). Furthermore, a recent study by Afonin et al. showed that siRNA-functionalized RNA cubes are capable of triggering RNAi in GFP-expressing MDA-MB-231 breast cancer cells and reducing HIV-1 production in HeLa cells.168
Finally, another promising platform for drug delivery has come from research on pRNA and its thermodynamically stable 3WJ.60,61 The phi29 pRNA 3WJ has been engineered to deliver a variety of therapeutic agents, including small molecules,108 miRNAs,109 anti-miRNAs,110,111 and siRNAs.99,112,113 In a recent example, Xu et al. combined the phi29 pRNA 3WJ with a four-way junction derived from the hairpin ribozyme to generate 2′-deoxyfluoro U/C modified RNA pyramids (Fig. 5F).169 These pyramids were functionalized with the chemotherapeutic drug paclitaxel via photocleavable spacers, and irradiation with ultraviolet light induced drug release and cytotoxicity in MDA-MB-231 breast cancer cells.169 In a different example, Li et al. reported the self-assembly of a nuclease-resistant, 2′-deoxyfluoro U/C modified RNA tetrahedron based on the phi29 pRNA 3WJ (Fig. 5G).106 Functionalization of the tetrahedron with ribozymes, aptamers, and siRNAs did not disrupt its structure, and the functional modules retained the capacity for ribozymatic cleavage, ligand binding, and gene knockdown in vitro.106 Furthermore, this study showed that tetrahedra functionalized with an aptamer against epidermal growth factor receptor (EGFR) and an siRNA were capable of internalizing into tumor tissue and silencing a luciferase reporter in a mouse model of breast cancer.106
Outlook
From the development of DNA junctions, tiles, and origami in DNA nanotechnology to the development of tectoRNAs, engineered pRNA, and co-transcriptionally folded structures in RNA nanotechnology, research over the past 40 years has expanded the possibilities for creating supramolecular nucleic acid architectures with precise structural and dynamic control. Today, high-order DNA and RNA structures are taking their place as powerful tools with promising biomedical applications. In biosensing, nano-structured DNA platforms boost sensitivity for the detection of biological analytes while preventing agglomeration. In drug delivery, DNA- and RNA-based platforms offer potential solutions to formidable in vivo challenges, including nuclease protection, systemic delivery, and targeted cellular uptake for oligonucleotide drugs. Many of the achievements in nucleic acid nanotechnology would not be possible without parallel achievements in chemical DNA and RNA synthesis, which have given ready access to the oligonucleotide sequences necessary for constructing nucleic acid assemblies.1 Still, for bottom-up fabrication using RNA, sequence length and yield are considerable limitations. RNA assemblies made by gene expression may circumvent these issues and facilitate applications in vivo. Additionally, the cost of oligonucleotide synthesis is decreasing. In fact, a relative of Moore's law has been reported for the effective cost of DNA, which is halved every 30 months.170 Continued developments in oligonucleotide synthesis, including in the production of longer sequences for lower costs, will be important for supporting efforts in DNA and RNA nanotechnology and for realizing the extraordinary potential of high-order nucleic acid structures for biomedical applications and beyond.Conflicts of interest
A. C. H. is a co-inventor and applicant on a patent for viral pRNA 3WJ sequences.Acknowledgements
A. C. H. was supported by a Novartis FreeNovation grant from the Novartis Research Foundation (Novartis Forschungsstiftung).References
- N. C. Seeman and H. F. Sleiman, DNA nanotechnology, Nat. Rev. Mater., 2017, 3, 17068 CrossRef.
- J. D. Watson and F. H. Crick, Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid, Nature, 1953, 171, 737–738 CrossRef CAS PubMed.
- N. C. Seeman, DNA in a material world, Nature, 2003, 421, 427–431 CrossRef PubMed.
- N. C. Seeman, Nucleic acid junctions and lattices, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- H. Qiu, J. C. Dewan and N. C. Seeman, A DNA decamer with a sticky end: The crystal structure of d-CGACGATCGT, J. Mol. Biol., 1997, 267, 881–898 CrossRef CAS PubMed.
- N. R. Kallenbach, R.-I. Ma and N. C. Seeman, An immobile nucleic acid junction constructed from oligonucleotides, Nature, 1983, 305, 829–831 CrossRef CAS.
- Y. L. Wang, J. E. Mueller, B. Kemper and N. C. Seeman, Assembly and characterization of five-arm and six-arm DNA branched junctions, Biochemistry, 1991, 30, 5667–5674 CrossRef CAS PubMed.
- J. H. Chen, N. R. Kallenbach and N. C. Seeman, A specific quadrilateral synthesized from DNA branched junctions, J. Am. Chem. Soc., 1989, 111, 6402–6407 CrossRef CAS.
- J. E. Mueller, S. M. Du and N. C. Seeman, Design and synthesis of a knot from single-stranded DNA, J. Am. Chem. Soc., 1991, 113, 6306–6308 CrossRef CAS.
- N. C. Seeman, The design of single-stranded nucleic acid knots, Mol. Eng., 1992, 2, 297–307 CrossRef CAS.
- C. Mao, W. Sun and N. C. Seeman, Assembly of Borromean rings from DNA, Nature, 1997, 386, 137–138 CrossRef CAS PubMed.
- J. Chen and N. C. Seeman, Synthesis from DNA of a molecule with the connectivity of a cube, Nature, 1991, 350, 631–633 CrossRef CAS PubMed.
- Y. Zhang and N. C. Seeman, Construction of a DNA-truncated octahedron, J. Am. Chem. Soc., 1994, 116, 1661–1669 CrossRef CAS.
- N. C. Seeman, DNA engineering and its application to nanotechnology, Trends Biotechnol., 1999, 17, 437–443 CrossRef CAS PubMed.
- N. C. Seeman, An overview of structural DNA nanotechnology, Mol. Biotechnol., 2007, 37, 246–257 CrossRef CAS PubMed.
- A. R. Chandrasekaran and R. Zhuo, A ‘tile’ tale: Hierarchical self-assembly of DNA lattices, Appl. Mater. Today, 2016, 2, 7–16 CrossRef.
- T. J. Fu and N. C. Seeman, DNA double-crossover molecules, Biochemistry, 1993, 32, 3211–3220 CrossRef CAS PubMed.
- P. Sa-Ardyen, A. V. Vologodskii and N. C. Seeman, The flexibility of DNA double crossover molecules, Biophys. J., 2003, 84, 3829–3837 CrossRef CAS PubMed.
- E. Winfree, F. Liu, L. A. Wenzler and N. C. Seeman, Design and self-assembly of two-dimensional DNA crystals, Nature, 1998, 394, 539–544 CrossRef CAS PubMed.
- H. Yan, T. H. LaBean, L. Feng and J. H. Reif, Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 8103 CrossRef CAS PubMed.
- T. H. LaBean, H. Yan, J. Kopatsch, F. Liu, E. Winfree, J. H. Reif and N. C. Seeman, Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes, J. Am. Chem. Soc., 2000, 122, 1848–1860 CrossRef CAS.
- Z. Shen, H. Yan, T. Wang and N. C. Seeman, Paranemic crossover DNA: A generalized Holliday structure with applications in nanotechnology, J. Am. Chem. Soc., 2004, 126, 1666–1674 CrossRef CAS PubMed.
- H. Yan, S. H. Park, G. Finkelstein, J. H. Reif and T. H. LaBean, DNA-templated self-assembly of protein arrays and highly conductive nanowires, Science, 2003, 301, 1882–1884 CrossRef CAS PubMed.
- Y. He, Y. Chen, H. Liu, A. E. Ribbe and C. Mao, Self-assembly of hexagonal DNA two-dimensional (2D) arrays, J. Am. Chem. Soc., 2005, 127, 12202–12203 CrossRef CAS PubMed.
- Y. He, Y. Tian, A. E. Ribbe and C. Mao, Highly connected two-dimensional crystals of DNA six-point-stars, J. Am. Chem. Soc., 2006, 128, 15978–15979 CrossRef CAS PubMed.
- U. Majumder, A. Rangnekar, K. V. Gothelf, J. H. Reif and T. H. LaBean, Design and construction of double-decker tile as a route to three-dimensional periodic assembly of DNA, J. Am. Chem. Soc., 2011, 133, 3843–3845 CrossRef CAS PubMed.
- S. Hamada and S. Murata, Substrate-assisted assembly of interconnected single-duplex DNA nanostructures, Angew. Chem., Int. Ed., 2009, 48, 6820–6823 CrossRef CAS PubMed.
- P. W. K. Rothemund, N. Papadakis and E. Winfree, Algorithmic self-assembly of DNA Sierpinski triangles, PLoS Biol., 2004, 2, e424 CrossRef PubMed.
- Y. He, T. Ye, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra, Nature, 2008, 452, 198 CrossRef CAS PubMed.
- C. Mao, W. Sun, Z. Shen and N. C. Seeman, A nanomechanical device based on the B-Z transition of DNA, Nature, 1999, 397, 144–146 CrossRef CAS PubMed.
- H. Yan, X. Zhang, Z. Shen and N. C. Seeman, A robust DNA mechanical device controlled by hybridization topology, Nature, 2002, 415, 62–65 CrossRef CAS PubMed.
- D. Liu, M. Wang, Z. Deng, R. Walulu and C. Mao, Tensegrity:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Construction of rigid DNA triangles with flexible four-arm DNA junctions, J. Am. Chem. Soc., 2004, 126, 2324–2325 CrossRef CAS PubMed.
Construction of rigid DNA triangles with flexible four-arm DNA junctions, J. Am. Chem. Soc., 2004, 126, 2324–2325 CrossRef CAS PubMed. - J. Zheng, J. J. Birktoft, Y. Chen, T. Wang, R. Sha, P. E. Constantinou, S. L. Ginell, C. Mao and N. C. Seeman, From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal, Nature, 2009, 461, 74 CrossRef CAS PubMed.
- T. Wang, R. Sha, J. Birktoft, J. Zheng, C. Mao and N. C. Seeman, A DNA crystal designed to contain two molecules per asymmetric unit, J. Am. Chem. Soc., 2010, 132, 15471–15473 CrossRef CAS PubMed.
- R. Sha, J. J. Birktoft, N. Nguyen, A. R. Chandrasekaran, J. Zheng, X. Zhao, C. Mao and N. C. Seeman, Self-assembled DNA crystals: The impact on resolution of 5′-phosphates and the DNA source, Nano Lett., 2013, 13, 793–797 CrossRef CAS PubMed.
- Y. P. Ohayon, A. R. Chandrasekaran, C. Hernandez, J. J. Birktoft, R. Sha, S. Ginell, P. Lukeman, C. Mao, P. M. Chaikin and N. C. Seeman, Programmable crystal contacts used to improve the resolution of self-assembled 3D DNA crystals, J. Biomol. Struct. Dyn., 2015, 33, 50–51 CrossRef.
- E. Stahl, F. Praetorius, C. C. de Oliveira Mann, K. P. Hopfner and H. Dietz, Impact of heterogeneity and lattice bond strength on DNA triangle crystal growth, ACS Nano, 2016, 10, 9156–9164 CrossRef CAS PubMed.
- J. Zhao, A. R. Chandrasekaran, Q. Li, X. Li, R. Sha, N. C. Seeman and C. Mao, Post-assembly stabilization of rationally designed DNA crystals, Angew. Chem., Int. Ed., 2015, 54, 9936–9939 CrossRef CAS PubMed.
- H. O. Abdallah, Y. P. Ohayon, A. R. Chandrasekaran, R. Sha, K. R. Fox, T. Brown, D. A. Rusling, C. Mao and N. C. Seeman, Stabilisation of self-assembled DNA crystals by triplex-directed photo-cross-linking, Chem. Commun., 2016, 52, 8014–8017 RSC.
- Z. Li, L. Liu, M. Zheng, J. Zhao, N. C. Seeman and C. Mao, Making engineered 3D DNA crystals robust, J. Am. Chem. Soc., 2019, 141, 15850–15855 CrossRef CAS PubMed.
- N. C. Seeman, Structural DNA nanotechnology: An overview, Methods Mol. Biol., 2005, 303, 143–166 CAS.
- W. M. Shih, J. D. Quispe and G. F. Joyce, A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron, Nature, 2004, 427, 618–621 CrossRef CAS PubMed.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- Y. Ke, J. Sharma, M. Liu, K. Jahn, Y. Liu and H. Yan, Scaffolded DNA origami of a DNA tetrahedron molecular container, Nano Lett., 2009, 9, 2445–2447 CrossRef CAS PubMed.
- S. M. Douglas, H. Dietz, T. Liedl, B. Högberg, F. Graf and W. M. Shih, Self-assembly of DNA into nanoscale three-dimensional shapes, Nature, 2009, 459, 414 CrossRef CAS PubMed.
- H. Dietz, S. M. Douglas and W. M. Shih, Folding DNA into twisted and curved nanoscale shapes, Science, 2009, 325, 725 CrossRef CAS PubMed.
- D. Han, S. Pal, J. Nangreave, Z. Deng, Y. Liu and H. Yan, DNA origami with complex curvatures in three-dimensional space, Science, 2011, 332, 342 CrossRef CAS PubMed.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. P. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Self-assembly of a nanoscale DNA box with a controllable lid, Nature, 2009, 459, 73 CrossRef CAS PubMed.
- B. Wei, M. Dai and P. Yin, Complex shapes self-assembled from single-stranded DNA tiles, Nature, 2012, 485, 623–626 CrossRef CAS PubMed.
- Y. Ke, L. L. Ong, W. M. Shih and P. Yin, Three-dimensional structures self-assembled from DNA bricks, Science, 2012, 338, 1177 CrossRef CAS PubMed.
- L. L. Ong, N. Hanikel, O. K. Yaghi, C. Grun, M. T. Strauss, P. Bron, J. Lai-Kee-Him, F. Schueder, B. Wang, P. Wang, J. Y. Kishi, C. Myhrvold, A. Zhu, R. Jungmann, G. Bellot, Y. Ke and P. Yin, Programmable self-assembly of three-dimensional nanostructures from 10000 unique components, Nature, 2017, 552, 72–77 CrossRef CAS PubMed.
- K. F. Wagenbauer, C. Sigl and H. Dietz, Gigadalton-scale shape-programmable DNA assemblies, Nature, 2017, 552, 78–83 CrossRef CAS PubMed.
- D. Han, X. Qi, C. Myhrvold, B. Wang, M. Dai, S. Jiang, M. Bates, Y. Liu, B. An, F. Zhang, H. Yan and P. Yin, Single-stranded DNA and RNA origami, Science, 2017, 358, eaao2648 CrossRef PubMed.
- X. Qi, F. Zhang, Z. Su, S. Jiang, D. Han, B. Ding, Y. Liu, W. Chiu, P. Yin and H. Yan, Programming molecular topologies from single-stranded nucleic acids, Nat. Commun., 2018, 9, 4579 CrossRef PubMed.
- E. Benson, A. Mohammed, J. Gardell, S. Masich, E. Czeizler, P. Orponen and B. Högberg, DNA rendering of polyhedral meshes at the nanoscale, Nature, 2015, 523, 441 CrossRef CAS PubMed.
- E. Benson, A. Mohammed, A. Bosco, A. I. Teixeira, P. Orponen and B. Högberg, Computer-aided production of scaffolded DNA nanostructures from flat sheet meshes, Angew. Chem., Int. Ed., 2016, 55, 8869–8872 CrossRef CAS PubMed.
- R. Veneziano, S. Ratanalert, K. Zhang, F. Zhang, H. Yan, W. Chiu and M. Bathe, Designer nanoscale DNA assemblies programmed from the top down, Science, 2016, 352, 1534 CrossRef CAS PubMed.
- H. Jun, F. Zhang, T. Shepherd, S. Ratanalert, X. Qi, H. Yan and M. Bathe, Autonomously designed free-form 2D DNA origami, Sci. Adv., 2019, 5, eaav0655 CrossRef PubMed.
- R. A. Hughes and A. D. Ellington, Synthetic DNA synthesis and assembly: Putting the synthetic in synthetic biology, Cold Spring Harbor Perspect. Biol., 2017, 9, a023812 CrossRef PubMed.
- P. Guo, The emerging field of RNA nanotechnology, Nat. Nanotechnol., 2010, 5, 833–842 CrossRef CAS PubMed.
- D. Jasinski, F. Haque, D. W. Binzel and P. Guo, Advancement of the emerging field of RNA nanotechnology, ACS Nano, 2017, 11, 1142–1164 CrossRef CAS PubMed.
- G. A. Soukup and R. R. Breaker, Relationship between internucleotide linkage geometry and the stability of RNA, RNA, 1999, 5, 1308–1325 CrossRef CAS PubMed.
- N. B. Leontis and E. Westhof, Geometric nomenclature and classification of RNA base pairs, RNA, 2001, 7, 499–512 CrossRef CAS PubMed.
- N. B. Leontis, J. Stombaugh and E. Westhof, The non-Watson-Crick base pairs and their associated isostericity matrices, Nucleic Acids Res., 2002, 30, 3497–3531 CrossRef CAS PubMed.
- J. A. Cruz and E. Westhof, The dynamic landscapes of RNA architecture, Cell, 2009, 136, 604–609 CrossRef CAS PubMed.
- E. J. Strobel, K. E. Watters, D. Loughrey and J. B. Lucks, RNA systems biology: Uniting functional discoveries and structural tools to understand global roles of RNAs, Curr. Opin. Biotechnol., 2016, 39, 182–191 CrossRef CAS PubMed.
- F. H. Crick, On protein synthesis, Symp. Soc. Exp. Biol., 1958, 12, 138–163 CAS.
- F. H. Crick, Central dogma of molecular biology, Nature, 1970, 227, 561–563 CrossRef CAS PubMed.
- N. Ban, P. Nissen, J. Hansen, P. B. Moore and T. A. Steitz, The complete atomic structure of the large ribosomal subunit at 2.4 A resolution, Science, 2000, 289, 905–920 CrossRef CAS PubMed.
- B. T. Wimberly, D. E. Brodersen, W. M. Clemons Jr, R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch and V. Ramakrishnan, Structure of the 30S ribosomal subunit, Nature, 2000, 407, 327–339 CrossRef CAS PubMed.
- R. T. Batey, R. P. Rambo and J. A. Doudna, Tertiary motifs in RNA structure and folding, Angew. Chem., Int. Ed., 1999, 38, 2326–2343 CrossRef PubMed.
- I. Tinoco and C. Bustamante, How RNA folds, J. Mol. Biol., 1999, 293, 271–281 CrossRef CAS PubMed.
- W. Grabow and L. Jaeger, RNA modularity for synthetic biology, F1000Prime Rep., 2013, 5, 46 Search PubMed.
- M. Sekine, in Synthesis of Therapeutic Oligonucleotides, ed. S. Obika and M. Sekine, Springer Singapore, Singapore, 2018, pp. 41–65 DOI:10.1007/978-981-13-1912-9_3.
- E. Westhof, B. Masquida and L. Jaeger, RNA tectonics: Towards RNA design, Folding Des., 1996, 1, R78–R88 CrossRef CAS PubMed.
- L. Jaeger and N. B. Leontis, Tecto-RNA: One-dimensional self-assembly through tertiary interactions, Angew. Chem., Int. Ed., 2000, 39, 2521–2524 CrossRef CAS PubMed.
- L. Jaeger, E. Westhof and N. B. Leontis, TectoRNA: Modular assembly units for the construction of RNA nano-objects, Nucleic Acids Res., 2001, 29, 455–463 CrossRef CAS PubMed.
- L. Nasalean, S. Baudrey, N. B. Leontis and L. Jaeger, Controlling RNA self-assembly to form filaments, Nucleic Acids Res., 2006, 34, 1381–1392 CrossRef CAS PubMed.
- S. Horiya, X. Li, G. Kawai, R. Saito, A. Katoh, K. Kobayashi and K. Harada, RNA LEGO: Magnesium-dependent formation of specific RNA assemblies through kissing interactions, Chem. Biol., 2003, 10, 645–654 CrossRef CAS.
- A. Chworos, I. Severcan, A. Y. Koyfman, P. Weinkam, E. Oroudjev, H. G. Hansma and L. Jaeger, Building programmable jigsaw puzzles with RNA, Science, 2004, 306, 2068 CrossRef CAS.
- I. Severcan, C. Geary, A. Chworos, N. Voss, E. Jacovetty and L. Jaeger, A polyhedron made of tRNAs, Nat. Chem., 2010, 2, 772–779 CrossRef CAS PubMed.
- Y. G. Yingling and B. A. Shapiro, Computational design of an RNA hexagonal nanoring and an RNA nanotube, Nano Lett., 2007, 7, 2328–2334 CrossRef CAS PubMed.
- M. Paliy, R. Melnik and B. A. Shapiro, Molecular dynamics study of the RNA ring nanostructure: a phenomenon of self-stabilization, Phys. Biol., 2009, 6, 046003 CrossRef PubMed.
- W. W. Grabow, P. Zakrevsky, K. A. Afonin, A. Chworos, B. A. Shapiro and L. Jaeger, Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes, Nano Lett., 2011, 11, 878–887 CrossRef CAS PubMed.
- C. Geary, A. Chworos, E. Verzemnieks, N. R. Voss and L. Jaeger, Composing RNA nanostructures from a syntax of RNA structural modules, Nano Lett., 2017, 17, 7095–7101 CrossRef CAS PubMed.
- A. C. Hill, L. E. Bartley and S. J. Schroeder, Prohead RNA: A noncoding viral RNA of novel structure and function, Wiley Interdiscip. Rev.: RNA, 2016, 7, 428–437 CrossRef CAS PubMed.
- S. Bailey, J. Wichitwechkarn, D. Johnson, B. E. Reilly and D. L. Anderson, and J. W. Bodley, Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging, J. Biol. Chem., 1990, 265, 22365–22370 CAS.
- P. X. Guo, S. Erickson and D. Anderson, A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA, Science, 1987, 236, 690–694 CrossRef CAS PubMed.
- C. Chen, C. Zhang and P. Guo, Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor, RNA, 1999, 5, 805–818 CrossRef CAS PubMed.
- P. Guo, C. Zhang, C. Chen, K. Garver and M. Trottier, Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation, Mol. Cell, 1998, 2, 149–155 CrossRef CAS PubMed.
- X. Gu and S. J. Schroeder, Different sequences show similar quaternary interaction stabilities in prohead viral RNA self-assembly, J. Biol. Chem., 2011, 286, 14419–14426 CrossRef CAS.
- Y. Hao and J. S. Kieft, Diverse self-association properties within a family of phage packaging RNAs, RNA, 2014, 20, 1759–1774 CrossRef CAS PubMed.
- Y. Hao and J. S. Kieft, Three-way junction conformation dictates self-association of phage packaging RNAs, RNA Biol., 2016, 13, 635–645 CrossRef PubMed.
- D. Shu, L. P. Huang, S. Hoeprich and P. Guo, Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices, J. Nanosci. Nanotechnol., 2003, 3, 295–302 CrossRef CAS PubMed.
- D. Shu, W. D. Moll, Z. Deng, C. Mao and P. Guo, Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology, Nano Lett., 2004, 4, 1717–1723 CrossRef CAS PubMed.
- C. Hao, X. Li, C. Tian, W. Jiang, G. Wang and C. Mao, Construction of RNA nanocages by re-engineering the packaging RNA of phi29 bacteriophage, Nat. Commun., 2014, 5, 3890 CrossRef CAS.
- S. Guo, N. Tschammer, S. Mohammed and P. Guo, Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA, Hum. Gene Ther., 2005, 16, 1097–1109 CrossRef CAS PubMed.
- A. Khaled, S. Guo, F. Li and P. Guo, Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology, Nano Lett., 2005, 5, 1797–1808 CrossRef CAS PubMed.
- D. Shu, Y. Shu, F. Haque, S. Abdelmawla and P. Guo, Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat. Nanotechnol., 2011, 6, 658 CrossRef CAS PubMed.
- A. C. Hill and S. J. Schroeder, Thermodynamic stabilities of three-way junction nanomotifs in prohead RNA, RNA, 2017, 23, 521–529 CrossRef CAS PubMed.
- X. Piao, H. Wang, D. W. Binzel and P. Guo, Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2′-F RNA, and LNA in the context of phi29 pRNA 3WJ, RNA, 2018, 24, 67–76 CrossRef CAS PubMed.
- E. F. Khisamutdinov, D. L. Jasinski and P. Guo, RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry, ACS Nano, 2014, 8, 4771–4781 CrossRef CAS PubMed.
- D. L. Jasinski, E. F. Khisamutdinov, Y. L. Lyubchenko and P. Guo, Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties, ACS Nano, 2014, 8, 7620–7629 CrossRef CAS PubMed.
- E. F. Khisamutdinov, M. N. Bui, D. Jasinski, Z. Zhao, Z. Cui and P. Guo, Simple method for constructing RNA triangle, square, pentagon by tuning interior RNA 3WJ angle from 60 degrees to 90 degrees or 108 degrees, Methods Mol. Biol., 2015, 1316, 181–193 CrossRef PubMed.
- D. L. Jasinski, H. Li and P. Guo, The effect of size and shape of RNA nanoparticles on biodistribution, Mol. Ther., 2018, 26, 784–792 CrossRef CAS PubMed.
- H. Li, K. Zhang, F. Pi, S. Guo, L. Shlyakhtenko, W. Chiu, D. Shu and P. Guo, Controllable self-assembly of RNA tetrahedrons with precise shape and size for cancer targeting, Adv. Mater., 2016, 28, 7501–7507 CrossRef CAS PubMed.
- E. F. Khisamutdinov, D. L. Jasinski, H. Li, K. Zhang, W. Chiu and P. Guo, Fabrication of RNA 3D nanoprisms for loading and protection of small RNAs and model drugs, Adv. Mater., 2016, 28, 10079–10087 CrossRef CAS.
- F. Pi, H. Zhang, H. Li, V. Thiviyanathan, D. G. Gorenstein, A. K. Sood and P. Guo, RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery, Nanomedicine, 2017, 13, 1183–1193 CrossRef CAS PubMed.
- D. W. Binzel, Y. Shu, H. Li, M. Sun, Q. Zhang, D. Shu, B. Guo and P. Guo, Specific delivery of miRNA for high efficient inhibition of prostate cancer by RNA nanotechnology, Mol. Ther., 2016, 24, 1267–1277 CrossRef CAS PubMed.
- D. Shu, H. Li, Y. Shu, G. Xiong, W. E. Carson, 3rd, F. Haque, R. Xu and P. Guo, Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology, ACS Nano, 2015, 9, 9731–9740 CrossRef CAS PubMed.
- T. J. Lee, J. Y. Yoo, D. Shu, H. Li, J. Zhang, J. G. Yu, A. C. Jaime-Ramirez, M. Acunzo, G. Romano, R. Cui, H. L. Sun, Z. Luo, M. Old, B. Kaur, P. Guo and C. M. Croce, RNA nanoparticle-based targeted therapy for glioblastoma through inhibition of oncogenic miR-21, Mol. Ther., 2017, 25, 1544–1555 CrossRef CAS PubMed.
- T. J. Lee, F. Haque, D. Shu, J. Y. Yoo, H. Li, R. A. Yokel, C. Horbinski, T. H. Kim, S. H. Kim, C. H. Kwon, I. Nakano, B. Kaur, P. Guo and C. M. Croce, RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model, Oncotarget, 2015, 6, 14766–14776 Search PubMed.
- D. Cui, C. Zhang, B. Liu, Y. Shu, T. Du, D. Shu, K. Wang, F. Dai, Y. Liu, C. Li, F. Pan, Y. Yang, J. Ni, H. Li, B. Brand-Saberi and P. Guo, Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA, Sci. Rep., 2015, 5, 10726 CrossRef PubMed.
- K. A. Afonin, D. J. Cieply and N. B. Leontis, Specific RNA self-assembly with minimal paranemic motifs, J. Am. Chem. Soc., 2008, 130, 93–102 CrossRef CAS.
- J. M. Stewart, H. K. K. Subramanian and E. Franco, Self-assembly of multi-stranded RNA motifs into lattices and tubular structures, Nucleic Acids Res., 2017, 45, 5449–5457 CrossRef CAS PubMed.
- J. Yu, Z. Liu, W. Jiang, G. Wang and C. Mao, De novo design of an RNA tile that self-assembles into a homo-octameric nanoprism, Nat. Commun., 2015, 6, 5724 CrossRef CAS PubMed.
- G. Zemora and C. Waldsich, RNA folding in living cells, RNA Biol., 2010, 7, 634–641 CrossRef PubMed.
- K. A. Afonin, E. Bindewald, A. J. Yaghoubian, N. Voss, E. Jacovetty, B. A. Shapiro and L. Jaeger, In vitro assembly of cubic RNA-based scaffolds designed in silico, Nat. Nanotechnol., 2010, 5, 676–682 CrossRef CAS PubMed.
- K. A. Afonin, M. Kireeva, W. W. Grabow, M. Kashlev, L. Jaeger and B. A. Shapiro, Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs, Nano Lett., 2012, 12, 5192–5195 CrossRef CAS PubMed.
- C. Geary, P. W. K. Rothemund and E. S. Andersen, A single-stranded architecture for cotranscriptional folding of RNA nanostructures, Science, 2014, 345, 799 CrossRef CAS.
- C. J. Delebecque, A. B. Lindner, P. A. Silver and F. A. Aldaye, Organization of intracellular reactions with rationally designed RNA assemblies, Science, 2011, 333, 470 CrossRef CAS.
- M. Li, M. Zheng, S. Wu, C. Tian, D. Liu, Y. Weizmann, W. Jiang, G. Wang and C. Mao, In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs, Nat. Commun., 2018, 9, 2196 CrossRef.
- N. B. Leontis and E. Westhof, Self-assembled RNA nanostructures, Science, 2014, 345, 732 CrossRef CAS.
- S. Li, T. Tian, T. Zhang, X. Cai and Y. Lin, Advances in biological applications of self-assembled DNA tetrahedral nanostructures, Mater. Today, 2019, 24, 57–68 CrossRef CAS.
- R. P. Goodman, R. M. Berry and A. J. Turberfield, The single-step synthesis of a DNA tetrahedron, Chem. Commun., 2004, 1372–1373, 10.1039/b402293a,.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication, Science, 2005, 310, 1661 CrossRef CAS PubMed.
- H. Pei, N. Lu, Y. Wen, S. Song, Y. Liu, H. Yan and C. Fan, A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing, Adv. Mater., 2010, 22, 4754–4758 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, In vitro selection of RNA molecules that bind specific ligands, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- H. Pei, Y. Wan, J. Li, H. Hu, Y. Su, Q. Huang and C. Fan, Regenerable electrochemical immunological sensing at DNA nanostructure-decorated gold surfaces, Chem. Commun., 2011, 47, 6254–6256 RSC.
- X. Chen, G. Zhou, P. Song, J. Wang, J. Gao, J. Lu, C. Fan and X. Zuo, Ultrasensitive electrochemical detection of prostate-specific antigen by using antibodies anchored on a DNA nanostructural scaffold, Anal. Chem., 2014, 86, 7337–7342 CrossRef CAS PubMed.
- Y. Wen, H. Pei, Y. Shen, J. Xi, M. Lin, N. Lu, X. Shen, J. Li and C. Fan, DNA nanostructure-based interfacial engineering for PCR-free ultrasensitive electrochemical analysis of microRNA, Sci. Rep., 2012, 2, 867 CrossRef PubMed.
- Z. Ge, M. Lin, P. Wang, H. Pei, J. Yan, J. Shi, Q. Huang, D. He, C. Fan and X. Zuo, Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor, Anal. Chem., 2014, 86, 2124–2130 CrossRef CAS PubMed.
- M. Lin, Y. Wen, L. Li, H. Pei, G. Liu, H. Song, X. Zuo, C. Fan and Q. Huang, Target-responsive, DNA nanostructure-based E-DNA sensor for microRNA analysis, Anal. Chem., 2014, 86, 2285–2288 CrossRef CAS.
- S. Liu, W. Su, Z. Li and X. Ding, Electrochemical detection of lung cancer specific microRNAs using 3D DNA origami nanostructures, Biosens. Bioelectron., 2015, 71, 57–61 CrossRef CAS.
- J. Lu, J. Wang, X. Hu, E. Gyimah, S. Yakubu, K. Wang, X. Wu and Z. Zhang, Electrochemical biosensor based on tetrahedral DNA nanostructures and G-quadruplex–hemin conformation for the ultrasensitive detection of microRNA-21 in serum, Anal. Chem., 2019, 91, 7353–7359 CrossRef CAS.
- G. Zhou, M. Lin, P. Song, X. Chen, J. Chao, L. Wang, Q. Huang, W. Huang, C. Fan and X. Zuo, Multivalent capture and detection of cancer cells with DNA nanostructured biosensors and multibranched hybridization chain reaction amplification, Anal. Chem., 2014, 86, 7843–7848 CrossRef CAS PubMed.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi, X. Lou, J. Shi and C. Fan, Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform, Nat. Protoc., 2016, 11, 1244 CrossRef CAS PubMed.
- C. Y. Tay, L. Yuan and D. T. Leong, Nature-inspired DNA nanosensor for real-time in situ detection of mRNA in living cells, ACS Nano, 2015, 9, 5609–5617 CrossRef CAS PubMed.
- N. Xie, J. Huang, X. Yang, Y. Yang, K. Quan, H. Wang, L. Ying, M. Ou and K. Wang, A DNA tetrahedron-based molecular beacon for tumor-related mRNA detection in living cells, Chem. Commun., 2016, 52, 2346–2349 RSC.
- W. Zhou, D. Li, C. Xiong, R. Yuan and Y. Xiang, Multicolor-encoded reconfigurable DNA nanostructures enable multiplexed sensing of intracellular microRNAs in living cells, ACS Appl. Mater. Interfaces, 2016, 8, 13303–13308 CrossRef CAS PubMed.
- L. He, D. Q. Lu, H. Liang, S. Xie, C. Luo, M. Hu, L. Xu, X. Zhang and W. Tan, Fluorescence resonance energy transfer-based DNA tetrahedron nanotweezer for highly reliable detection of tumor-related mRNA in living cells, ACS Nano, 2017, 11, 4060–4066 CrossRef CAS PubMed.
- K. Zhang, W. Huang, Y. Huang, H. Li, K. Wang, X. Zhu and M. Xie, DNA tetrahedron based biosensor for Argonaute2 assay in single cells and human immunodeficiency virus type-1 related ribonuclease H detection in vitro, Anal. Chem., 2019, 91, 7086–7096 CrossRef CAS PubMed.
- H. Pei, L. Liang, G. Yao, J. Li, Q. Huang and C. Fan, Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors, Angew. Chem., Int. Ed., 2012, 51, 9020–9024 CrossRef CAS PubMed.
- W. Zhou, W. Liang, D. Li, R. Yuan and Y. Xiang, Dual-color encoded DNAzyme nanostructures for multiplexed detection of intracellular metal ions in living cells, Biosens. Bioelectron., 2016, 85, 573–579 CrossRef CAS.
- C. M. Erben, R. P. Goodman and A. J. Turberfield, Single-molecule protein encapsulation in a rigid DNA cage, Angew. Chem., Int. Ed., 2006, 45, 7414–7417 CrossRef CAS PubMed.
- J.-W. Keum and H. Bermudez, Enhanced resistance of DNA nanostructures to enzymatic digestion, Chem. Commun., 2009, 7036–7038, 10.1039/B917661F.
- R. P. Goodman, M. Heilemann, S. Doose, C. M. Erben, A. N. Kapanidis and A. J. Turberfield, Reconfigurable, braced, three-dimensional DNA nanostructures, Nat. Nanotechnol., 2008, 3, 93–96 CrossRef CAS PubMed.
- A. Banerjee, D. Bhatia, A. Saminathan, S. Chakraborty, S. Kar and Y. Krishnan, Controlled release of encapsulated cargo from a DNA icosahedron using a chemical trigger, Angew. Chem., Int. Ed., 2013, 52, 6854–6857 CrossRef CAS PubMed.
- S. Juul, F. Iacovelli, M. Falconi, S. L. Kragh, B. Christensen, R. Frøhlich, O. Franch, E. L. Kristoffersen, M. Stougaard, K. W. Leong, Y.-P. Ho, E. S. Sørensen, V. Birkedal, A. Desideri and B. R. Knudsen, Temperature-controlled encapsulation and release of an active enzyme in the cavity of a self-assembled DNA nanocage, ACS Nano, 2013, 7, 9724–9734 CrossRef CAS PubMed.
- A. S. Walsh, H. Yin, C. M. Erben, M. J. Wood and A. J. Turberfield, DNA cage delivery to mammalian cells, ACS Nano, 2011, 5, 5427–5432 CrossRef CAS.
- J. Li, H. Pei, B. Zhu, L. Liang, M. Wei, Y. He, N. Chen, D. Li, Q. Huang and C. Fan, Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides, ACS Nano, 2011, 5, 8783–8789 CrossRef CAS PubMed.
- L. Liang, J. Li, Q. Li, Q. Huang, J. Shi, H. Yan and C. Fan, Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells, Angew. Chem., Int. Ed., 2014, 53, 7745–7750 CrossRef CAS PubMed.
- S. Raniolo, S. Croce, R. P. Thomsen, A. H. Okholm, V. Unida, F. Iacovelli, A. Manetto, J. Kjems, A. Desideri and S. Biocca, Cellular uptake of covalent and non-covalent DNA nanostructures with different sizes and geometries, Nanoscale, 2019, 11, 10808–10818 RSC.
- C. Wiraja, Y. Zhu, D. C. S. Lio, D. C. Yeo, M. Xie, W. Fang, Q. Li, M. Zheng, M. Van Steensel, L. Wang, C. Fan and C. Xu, Framework nucleic acids as programmable carrier for transdermal drug delivery, Nat. Commun., 2019, 10, 1147 CrossRef PubMed.
- M. Chang, C.-S. Yang and D.-M. Huang, Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy, ACS Nano, 2011, 5, 6156–6163 CrossRef CAS PubMed.
- H. Lee, A. K. R. Lytton-Jean, Y. Chen, K. T. Love, A. I. Park, E. D. Karagiannis, A. Sehgal, W. Querbes, C. S. Zurenko, M. Jayaraman, C. G. Peng, K. Charisse, A. Borodovsky, M. Manoharan, J. S. Donahoe, J. Truelove, M. Nahrendorf, R. Langer and D. G. Anderson, Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery, Nat. Nanotechnol., 2012, 7, 389 CrossRef CAS PubMed.
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- S. T. Crooke, J. L. Witztum, C. F. Bennett and B. F. Baker, RNA-targeted therapeutics, Cell Metab., 2018, 27, 714–739 CrossRef CAS PubMed.
- P. Charoenphol and H. Bermudez, Aptamer-targeted DNA nanostructures for therapeutic delivery, Mol. Pharmaceutics, 2014, 11, 1721–1725 CrossRef CAS PubMed.
- J. J. Fakhoury, C. K. McLaughlin, T. W. Edwardson, J. W. Conway and H. F. Sleiman, Development and characterization of gene silencing DNA cages, Biomacromolecules, 2014, 15, 276–282 CrossRef CAS PubMed.
- K. E. Bujold, J. C. C. Hsu and H. F. Sleiman, Optimized DNA “nanosuitcases” for encapsulation and conditional release of siRNA, J. Am. Chem. Soc., 2016, 138, 14030–14038 CrossRef CAS PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, A logic-gated nanorobot for targeted transport of molecular payloads, Science, 2012, 335, 831 CrossRef CAS PubMed.
- S. Li, Q. Jiang, S. Liu, Y. Zhang, Y. Tian, C. Song, J. Wang, Y. Zou, G. J. Anderson, J.-Y. Han, Y. Chang, Y. Liu, C. Zhang, L. Chen, G. Zhou, G. Nie, H. Yan, B. Ding and Y. Zhao, A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo, Nat. Biotechnol., 2018, 36, 258 CrossRef CAS PubMed.
- Y. Huang, H. Shi, H. Zhou, X. Song, S. Yuan and Y. Luo, The angiogenic function of nucleolin is mediated by vascular endothelial growth factor and nonmuscle myosin, Blood, 2006, 107, 3564–3571 CrossRef CAS PubMed.
- K. A. Afonin, W. Kasprzak, E. Bindewald, P. S. Puppala, A. R. Diehl, K. T. Hall, T. J. Kim, M. T. Zimmermann, R. L. Jernigan, L. Jaeger and B. A. Shapiro, Computational and experimental characterization of RNA cubic nanoscaffolds, Methods, 2014, 67, 256–265 CrossRef CAS PubMed.
- K. A. Afonin, W. W. Grabow, F. M. Walker, E. Bindewald, M. A. Dobrovolskaia, B. A. Shapiro and L. Jaeger, Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine, Nat. Protoc., 2011, 6, 2022–2034 CrossRef CAS PubMed.
- K. A. Afonin, M. Viard, I. Kagiampakis, C. L. Case, M. A. Dobrovolskaia, J. Hofmann, A. Vrzak, M. Kireeva, W. K. Kasprzak, V. N. KewalRamani and B. A. Shapiro, Triggering of RNA interference with RNA-RNA, RNA-DNA, and DNA-RNA nanoparticles, ACS Nano, 2015, 9, 251–259 CrossRef CAS PubMed.
- C. Xu, H. Li, K. Zhang, D. W. Binzel, H. Yin, W. Chiu and P. Guo, Photo-controlled release of paclitaxel and model drugs from RNA pyramids, Nano Res., 2019, 12, 41–48 CrossRef CAS.
- N. C. Seeman, Structural DNA Nanotechnology, Cambridge University Press, Cambridge, 2016 Search PubMed.
| This journal is © the Partner Organisations 2020 |


