Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Online tracing of molecular weight evolution during radical polymerization via high-resolution FlowNMR spectroscopy†
Jeroen H.
Vrijsen
ab,
Isabel A.
Thomlinson
 c,
Martin E.
Levere
d,
Catherine L.
Lyall
d,
Matthew G.
Davidson
c,
Ulrich
Hintermair
*cd and
Tanja
Junkers
c,
Martin E.
Levere
d,
Catherine L.
Lyall
d,
Matthew G.
Davidson
c,
Ulrich
Hintermair
*cd and
Tanja
Junkers
 *ab
*ab
aHasselt University, Martelarenlaan 42, 3500 Hasselt, Belgium. E-mail: tanja.junkers@monash.edu
bPolymer Reaction Design Group, School of Chemistry, Monash University, 19 Rainforest Walk, Building 23, Clayton, Vic 3800, Australia
cCentre for Sustainable and Circular Technologies, University of Bath, Claverton Down, Bath BA2 7AY, UK. E-mail: u.hintermair@bath.ac.uk
dDynamic Reaction Monitoring Facility, University of Bath, Claverton Down, Bath BA2 7AY, UK
First published on 6th May 2020
Abstract
High-resolution FlowNMR was coupled to a continuous flow reactor to monitor polymer molecular weight evolution online by diffusion ordered NMR spectroscopy. Polymers were synthesized by reversible addition fragmentation chain transfer polymerization in continuous flow. The setup allows to target various polymer chain lengths in a dynamic manner without requiring additional purification or sample preparation. Obtaining molecular weight information in this manner is shown to be more accurate than classical SEC analysis at comparable measurement times, with relative errors around 5%.
Online monitoring and in situ measurements of chemical reactions are becoming increasingly important in synthetic chemistry. With traditional concepts still prevailing, operando measurements, data science and machine learning are rapidly developing areas that hold significant potential to have a profound impact on chemical manufacturing in the near future.1–5 Especially in the realm of polymerization, the concept of following the reaction progress with high temporal resolution and specificity is appealing due to the intimate link of the reaction kinetics with structure that ultimately determines the physical properties of the product. Several monitoring techniques have been employed in the past to explore this. In addition to for example infrared6–9 and Raman spectroscopy10,11 also NMR spectroscopy has entered the scene recently.2,3,12–14 These methods give important access to information that allows to follow monomer conversion, while methods such as UV Vis for example provide concentration information.15 Recently, mass spectrometry was introduced as online monitoring tool, giving access to time-resolved data on polymer end groups and defect structures.16
Direct, non-invasive reaction monitoring unfolds its potential in combination with continuous flow reactors. It allows to move away from passive in situ monitoring towards creation of dynamic feedback loops using analysis data for process control and parameter optimization within a single experiment.1,17–20
One of the most important parameters to assess during a polymerization is the average molecular weight of the product mixture. Polymer molecular weights are classically assessed via size exclusion chromatography (SEC). While fairly reliable and easy to use, SEC isn't very well suited to online monitoring as it requires sample injections at defined moments in time, followed by relatively long analysis times. Also, SEC is typically associated with large experimental errors, and even carefully calibrated apparatuses are believed to yield relative errors of up to 20%.21–24 Thus, time resolution and precision are rather low. This relatively large error in SEC might surprise as SEC is in fact able to discern smaller differences between polymer samples. This is, however, only true when samples are compared on a specific set of columns and calibration, and comparison of samples measured between different setups and laboratories is much more difficult. The error of 20% is the recommended assumption on SEC accuracy by the IUPAC working party on kinetic and mechanisms in radical polymerization.25,26 Nevertheless, several successful approaches have been reported to coupling SECs online to polymerization reactors. Coupling online SEC to polymerizations is quite successful with an analysis time of 4 minutes.27,28 Using sample loops, reaction aliquots can be injected into SEC machines at defined time intervals. A good example for such approach is the CORSEMP system.29 Rubens et al. have demonstrated the use of a flow reactor coupled to SEC for machine-assisted synthesis, allowing for self-optimization of reaction conditions in order to produce target molecular weights with high fidelity.1
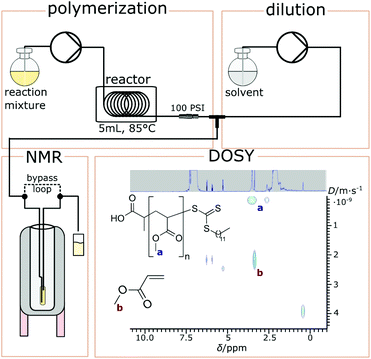
Yet, the development of true online analysis of molecular weights is still outstanding. In principle, viscometry could be a suitable tool (see the ACOMP system), or static light scattering could be used for this purpose,30 but this wasn't implemented for flow polymerizations to date. In this work we present the use of high-resolution FlowNMR spectroscopy for monitoring the progress of a polymerisation reaction in continuous flow mode. End group analysis by way of quantitative NMR spectroscopy (qNMR) can give access to average molecular weights in some cases, but is generally too imprecise as a general method for following polymerisations over the course of a reaction. Li et al. have shown that diffusion ordered NMR spectroscopy (DOSY) may be used to obtain molecular weight data on polymeric mixtures.31 Through Einstein–Smoluchowski relationship the diffusion coefficient of a molecule is inversely proportional to the viscosity of the medium and the size of the diffusing compound from which its molecular weight can be derived.32 While the determination of absolute diffusion coefficients from DOSY data can be challenging due to experimental disturbances (mainly convection) and assumptions on molecular shape, a relative size correlation of similar compounds in a mixture (such as growing polymer chains) is typically very precise. Further, the current method relies on relatively sharp diffusion profiles, and hence polymers of low dispersity and ideally symmetric distributions. RAFT does, however, provide such polymers in good proximity. When correlated with an absolute molecular weight measurement from SEC for example, it should be possible to follow polymerisations online with a combination of DOSY and traditional NMR experiments to derive a wealth of information on the growing polymer architecture. Using a high-resolution FlowNMR setup33 interfaced with a continuous flow polymerisation reactor34 we tested this hypothesis by following the reversible addition fragmentation chain transfer polymerization (RAFT) of methyl acrylate as a proof-of-concept. A schematic of the experimental set-up is shown in Fig. 1.
Continuous flow reactors enable convenient screening of reaction conditions via adjustment of residence times by simply varying the flowrate of the reaction mixture. Since RAFT is used as polymerization technique various polymer lengths can be conveniently targeted and analyzed in line with the presented set-up. Monomer conversion data was obtained from 1H qNMR measurements after stabilization of the reactor (typically 1.5 residence times after each parameter change). Diffusion coefficients of polymers from DOSY experiments are strongly influenced by variables that impact on solvent viscosity (including temperature but also polymer concentration) as well as acquisition parameters such as pulse strength, gradient range and diffusion delay. For an overview and the influence of various parameters on the measurement we refer to a comprehensive review by Groves.35 To ensure optimal polymer concentrations (1–4 mM) the reaction mixture was diluted in-line after exiting the reactor prior DOSY measurements. Once delivered into the tip of the FlowNMR tube the mixture was isolated from the flow by switching the bypass valve to ensure static conditions for the diffusion measurement. Diffusion coefficients were then derived from DOSY traces of the proton resonances of the methyl esters (see Fig. 1) which are clearly different in polymer versus monomer form (Fig. 1 a versus b). Samples were also collected and analyzed by SEC-multiangle laser light scattering (SEC-MALS). Various residence times and henceforth polymer chain lengths could thus be targeted in a dynamic manner with this continuous flow set-up without requiring any purification or sample preparation. With around 20 minutes per DOSY measurement 6 residence times/RAFT polymerization could be screened within 2–3 hours, providing a wealth of information with high correlation. While this approach does not allow for true in-line measurements due to the inherent requirements of DOSY, all advantages of flow chemistry (simple adjustment of residence times, ability to move from high to low residence times within an experiment and high reactor stability and thus reproducibility) still prevail with this on-line setup.
In a first step, we correlated the evolution of polymer diffusion coefficient with monomer conversion (see Fig. 2). Notably, both data have been directly obtained from non-invasive online FlowNMR experiments on the same reaction aliquot. Polymerizations proceeded quickly and reached conversions above 50% within 20 min of reaction time. At the same time, the diffusion coefficient decreased from 0.42 × 10−9 m2 s−1 to about 0.28 × 10−9 m2 s−1. The decline is seemingly exponential and a very good correlation is seen. In the next step, we tested for the accuracy of the data. Fig. 3 (l.h.s.) gives the same data as a function of molecular weight in combination with datasets obtained from RAFT polymerizations with a higher target DP (thus lower RAFT agent concentration). The datasets overlap nicely, allowing for the direct conclusion that the method yields accurate and reproducible results. It can also be seen that the error associated with the DOSY analysis is very small (in the order of few percent) for most data points, highlighting again the statistical robustness of the method. In Fig. 3, all molecular weight data have been determined by SEC-MALS for highest accuracy. The determined molecular weights match the theoretically expected molecular weight relatively well. In this case, MALS is used to benchmark the data in an absolute calibration of the data. The weight-average molecular weight Mw was chosen for representation, as this parameter scales directly with the physical properties of a polymer.
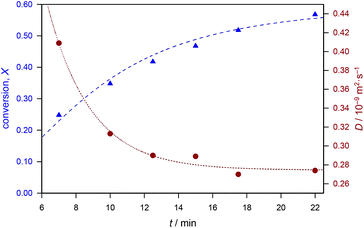 | ||
| Fig. 2 Monomer conversion as a function of time (blue) and diffusion coefficients determined by DOSY in a continuous flow process for MA RAFT polymerization. | ||
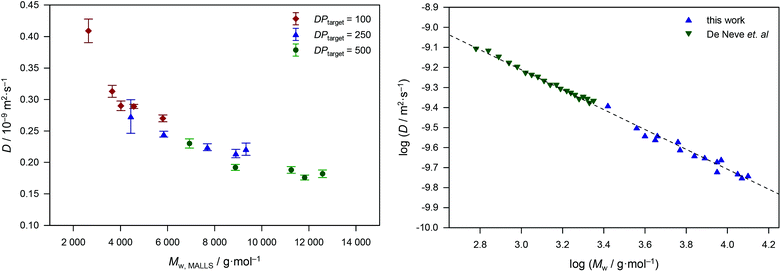 | ||
| Fig. 3 Diffusion coefficients as a function of weight-average molecular weight as benchmarked by SEC-MALS for MA RAFT polymerizations with varying target degree of polymerization (left) and linearized data with best fit to the data in conjunction with data from discrete RAFT MA oligomers from De Neve et al.36 | ||
As mentioned above, the diffusion coefficient follows an inverse relation with bulk viscosity and the hydrodynamic radius of the polymer particle, hence the size of the chain. Since the presented DOSY measurements have been carried out under dilute conditions one does not need to account for changes in bulk viscosity due to polymer growth. The hydrodynamic radius Rh can thus be correlated with molecular weight via a Mark–Houwink–Sakurada-like equation, following a generic Rh = a·Mbw. Hence when the logarithm of the diffusion coefficient is plotted against the logarithmic molecular weight a linear relationship with a negative slope should be obtained, meaning that the observed change in D directly correlates with changes in Rh. This approach represents an interesting alternative to determining the radius of gyration, which otherwise requires different offline analysis techniques such as viscometry and light scattering. On the r.h.s. of Fig. 3 the combined data in conjunction with literature data on discrete RAFT-made oligo(methyl acrylates) are shown.36 Both datasets fit nicely demonstrating the high accuracy of the DOSY method again. Since the literature data represent discrete oligomers they are essentially error-free in molecular weight determination, and hence confirm the accuracy of the MALS measurements of our data. The combined dataset can be fitted to:
| log(D/m2 s−1) = −7.72 − 0.497 log(Mw/g mol−1) |
Surprisingly, the full data range from a few hundred to over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 are accurately represented by a single fit. Polymer chains coil from a certain critical chain length on, which for acrylates is believed to be around 30–50 repeat units (or 4300 g mol−1).37 Due to coil dynamics, a change in the exponent could be expected along the molecular weight axis.38 Since this was not observed, one can conclude that already small oligomers behave – with respect to their translational diffusion – similar to larger chains in this case. Within a 95% confidence level, the slope of the fit is associated with an error of only 2.5%. Since the determination of D from DOSY has a similar level of accuracy, a total relative error of 5% can be estimated. Even though only yielding a molecular weight distribution average, this form of Mw determination is thus much more accurate than classical SEC analysis. While the size range covered in this study is not as large as a conventional SEC calibration, it is still sufficient for kinetic screening of most controlled polymerizations and determination of molecular weights in the relevant mass range. Yet, in principle DOSY is applicable in the whole molecular weight range and avoids difficulties with oligomers as typically seen with SEC. Strictly speaking only interpolation should be done, yet extrapolation towards higher molecular weights could be feasible within certain limits under dilute conditions. Another interesting observation is that the residual fit parameters are very close to the ones that have previously been determined offline for polystyrene as log(D/m2 s−1) = −7.697 − 0.537 log(Mw/g mol−1),31 and for poly(methyl methacrylate) as log(D/m2 s−1) = −7.525 − 0.557 log(Mw/g mol−1).39 This raises the question if Rh may be used as a universal (absolute) calibration tool. Future experiments in our laboratories are underway to test this hypothesis.
000 g mol−1 are accurately represented by a single fit. Polymer chains coil from a certain critical chain length on, which for acrylates is believed to be around 30–50 repeat units (or 4300 g mol−1).37 Due to coil dynamics, a change in the exponent could be expected along the molecular weight axis.38 Since this was not observed, one can conclude that already small oligomers behave – with respect to their translational diffusion – similar to larger chains in this case. Within a 95% confidence level, the slope of the fit is associated with an error of only 2.5%. Since the determination of D from DOSY has a similar level of accuracy, a total relative error of 5% can be estimated. Even though only yielding a molecular weight distribution average, this form of Mw determination is thus much more accurate than classical SEC analysis. While the size range covered in this study is not as large as a conventional SEC calibration, it is still sufficient for kinetic screening of most controlled polymerizations and determination of molecular weights in the relevant mass range. Yet, in principle DOSY is applicable in the whole molecular weight range and avoids difficulties with oligomers as typically seen with SEC. Strictly speaking only interpolation should be done, yet extrapolation towards higher molecular weights could be feasible within certain limits under dilute conditions. Another interesting observation is that the residual fit parameters are very close to the ones that have previously been determined offline for polystyrene as log(D/m2 s−1) = −7.697 − 0.537 log(Mw/g mol−1),31 and for poly(methyl methacrylate) as log(D/m2 s−1) = −7.525 − 0.557 log(Mw/g mol−1).39 This raises the question if Rh may be used as a universal (absolute) calibration tool. Future experiments in our laboratories are underway to test this hypothesis.
Conclusions
We have demonstrated the use of high-resolution FlowNMR spectroscopy for monitoring monomer conversion and molecular weight distributions of a growing polymer during a methyl acrylate RAFT polymerization. With increasing reactor residence times (and hence increasing conversion) decreasing diffusion coefficients of the translational movement of polymer chains could be observed by DOSY under dilute stopped-flow conditions. Data from several reactions under overlapping conditions showed excellent correlation, and the dependence of the diffusion coefficient on the weight-average molecular weight could be accurately represented with a single double logarithmic fit. Determination of molecular weights in this manner is shown to be more accurate than classical SEC analysis, with relative errors around 5%. At the same time, the measurements are relatively quick, and can be carried on the same timescale as classical SEC, if not faster. Combined with the benefits of tight reaction control and high reproducibility offered by continuous flow reactors, we anticipate that this method will allow for fast and reliable determination of molecular weights in a continuous fashion. It will be interesting to see this method extended to the investigation of more complex polymer architectures such as nanoparticles, and utilise it for autonomous self-regulating and self-optimising polymerisation reactors in the future.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful to the Fonds Wetenschappelijk Onderzoek (FWO) for providing a SB scholarship for JHV, the Royal Society for a University Research Fellowship to UH (UF160458), and EPSRC for funding the Dynamic Reaction Monitoring Facility at the University of Bath (EP/P001475/1) and for the EPSRC Centre for Doctoral Training in Sustainable Chemical Technologies (EP/L016354/1, PhD studentship for IAT). We thank Joris Haven (Monash University), Maciek Kopec and John Lowe (University of Bath) for stimulating discussions.Notes and references
- M. Rubens, J. H. Vrijsen, J. Laun and T. Junkers, Angew. Chem., Int. Ed., 2019, 58, 3183–3187 CrossRef CAS PubMed.
- M. Rubens, J. Van Herck and T. Junkers, ACS Macro Lett., 2019, 8, 1437–1441 CrossRef CAS.
- S. T. Knox, S. Parkinson, R. Stone and N. J. Warren, Polym. Chem., 2019, 10, 4774–4778 RSC.
- T. McAfee, C. W. Jarand, T. Zekoski, R. Montgomery and W. F. Reed, Macromol. React. Eng., 2019, 13, 1900039 CrossRef CAS.
- J. M. Granda, L. Donina, V. Dragone, D.-L. Long and L. Cronin, Nature, 2018, 559, 377–381 CrossRef CAS PubMed.
- C. J. Chuck, M. G. Davidson, G. Gobius du Sart, P. K. Ivanova-Mitseva, G. I. Kociok-Köhn and L. B. Manton, Inorg. Chem., 2013, 52, 10804–10811 CrossRef CAS PubMed.
- J. Langanke, A. Wolf, J. Hofmann, K. Böhm, M. A. Subhani, T. E. Müller, W. Leitner and C. Gürtler, Green Chem., 2014, 16, 1865–1870 RSC.
- Y.-M. Chuang, A. Ethirajan and T. Junkers, ACS Macro Lett., 2014, 3, 732–737 CrossRef CAS.
- Y.-M. Chuang, B. Wenn, S. Gielen, A. Ethirajan and T. Junkers, Polym. Chem., 2015, 6, 6488–6497 RSC.
- P. M. Schäfer, M. Fuchs, A. Ohligschläger, R. Rittinghaus, P. McKeown, E. Akin, M. Schmidt, A. Hoffmann, M. A. Liauw, M. D. Jones and S. Herres-Pawlis, ChemSusChem, 2017, 10, 3547–3556 CrossRef PubMed.
- R. D. Rittinghaus, P. M. Schäfer, P. Albrecht, C. Conrads, A. Hoffmann, A. N. Ksiazkiewicz, O. Bienemann, A. Pich and S. Herres-Pawlis, ChemSusChem, 2019, 12, 2161–2165 CrossRef CAS PubMed.
- S. Roberge and M. A. Dubé, J. Appl. Polym. Sci., 2016, 133, 43574 CrossRef.
- C. Botha, J. Höpfner, B. Mayerhöfer and M. Wilhelm, Polym. Chem., 2019, 10, 2230–2246 RSC.
- M. Hehn, T. Wagner and W. Hiller, Anal. Chem., 2014, 86, 490–497 CrossRef CAS.
- F. Lauterbach and V. Abetz, Macromol. Rapid Commun., 2020 DOI:10.1002/marc.202000029.
- J. J. Haven, J. Vandenbergh and T. Junkers, ChemComm, 2015, 51, 4611–4614 RSC.
- N. Holmes, G. R. Akien, R. J. Savage, C. Stanetty, I. R. Baxendale, A. J. Blacker, B. A. Taylor, R. L. Woodward, R. E. Meadows and R. A. Bourne, React. Chem. Eng., 2016, 1, 96–100 RSC.
- E. Wimmer, D. Cortés-Borda, S. Brochard, E. Barré, C. Truchet and F.-X. Felpin, React. Chem. Eng., 2019, 4, 1608–1615 RSC.
- P. Sagmeister, J. D. Williams, C. A. Hone and C. O. Kappe, React. Chem. Eng., 2019, 4, 1571–1578 RSC.
- E. M. Barreiro, Z. Hao, L. A. Adrio, J. R. van Ommen, K. Hellgardt and K. K. Hii, Catal. Today, 2018, 308, 64–70 CrossRef CAS.
- L. Letot, J. Lesec and C. Quivoron, J. Liq. Chromatogr., 1980, 3, 1637–1655 CrossRef CAS.
- L. Andersson, J. Chromatogr. A, 1985, 325, 37–42 CrossRef CAS.
- L. Hagel, J. Chromatogr. A, 1993, 648, 19–25 CrossRef CAS.
- R. J. Bruessau, Macromol. Symp., 1996, 110, 15–32 CrossRef CAS.
- M. Buback, R. G. Gilbert, R. A. Hutchinson, B. Klumperman, F.-D. Kuchta, B. G. Manders, K. F. O'Driscoll, G. T. Russell and J. Schweer, Macromol. Chem. Phys., 1995, 196, 3267–3280 CrossRef CAS.
- C. Barner-Kowollik, S. Beuermann, M. Buback, P. Castignolles, B. Charleux, M. L. Coote, R. A. Hutchinson, T. Junkers, I. Lacík, G. T. Russell, M. Stach and A. M. van Herk, Polym. Chem., 2014, 5, 204 RSC.
- M. E. Levere, I. Willoughby, S. O'Donohue, A. de Cuendias, A. J. Grice, C. Fidge, C. R. Becer and D. M. Haddleton, Polym. Chem., 2010, 1, 1086–1094 RSC.
- M. E. Levere, I. Willoughby, S. O'Donohue, P. M. Wright, A. J. Grice, C. Fidge, C. Remzi Becer and D. M. Haddleton, J. Polym. Sci., Part A: Polym. Chem., 2011, 49, 1753–1763 CrossRef CAS.
- C. Rosenfeld, C. Serra, S. O'Donohue and G. Hadziioannou, Macromol. React. Eng., 2007, 1, 547–552 CrossRef CAS.
- A. M. Alb, M. F. Drenski and W. F. Reed, Polym. Int., 2008, 57, 390–396 CrossRef CAS.
- W. Li, H. Chung, C. Daeffler, J. A. Johnson and R. H. Grubbs, Macromolecules, 2012, 45, 9595–9603 CrossRef CAS PubMed.
- A. Chen, D. Wu and C. S. Johnson, J. Am. Chem. Soc., 1995, 117, 7965–7970 CrossRef CAS.
- A. M. R. Hall, J. C. Chouler, A. Codina, P. T. Gierth, J. P. Lowe and U. Hintermair, Catal. Sci. Technol., 2016, 6, 8406–8417 RSC.
- Dynamic Reaction Monitoring Facility, https://www.bath.ac.uk/research-facilities/dynamic-reaction-monitoring-facility/.
- P. Groves, Polym. Chem., 2017, 8, 6700–6708 RSC.
- J. De Neve, J. J. Haven, S. Harrisson and T. Junkers, Angew. Chem., Int. Ed., 2019, 58, 13869–13873 CrossRef CAS PubMed.
- M. Buback, P. Hesse, T. Junkers, T. Theis and P. Vana, Aust. J. Chem., 2007, 60, 779–787 CrossRef CAS.
- G. B. Smith, G. T. Russell and J. P. A. Heuts, Macromol. Theory Simul., 2003, 12, 299–314 CrossRef CAS.
- N. Cherifi, A. Khoukh, A. Benaboura and L. Billon, Polym. Chem., 2016, 7, 5249–5257 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0py00475h |
| This journal is © The Royal Society of Chemistry 2020 |